Integrated Analysis of Proteomics and Metabolomics for Heat Stress in Chinese Holstein Cows
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Dairy Cows and Sample Collections
2.2. Proteomics Sequence Procedures
2.3. Mass Spectrometry Analysis for Proteomics and Data Processing
2.4. Subcellular Localization, Structural Domain, and Transcription Factor Prediction
2.5. Protein–Protein Interaction (PPI) Network
2.6. Metabolite Extraction and LC-MS Analysis and Data Processing
2.7. Cell Culture and Transfection
2.8. Quantitative Reverse Transcription Polymerase Chain Reaction
2.9. Western Blot
2.10. Bioinformatics Analysis
3. Results
3.1. Protein Quality, Characterization, and Differentially Expressed Protein
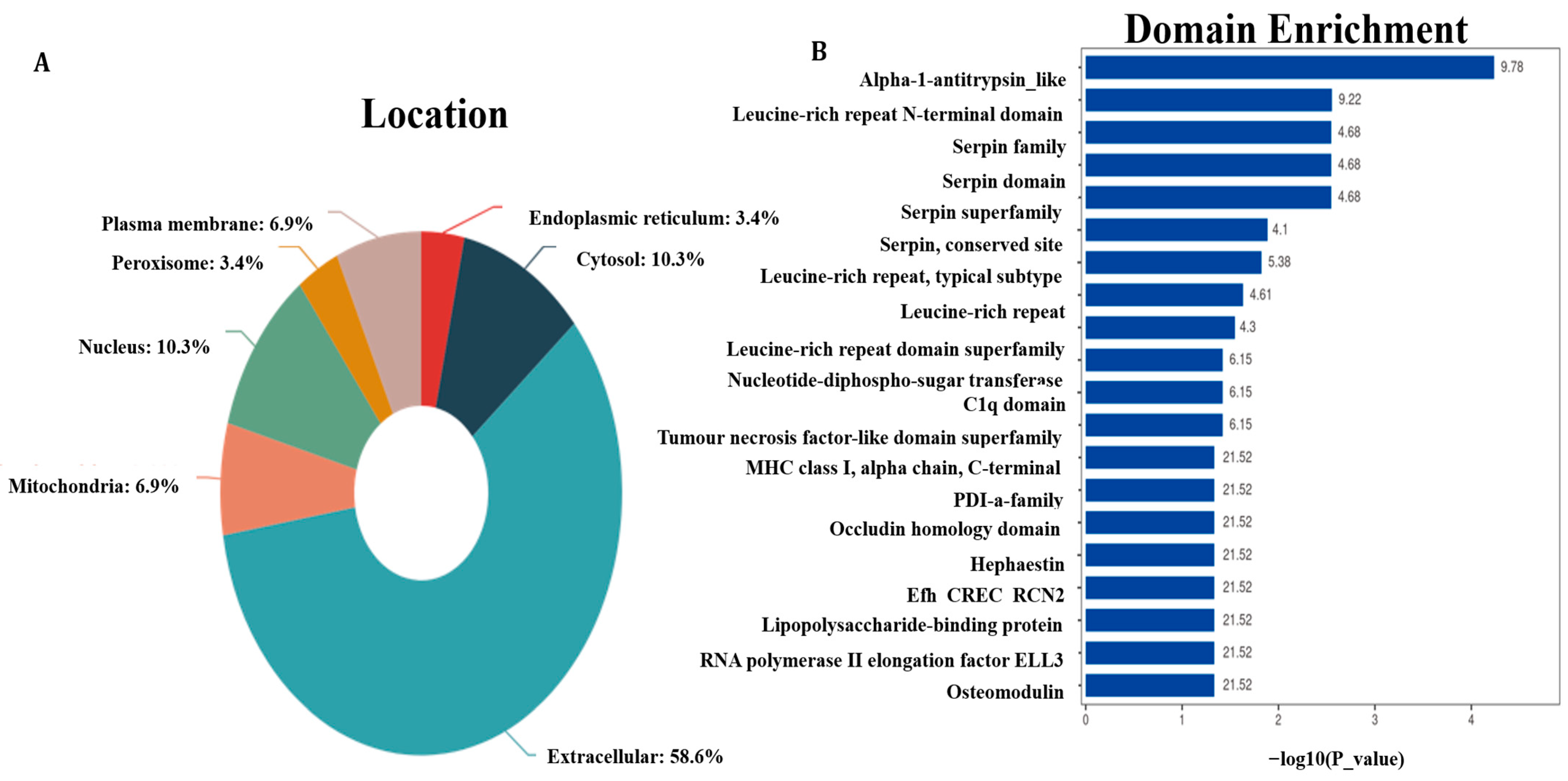
3.2. Proteins Subcellular Localization and Structural Domain
3.3. Functional Enrichment Analysis
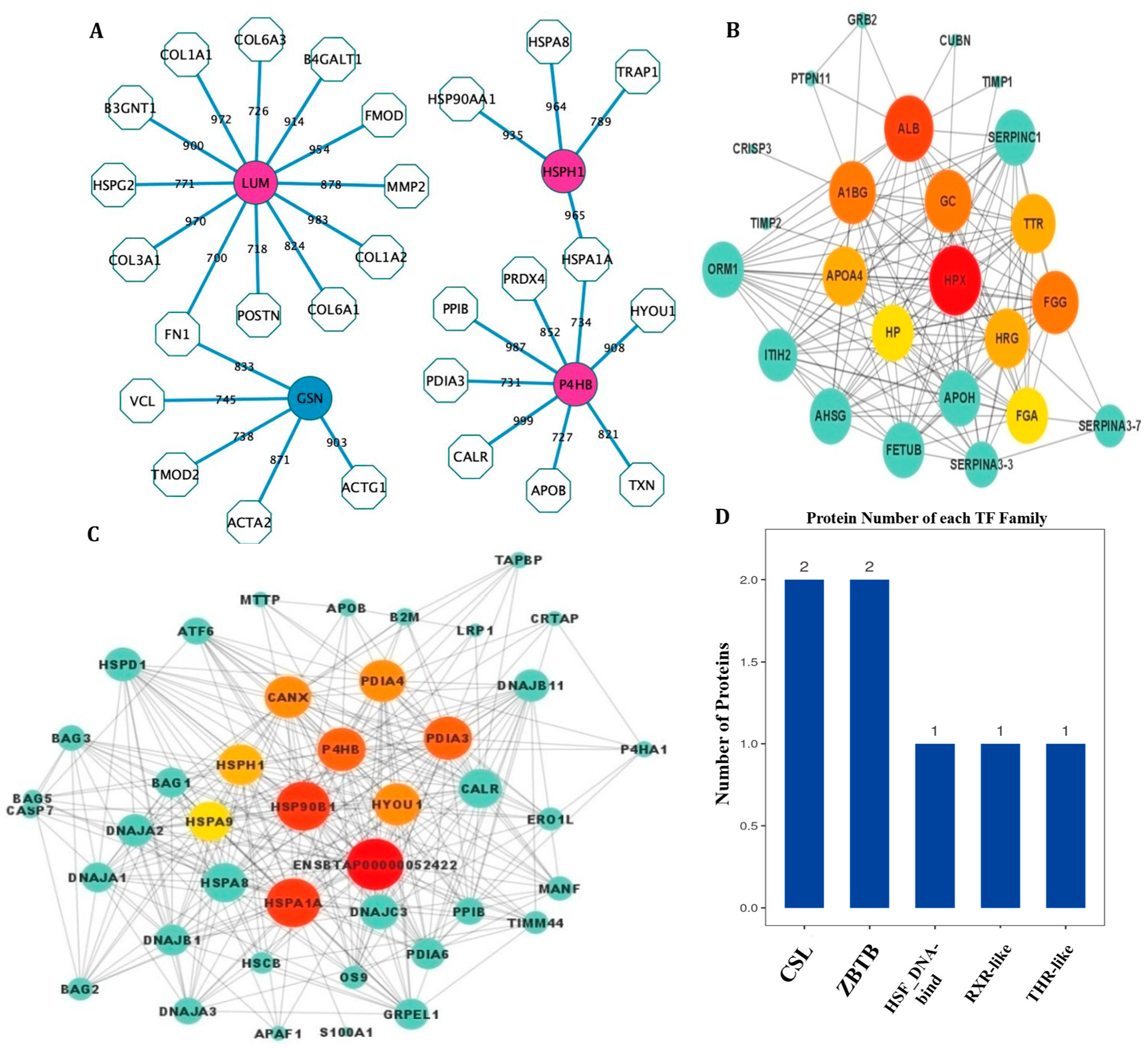
3.4. PPI Network Analysis and Transcription Factors
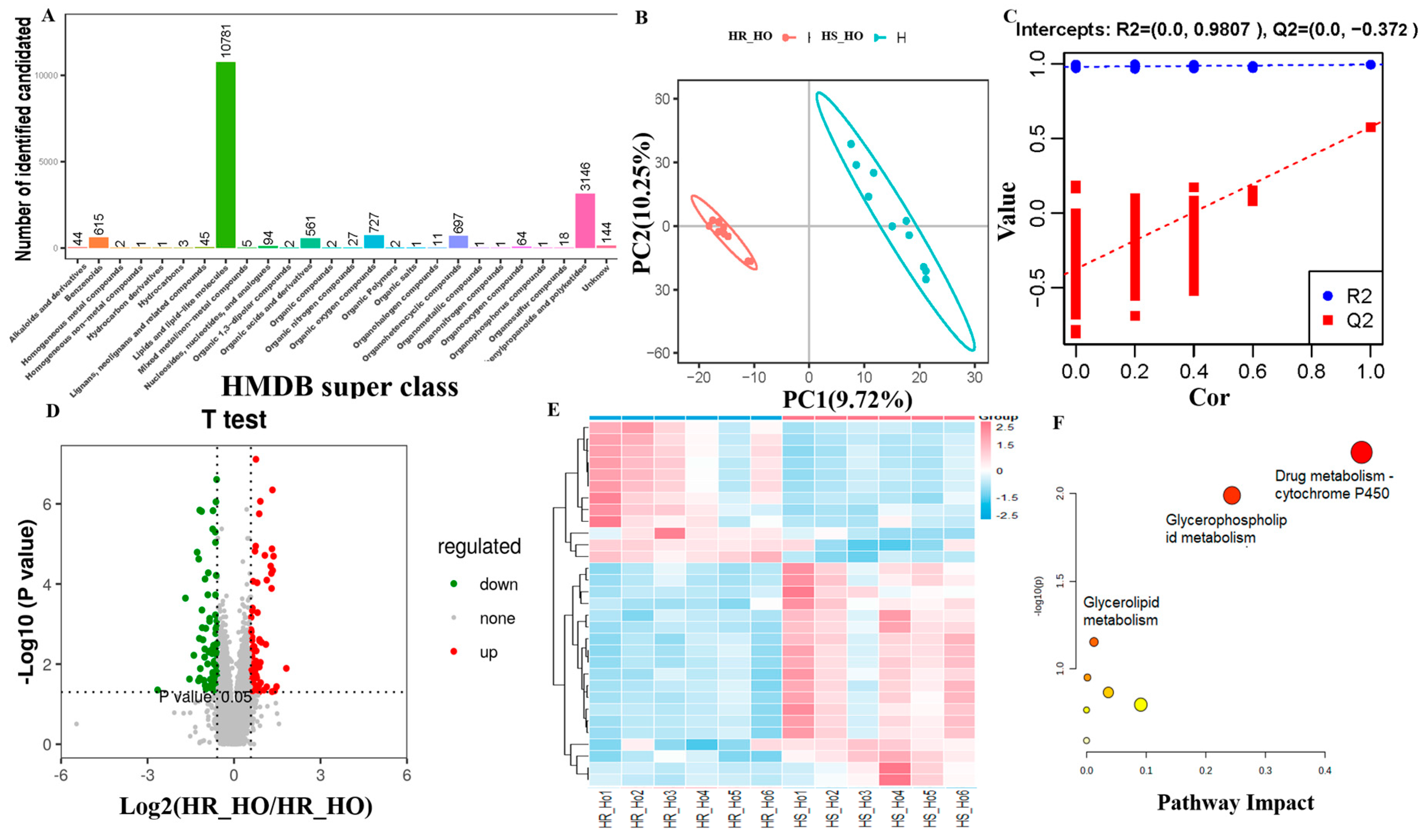
3.5. Metabolite Identification
3.6. Differential Positive and Negative Metabolites
3.7. Integrated Analysis for Proteomics and Metabolomics
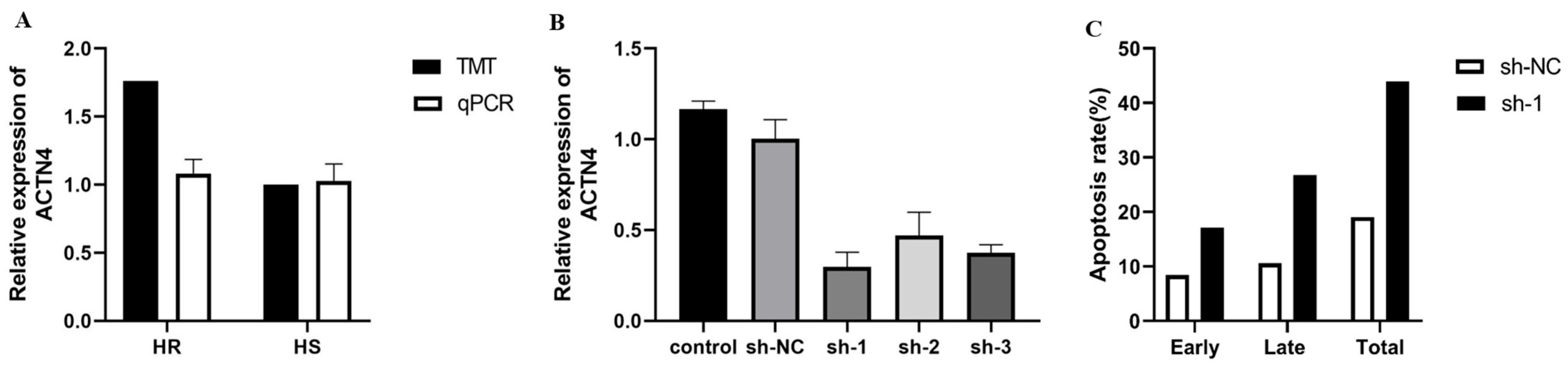
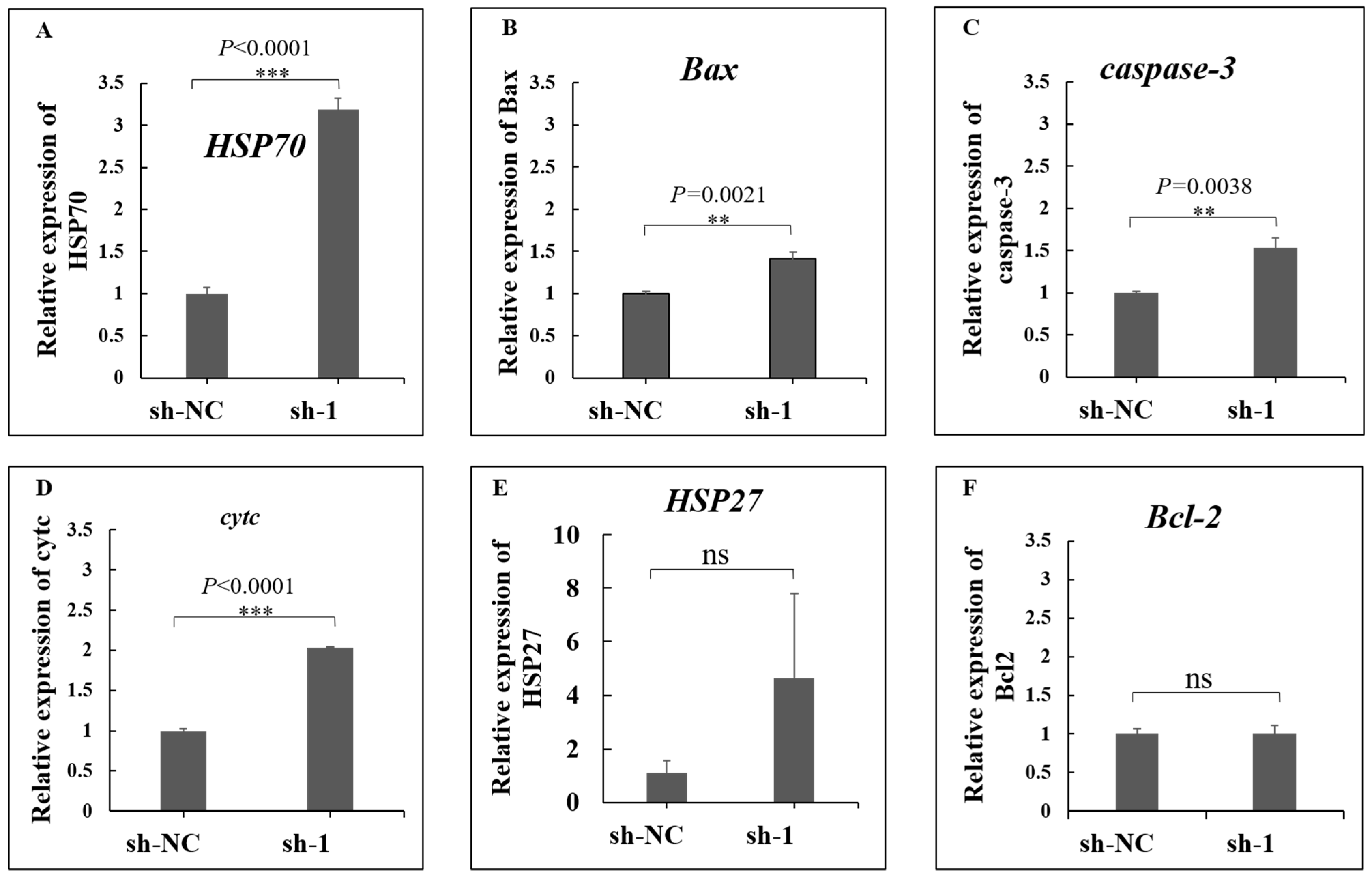
3.8. Interference of ACTN4 Promotes Apoptosis in Epithelial Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Polsky, L.; Von Keyserlingk, M.A. Invited review: Effects of heat stress on dairy cattle welfare. J. Dairy Sci. 2017, 100, 8645–8657. [Google Scholar] [CrossRef]
- Chen, L.; Thorup, V.M.; Kudahl, A.B.; Østergaard, S. Effects of heat stress on feed intake, milk yield, milk composition, and feed efficiency in dairy cows: A meta-analysis. J. Dairy Sci. 2024, 107, 3207–3218. [Google Scholar] [CrossRef]
- Lovarelli, D.; Minozzi, G.; Arazi, A.; Guarino, M.; Tiezzi, F. Effect of extended heat stress in dairy cows on productive and behavioral traits. Animal 2024, 18, 101089. [Google Scholar] [CrossRef] [PubMed]
- Perano, K.M.; Usack, J.G.; Angenent, L.T.; Gebremedhin, K.G. Production and physiological responses of heat-stressed lactating dairy cattle to conductive cooling. J. Dairy Sci. 2015, 98, 5252–5261. [Google Scholar] [CrossRef]
- Li, R.; Ahmad, M.J.; Hou, M.; Wang, X.; Liu, S.; Li, J.; Jiang, Q.; Huang, J.; Yang, L. Identification of target genes and pathways related to heat tolerance in Chinese Holstein cows. Livest. Sci. 2023, 271, 105213. [Google Scholar] [CrossRef]
- Min, L.; Zheng, N.; Zhao, S.; Cheng, J.; Yang, Y.; Zhang, Y.; Yang, H.; Wang, J. Long-term heat stress induces the inflammatory response in dairy cows revealed by plasma proteome analysis. Biochem. Biophys. Res. Commun. 2016, 471, 296–302. [Google Scholar] [CrossRef]
- Cheng, J.; Min, L.; Zheng, N.; Fan, C.; Zhao, S.; Zhang, Y.; Wang, J. Strong, sudden cooling alleviates the inflammatory responses in heat-stressed dairy cows based on iTRAQ proteomic analysis. Int. J. Biometeorol. 2018, 62, 177–182. [Google Scholar] [CrossRef]
- Reolon, H.G.; Abduch, N.G.; Freitas, A.C.d.; Silva, R.M.d.O.; Fragomeni, B.d.O.; Lourenco, D.; Baldi, F.; Paz, C.C.P.d.; Stafuzza, N.B. Proteomic changes of the bovine blood plasma in response to heat stress in a tropically adapted cattle breed. Front. Genet. 2024, 15, 1392670. [Google Scholar] [CrossRef]
- Skibiel, A.L.; Zachut, M.; do Amaral, B.C.; Levin, Y.; Dahl, G.E. Liver proteomic analysis of postpartum Holstein cows exposed to heat stress or cooling conditions during the dry period. J. Dairy Sci. 2018, 101, 705–716. [Google Scholar] [CrossRef]
- Zachut, M.; Kra, G.; Livshitz, L.; Portnick, Y.; Yakoby, S.; Friedlander, G.; Levin, Y. Seasonal heat stress affects adipose tissue proteome toward enrichment of the Nrf2-mediated oxidative stress response in late-pregnant dairy cows. J. Proteom. 2017, 158, 52–61. [Google Scholar] [CrossRef]
- Min, L.; Zhao, S.; Tian, H.; Zhou, X.; Zhang, Y.; Li, S.; Yang, H.; Zheng, N.; Wang, J. Metabolic responses and “omics” technologies for elucidating the effects of heat stress in dairy cows. Int. J. Biometeorol. 2017, 61, 1149–1158. [Google Scholar] [CrossRef]
- Grelet, C.; Vanden Dries, V.; Leblois, J.; Wavreille, J.; Mirabito, L.; Soyeurt, H.; Franceschini, S.; Gengler, N.; Brostaux, Y.; Dehareng, F. Identification of chronic stress biomarkers in dairy cows. Animal 2022, 16, 100502. [Google Scholar] [CrossRef]
- Wang, X.; Kadarmideen, H.N. Metabolite genome-wide association study (mGWAS) and gene-metabolite interaction network analysis reveal potential biomarkers for feed efficiency in pigs. Metabolites 2020, 10, 201. [Google Scholar] [CrossRef]
- Stefanska, B.; Sobolewska, P.; Fievez, V.; Pruszynska-Oszmałek, E.; Purwin, C.; Nowak, W. The effect of heat stress on performance, fertility, and adipokines involved in regulating systemic immune response during lipolysis of early lactating dairy cows. J. Dairy Sci. 2024, 107, 2111–2128. [Google Scholar] [CrossRef]
- Wheelock, J.B.; Rhoads, R.P.; VanBaale, M.J.; Sanders, S.R.; Baumgard, L.H. Effects of heat stress on energetic metabolism in lactating Holstein cows. J. Dairy Sci. 2010, 93, 644–655. [Google Scholar] [CrossRef]
- Baumgard, L.; Wheelock, J.; Sanders, S.; Moore, C.; Green, H.; Waldron, M.; Rhoads, R. Postabsorptive carbohydrate adaptations to heat stress and monensin supplementation in lactating Holstein cows. J. Dairy Sci. 2011, 94, 5620–5633. [Google Scholar] [CrossRef] [PubMed]
- Salama, A.A.; Contreras-Jodar, A.; Love, S.; Mehaba, N.; Such, X.; Caja, G. Milk yield, milk composition, and milk metabolomics of dairy goats intramammary-challenged with lipopolysaccharide under heat stress conditions. Sci. Rep. 2020, 10, 5055. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Guo, J.; Quan, S.; Nan, X.; Fernandez, M.S.; Baumgard, L.; Bu, D. The effects of heat stress on protein metabolism in lactating Holstein cows. J. Dairy Sci. 2017, 100, 5040–5049. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Gao, Y.; Canela-Xandri, O.; Wang, S.; Yu, Y.; Cai, W.; Li, B.; Xiang, R.; Chamberlain, A.J.; Pairo-Castineira, E. A multi-tissue atlas of regulatory variants in cattle. Nat. Genet. 2022, 54, 1438–1447. [Google Scholar] [CrossRef]
- Fang, L.; Teng, J.; Lin, Q.; Bai, Z.; Liu, S.; Guan, D.; Li, B.; Gao, Y.; Hou, Y.; Gong, M. The farm animal genotype–tissue expression (FarmGtex) project. Nat. Genet. 2025, 57, 786–796. [Google Scholar] [CrossRef]
- Kendall, P.; Webster, J. Season and physiological status affects the circadian body temperature rhythm of dairy cows. Livest. Sci. 2009, 125, 155–160. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Dönnes, P.; Höglund, A. Predicting protein subcellular localization: Past, present, and future. Genom. Proteom. Bioinform. 2004, 2, 209–215. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Shen, W.-K.; Chen, S.-Y.; Gan, Z.-Q.; Zhang, Y.-Z.; Yue, T.; Chen, M.-M.; Xue, Y.; Hu, H.; Guo, A.-Y. AnimalTFDB 4.0: A comprehensive animal transcription factor database updated with variation and expression annotations. Nucleic Acids Res. 2023, 51, D39–D45. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Kuhl, C.; Tautenhahn, R.; Bottcher, C.; Larson, T.R.; Neumann, S. CAMERA: An integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 2012, 84, 283–289. [Google Scholar] [CrossRef]
- Wen, B.; Mei, Z.; Zeng, C.; Liu, S. metaX: A flexible and comprehensive software for processing metabolomics data. BMC Bioinform. 2017, 18, 183. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Foroutan, A.; Fitzsimmons, C.; Mandal, R.; Piri-Moghadam, H.; Zheng, J.; Guo, A.; Li, C.; Guan, L.L.; Wishart, D.S. The bovine metabolome. Metabolites 2020, 10, 233. [Google Scholar] [CrossRef]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; MacDonald, P.E.; Wishart, D.S.; Li, S. MetaboAnalyst 6.0: Towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef]
- Jedrzejczak, M.; Szatkowska, I. Bovine mammary epithelial cell cultures for the study of mammary gland functions. In Vitro Cell. Dev. Biol. Anim. 2014, 50, 389–398. [Google Scholar] [CrossRef]
- Walker, A.K.; Yang, F.; Jiang, K.; Ji, J.-Y.; Watts, J.L.; Purushotham, A.; Boss, O.; Hirsch, M.L.; Ribich, S.; Smith, J.J. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010, 24, 1403–1417. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Wang, J.; Bu, D.; Wei, H.; Zhou, L.; Li, F.; Loor, J.J. In vitro culture and characterization of a mammary epithelial cell line from Chinese Holstein dairy cow. PLoS ONE 2009, 4, e7636. [Google Scholar] [CrossRef]
- Péré-Brissaud, A.; Blanchet, X.; Delourme, D.; Pélissier, P.; Forestier, L.; Delavaud, A.; Duprat, N.; Picard, B.; Maftah, A.; Brémaud, L. Expression of SERPINA3s in cattle: Focus on bovSERPINA3-7 reveals specific involvement in skeletal muscle. Open Biol. 2015, 5, 150071. [Google Scholar] [CrossRef]
- Herrera-Mendez, C.H.; Becila, S.; Blanchet, X.; Pelissier, P.; Delourme, D.; Coulis, G.; Sentandreu, M.A.; Boudjellal, A.; Bremaud, L.; Ouali, A. Inhibition of human initiator caspase 8 and effector caspase 3 by cross-class inhibitory bovSERPINA3-1 and A3-3. FEBS Lett. 2009, 583, 2743–2748. [Google Scholar] [CrossRef] [PubMed]
- Gettins, P.G. Serpin structure, mechanism, and function. Chem. Rev. 2002, 102, 4751–4804. [Google Scholar] [CrossRef]
- Mehla, K.; Magotra, A.; Choudhary, J.; Singh, A.; Mohanty, A.; Upadhyay, R.; Srinivasan, S.; Gupta, P.; Choudhary, N.; Antony, B. Genome-wide analysis of the heat stress response in Zebu (Sahiwal) cattle. Gene 2014, 533, 500–507. [Google Scholar] [CrossRef]
- Li, G.; Yu, X.; Portela Fontoura, A.B.; Javaid, A.; de la Maza-Escolà, V.S.; Salandy, N.S.; Fubini, S.L.; Grilli, E.; McFadden, J.W.; Duan, J.E. Transcriptomic regulations of heat stress response in the liver of lactating dairy cows. BMC Genom. 2023, 24, 410. [Google Scholar] [CrossRef]
- Khan, R.I.N.; Sahu, A.R.; Malla, W.A.; Praharaj, M.R.; Hosamani, N.; Kumar, S.; Gupta, S.; Sharma, S.; Saxena, A.; Varshney, A. Systems biology under heat stress in Indian cattle. Gene 2021, 805, 145908. [Google Scholar] [CrossRef]
- Qu, Y.; Wang, Y.; Hu, Z.; Su, C.; Qian, C.; Pan, J.; Zhu, Y.; Shi, A. Role of metabolomic profile as a potential marker to discriminate membranous nephropathy from IgA nephropathy. Int. Urol. Nephrol. 2024, 56, 635–651. [Google Scholar] [CrossRef]
- Wu, D.; Shu, T.; Yang, X.; Song, J.-X.; Zhang, M.; Yao, C.; Liu, W.; Huang, M.; Yu, Y.; Yang, Q. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 2020, 7, 1157–1168. [Google Scholar] [CrossRef]
- Yin, C.; Harms, A.C.; Hankemeier, T.; Kindt, A.; de Lange, E.C. Status of metabolomic measurement for insights in Alzheimer’s disease progression—What is missing? Int. J. Mol. Sci. 2023, 24, 4960. [Google Scholar] [CrossRef]
- Beyoğlu, D.; Popov, Y.V.; Idle, J.R. Metabolomic Hallmarks of Obesity and Metabolic Dysfunction-Associated Steatotic Liver Disease. Int. J. Mol. Sci. 2024, 25, 12809. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Xue, Y.; Ma, Y. Identification of N α-acetyl-α-lysine as a probable thermolyte and its accumulation mechanism in Salinicoccus halodurans H3B36. Sci. Rep. 2015, 5, 18518. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Zhang, Z.; Xiang, Z.; Yang, Z. Exogenous application of citric acid ameliorates the adverse effect of heat stress in tall fescue (Lolium arundinaceum). Front. Plant Sci. 2016, 7, 179. [Google Scholar] [CrossRef]
- Bai, X.; Shi, Y.; Tang, L.; Chen, L.; Fan, H.; Wang, H.; Wang, J.; Jia, X.; Chen, S.; Lai, S. Heat stress affects faecal microbial and metabolic alterations of rabbits. Front. Microbiol. 2022, 12, 817615. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, B.A.; Snoeren, N.; Samim, M.; Rosing, H.; de Vries, N.; Deenen, M.J.; Beijnen, J.H.; Schellens, J.H.; Koopman, M.; van Hillegersberg, R. The impact of liver resection on the dihydrouracil: Uracil plasma ratio in patients with colorectal liver metastases. Eur. J. Clin. Pharmacol. 2018, 74, 737–744. [Google Scholar] [CrossRef]
- Antonioli, L.; Pacher, P.; Vizi, E.S.; Haskó, G. CD39 and CD73 in immunity and inflammation. Trends Mol. Med. 2013, 19, 355–367. [Google Scholar] [CrossRef]
- Roncal-Jimenez, C.; García-Trabanino, R.; Barregard, L.; Lanaspa, M.A.; Wesseling, C.; Harra, T.; Aragón, A.; Grases, F.; Jarquin, E.R.; González, M.A. Heat stress nephropathy from exercise-induced uric acid crystalluria: A perspective on Mesoamerican nephropathy. Am. J. Kidney Dis. 2016, 67, 20–30. [Google Scholar] [CrossRef]
- Minamino, T.; Komuro, I.; Kitakaze, M. Endoplasmic reticulum stress as a therapeutic target in cardiovascular disease. Circ. Res. 2010, 107, 1071–1082. [Google Scholar] [CrossRef]
- Qi, Y.; Xu, R. Roles of PLODs in collagen synthesis and cancer progression. Front. Cell Dev. Biol. 2018, 6, 66. [Google Scholar] [CrossRef]
- Yuan, B.; Xu, Y.; Zheng, S. PLOD1 acts as a tumor promoter in glioma via activation of the HSF1 signaling pathway. Mol. Cell. Biochem. 2022, 477, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Frezarim, G.B.; Mota, L.F.; Fonseca, L.F.; Salatta, B.M.; Arikawa, L.M.; Schmidt, P.I.; Silva, D.B.; Albuquerque, L.G. Multi-omics integration identifies molecular markers and biological pathways for carcass and meat quality traits in Nellore cattle. Sci. Rep. 2025, 15, 10467. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Vega, A.; Arranz, J.J.; Pérez, V.; de la Fuente, L.; Mateo, J.; Gutiérrez-Gil, B. Early adipose deposits in sheep: Comparative analysis of the perirenal fat transcriptome of Assaf and Churra suckling lambs. Anim. Genet. 2018, 49, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Fang, H.; Abbas, Z.; Luo, H.; Brito, L.F.; Wang, Y.; Xu, Q. The HSP90AA1 gene is involved in heat stress responses and its functional genetic polymorphisms are associated with heat tolerance in Holstein cows. J. Dairy Sci. 2024, 107, 5132–5149. [Google Scholar] [CrossRef]
- Tian, H.; Wang, W.; Zheng, N.; Cheng, J.; Li, S.; Zhang, Y.; Wang, J. Identification of diagnostic biomarkers and metabolic pathway shifts of heat-stressed lactating dairy cows. J. Proteom. 2015, 125, 17–28. [Google Scholar] [CrossRef]
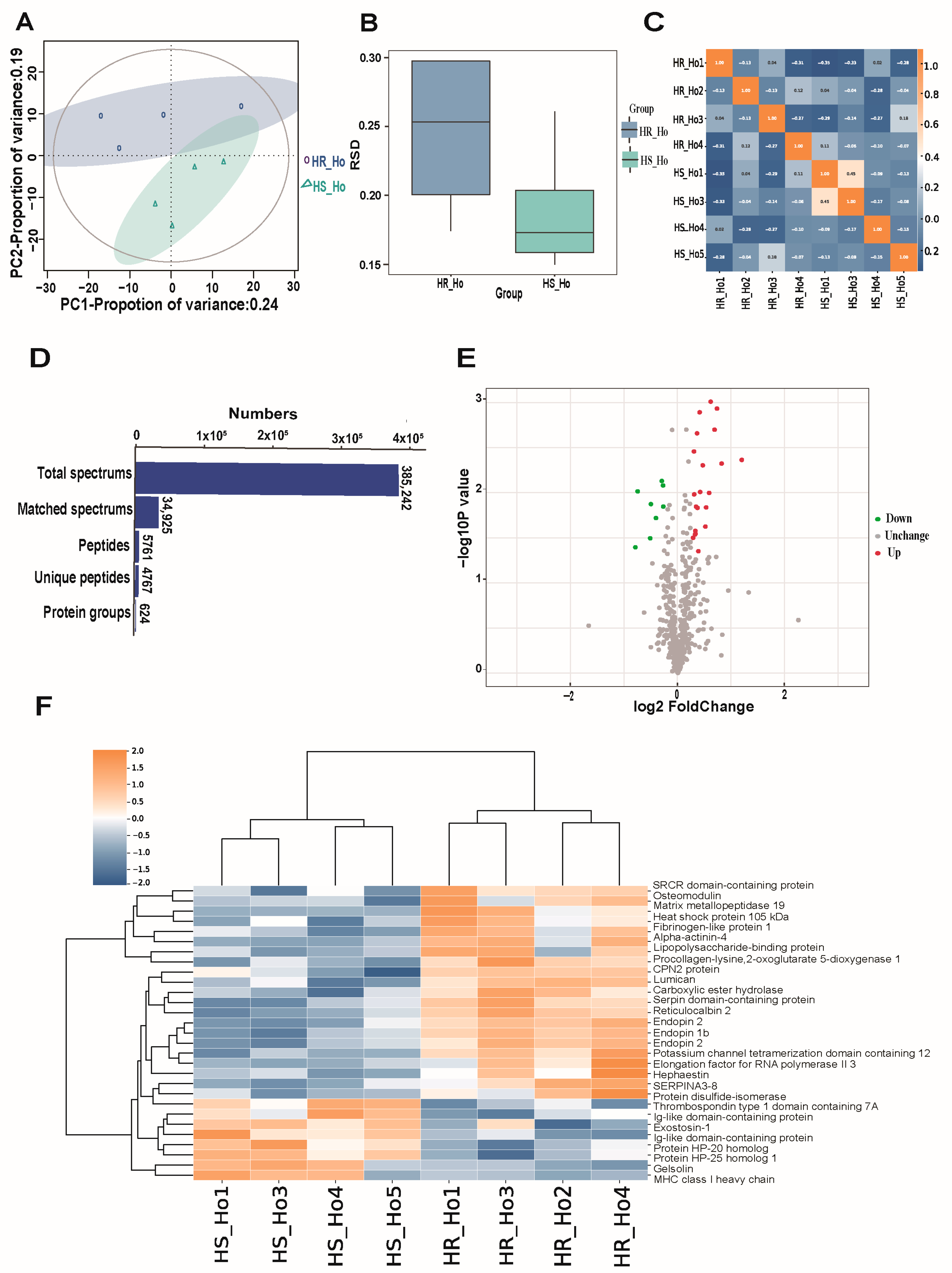


| UniProKB | GeneName | AAs | Coverage [%] | Peptides | PSMs | Unique Peptides | AAs | MW | calc. | Score | Regulation | p-Value | HR_HO/HS_HO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [kDa] | pI | Mascot | |||||||||||
| A2I7N3 | SERPINA3-7 | 417 | 49 | 22 | 394 | 13 | 417 | 46.9 | 6.01 | 6896 | UP | 0.0043325 | 2.29 |
| A5D7D1 | ACTN4 | 1032 | 1 | 1 | 1 | 1 | 1032 | 117 | 6.38 | 0 | UP | 0.0047489 | 1.76 |
| Q3ZEJ6 | SERPINA3-3 | 411 | 43 | 15 | 326 | 5 | 411 | 46.1 | 6.48 | 8029 | UP | 0.0011647 | 1.66 |
| A2I7N1 | SERPINA3-7 | 415 | 33 | 16 | 448 | 1 | 415 | 46.5 | 5.81 | 10,012 | UP | 0.0020006 | 1.61 |
| Q0VCQ9 | RCN2 | 317 | 3 | 1 | 1 | 1 | 317 | 36.9 | 4.4 | 0 | UP | 0.0009764 | 1.53 |
| F6PYF1 | MMP19 | 499 | 3 | 1 | 1 | 1 | 499 | 56.4 | 6.77 | 42 | UP | 0.0100722 | 1.50 |
| UPI0000616416 | MGC137014 | 191 | 66 | 10 | 113 | 10 | 191 | 20.6 | 8.59 | 2185 | DOWN | 0.0142715 | 0.83 |
| A0A4W2EXA9 | THSD7A | 1676 | 1 | 1 | 2 | 1 | 1676 | 186.8 | 7.43 | 0 | DOWN | 0.0083202 | 0.83 |
| UPI0000693DEA | 212 | 50 | 9 | 67 | 9 | 212 | 22.5 | 7.46 | 1662 | DOWN | 0.0074206 | 0.81 | |
| A5D7I4 | EXT1 | 746 | 2 | 1 | 1 | 1 | 746 | 86.2 | 9.04 | 30 | DOWN | 0.0191447 | 0.75 |
| F1N1I6 | GSN | 846 | 61 | 42 | 316 | 1 | 846 | 92 | 6.86 | 11,356 | DOWN | 0.0134129 | 0.71 |
| ID | Metabolites | Iron | p-Value | VIP Value | Up/Down-Regulated Status |
|---|---|---|---|---|---|
| M155T113 | Trigonelline | Positive | 7.57 × 10−9 | 4.00 | Up |
| M283T467 | 2-methyl-2-(4-methylpent-3-en-1-yl)-2H-chromen-8-ol | Positive | 9.03 × 10−6 | 4.07 | Down |
| M581T467 | Atorvastatin | Positive | 1.08 × 10−5 | 4.16 | Down |
| M607T467 | Phaeophorbide b | Positive | 2.57 × 10−5 | 4.12 | Down |
| M275T138 | Phenytoin | Positive | 2.73 × 10−5 | 3.03 | Up |
| M212T138 | Valganciclovir | Negative | 7.73 × 10−8 | 3.23 | Up |
| M256T65 | 5-Methylcytidine | Negative | 8.66 × 10−7 | 3.45 | Up |
| M383T380 | MG(0:0/20:1(11Z)/0:0) | Negative | 8.90 × 10−7 | 2.79 | Down |
| M333T96 | Penicillin G | Negative | 1.48 × 10−6 | 3.02 | Down |
| M279T95 | 6-[(2-carboxyacetyl)oxy]-3,4,5-trihydroxyoxane-2-carboxylic acid | Negative | 4.26 × 10−6 | 3.04 | Down |
| Pathways | Pathway Code | Protein-Coding Gene | Metabolites |
|---|---|---|---|
| Lysine degradation | ko00310 | Up-regulated PLOD1 | Down-regulated N6-Acetyl-L-lysine |
| Metabolic pathways | ko01100 | Up-regulated PLOD1 and down-regulated EXT1 | Down-regulated citric acid and down-regulated 4-Pyridoxic acid |
| Down-regulated uracil, down-regulated uric acid, down-regulated N6-Acetyl-L-lysine, and up-regulated arachidonic acid | |||
| Fc gamma R-mediated Phagocytosis | ko04666 | Down-regulated GSN | Up-regulated arachidonic acid |
| Amoebiasis | ko05146 | Up-regulated ACTN4 | Up-regulated arachidonic acid |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Yuan, Y.; Pei, F.; Yang, J.; Wang, C.; Bao, P.; Zhao, X.; Liu, H.; Gao, H.; Hou, M.; et al. Integrated Analysis of Proteomics and Metabolomics for Heat Stress in Chinese Holstein Cows. Animals 2025, 15, 3049. https://doi.org/10.3390/ani15203049
Wang X, Yuan Y, Pei F, Yang J, Wang C, Bao P, Zhao X, Liu H, Gao H, Hou M, et al. Integrated Analysis of Proteomics and Metabolomics for Heat Stress in Chinese Holstein Cows. Animals. 2025; 15(20):3049. https://doi.org/10.3390/ani15203049
Chicago/Turabian StyleWang, Xiao, Yinglin Yuan, Fen Pei, Jian Yang, Chenchen Wang, Peng Bao, Xiuxin Zhao, Huiming Liu, Hongding Gao, Minghai Hou, and et al. 2025. "Integrated Analysis of Proteomics and Metabolomics for Heat Stress in Chinese Holstein Cows" Animals 15, no. 20: 3049. https://doi.org/10.3390/ani15203049
APA StyleWang, X., Yuan, Y., Pei, F., Yang, J., Wang, C., Bao, P., Zhao, X., Liu, H., Gao, H., Hou, M., Gao, Y., Li, J., Hao, D., & Li, R. (2025). Integrated Analysis of Proteomics and Metabolomics for Heat Stress in Chinese Holstein Cows. Animals, 15(20), 3049. https://doi.org/10.3390/ani15203049






