Genomic Insights into Genetic Diversity and Adaptation of Nanyang Cattle: Implications for Conservation and Breeding
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Sequencing
2.3. Genetic Diversity Analysis
2.4. Population Structure Analysis
2.5. Selective Signature Analysis and Functional Enrichment
3. Results
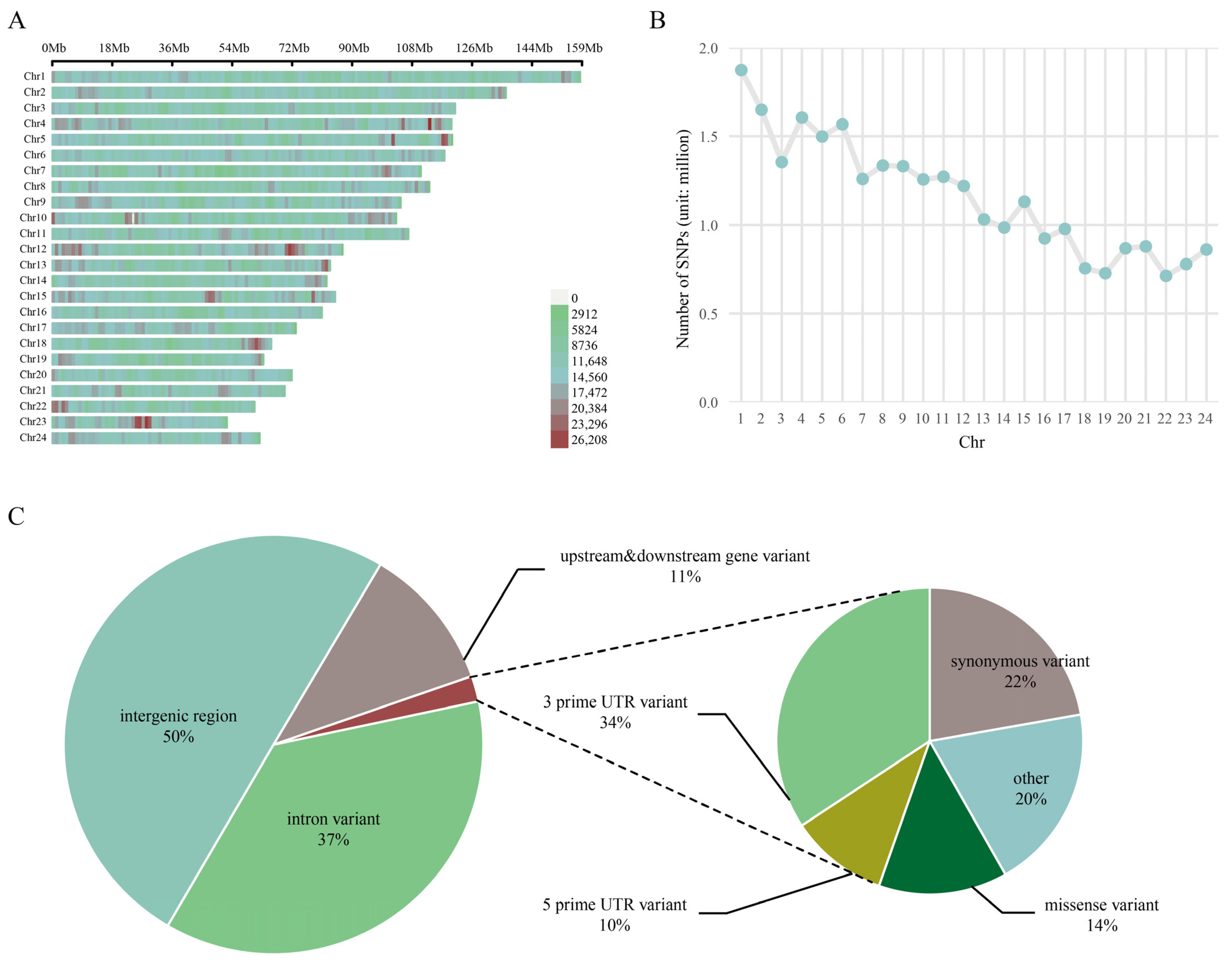
3.1. Descriptive Statistics of SNP Loci from Whole-Genome Resequencing Data
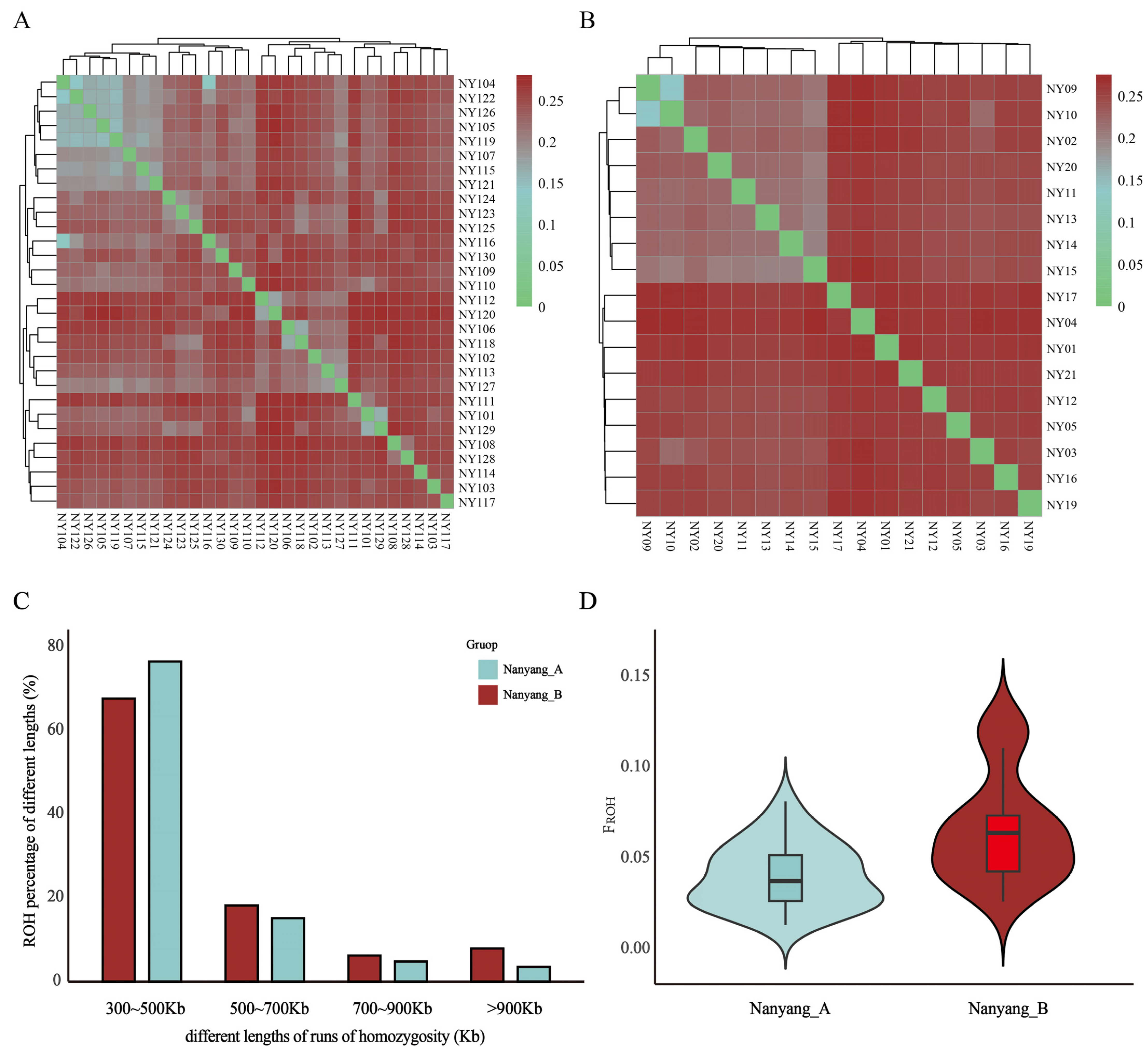
3.2. Genomic Genetic Diversity Statistics
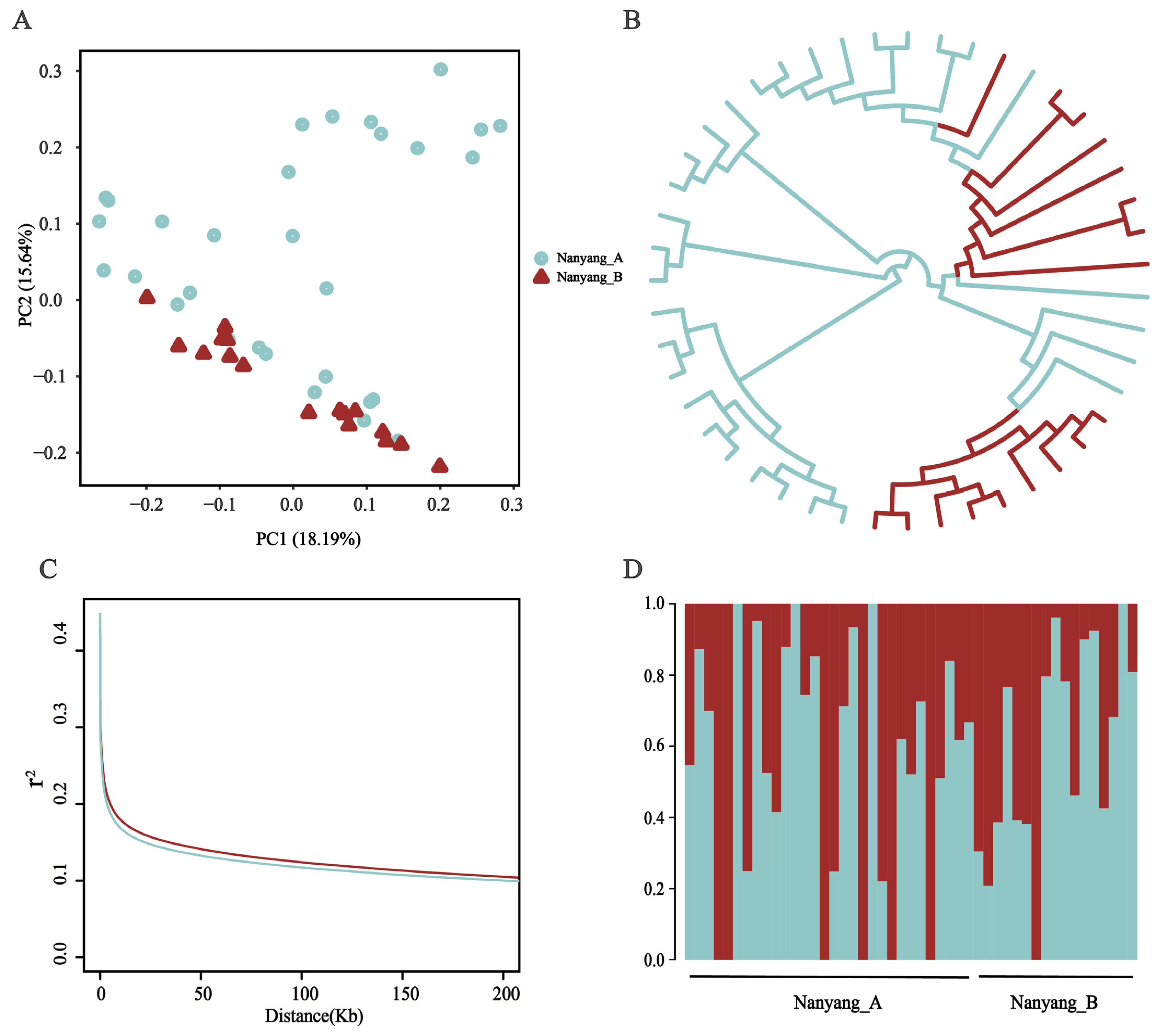
3.3. Population Structure Analysis
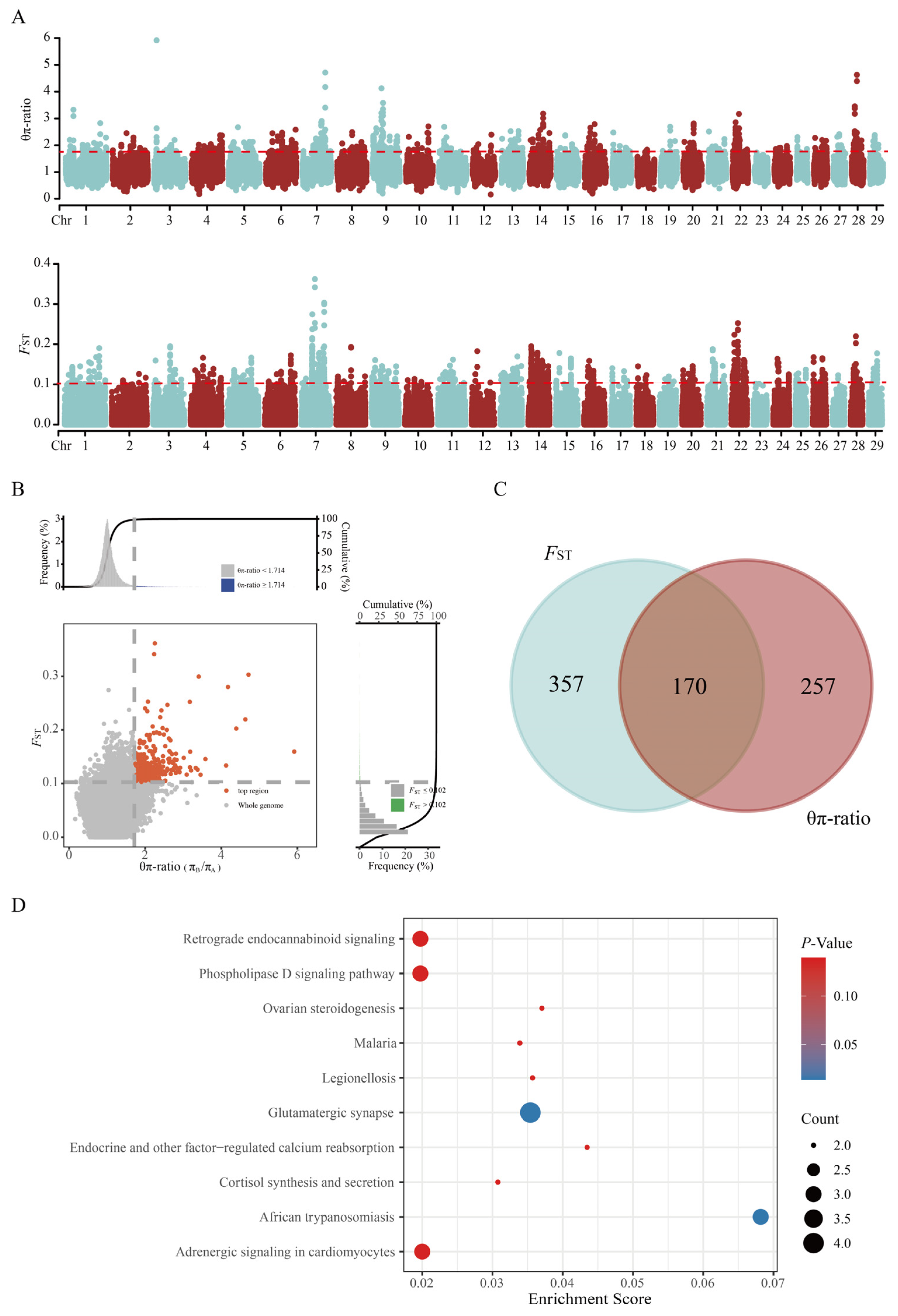
3.4. Selective Signature Analysis and Functional Enrichment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| WGS | Whole-genome sequencing |
| PCA | principal component analysis |
| NJ | Neighbor-joining |
| FST | Ixation index |
| θπ ratio | Nucleotide diversity ratio |
| IBS | Identity-by-state |
| ROH | Runs of homozygosity |
| FROH | Inbreeding coefficient |
| LD | Linkage disequilibrium |
References
- Chen, N.; Cai, Y.; Chen, Q.; Li, R.; Wang, K.; Huang, Y.; Hu, S.; Huang, S.; Zhang, H.; Zheng, Z.; et al. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat. Commun. 2018, 9, 2337. [Google Scholar] [CrossRef]
- Zhang, M.; Peng, W.F.; Hu, X.J.; Zhao, Y.X.; Lv, F.H.; Yang, J. Global genomic diversity and conservation priorities for domestic animals are associated with the economies of their regions of origin. Sci. Rep. 2018, 8, 11677. [Google Scholar] [CrossRef]
- Ramljak, J.; Bunevski, G.; Bytyqi, H.; Marković, B.; Brka, M.; Ivanković, A.; Kume, K.; Stojanović, S.; Nikolov, V.; Simčič, M.; et al. Conservation of a domestic metapopulation structured into related and partly admixed strains. Mol. Ecol. 2018, 27, 1633–1650. [Google Scholar] [CrossRef]
- Ginja, C.; Gama, L.T.; Cortés, O.; Burriel, I.M.; Vega-Pla, J.L.; Penedo, C.; Sponenberg, P.; Cañón, J.; Sanz, A.; do Egito, A.A.; et al. The genetic ancestry of American Creole cattle inferred from uniparental and autosomal genetic markers. Sci. Rep. 2019, 9, 11486. [Google Scholar] [CrossRef]
- Lyu, Y.; Wang, F.; Cheng, H.; Han, J.; Dang, R.; Xia, X.; Wang, H.; Zhong, J.; Lenstra, J.A.; Zhang, H.; et al. Recent selection and introgression facilitated high-altitude adaptation in cattle. Sci. Bull. 2024, 69, 3415–3424. [Google Scholar] [CrossRef] [PubMed]
- Mei, C.; Wang, H.; Liao, Q.; Wang, L.; Cheng, G.; Wang, H.; Zhao, C.; Zhao, S.; Song, J.; Guang, X.; et al. Genetic Architecture and Selection of Chinese Cattle Revealed by Whole Genome Resequencing. Mol. Biol. Evol. 2018, 35, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gao, X.; Zhang, Y.; Zhao, Y.; Zhang, J.; Jia, Y.; Zhu, B.; Xu, L.; Zhang, L.; Gao, H.; et al. Genome-wide assessment of genetic diversity and population structure insights into admixture and introgression in Chinese indigenous cattle. BMC Genet. 2018, 19, 114. [Google Scholar] [CrossRef]
- Zhang, L.; Jia, S.; Plath, M.; Huang, Y.; Li, C.; Lei, C.; Zhao, X.; Chen, H. Impact of Parental Bos taurus and Bos indicus Origins on Copy Number Variation in Traditional Chinese Cattle Breeds. Genome Biol. Evol. 2015, 7, 2352–2361. [Google Scholar] [CrossRef]
- Kosugi, S.; Momozawa, Y.; Liu, X.; Terao, C.; Kubo, M.; Kamatani, Y. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 2019, 20, 117. [Google Scholar] [CrossRef] [PubMed]
- Austin-Tse, C.A.; Jobanputra, V.; Perry, D.L.; Bick, D.; Taft, R.J.; Venner, E.; Gibbs, R.A.; Young, T.; Barnett, S.; Belmont, J.W.; et al. Best practices for the interpretation and reporting of clinical whole genome sequencing. npj Genom. Med. 2022, 7, 27. [Google Scholar] [CrossRef]
- Yan, C.L.; Lin, J.; Huang, Y.Y.; Gao, Q.S.; Piao, Z.Y.; Yuan, S.L.; Chen, L.; Ren, X.; Ye, R.C.; Dong, M.; et al. Population genomics reveals that natural variation in PRDM16 contributes to cold tolerance in domestic cattle. Zool. Res. 2022, 43, 275–284. [Google Scholar] [CrossRef]
- Xia, X.; Wang, F.; Luo, X.; Li, S.; Lyu, Y.; Zheng, Y.; Ma, Z.; Qu, K.; Song, R.; Liu, J.; et al. Structural Variations Associated with Adaptation and Coat Color in Qinghai-Tibetan Plateau Cattle. Adv. Sci. 2025, 12, e03258. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang le, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Ginestet, C. ggplot2: Elegant Graphics for Data Analysis. J. R. Stat. Soc. Ser. A Stat. Soc. 2011, 174, 245–246. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.-S.; Xu, J.-Y.; He, W.-M.; Yang, T.-L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2018, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Akash; Hoque, M.; Mondal, S.; Adusumilli, S. Chapter Four—Sustainable livestock production and food security. In Emerging Issues in Climate Smart Livestock Production; Mondal, S., Singh, R.L., Eds.; Academic Press: San Diego, CA, USA, 2022; pp. 71–90. [Google Scholar]
- Purfield, D.C.; Berry, D.P.; McParland, S.; Bradley, D.G. Runs of homozygosity and population history in cattle. BMC Genet. 2012, 13, 70. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.C.; Visscher, P.M.; Goddard, M.E. Quantification of inbreeding due to distant ancestors and its detection using dense single nucleotide polymorphism data. Genetics 2011, 189, 237–249. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, Z.; Zhang, M.; Wang, S.; Gao, T.; Huang, H.; Zhang, T.; Cai, H.; Liu, X.; Fu, T.; et al. Population Structure and Selection Signal Analysis of Nanyang Cattle Based on Whole-Genome Sequencing Data. Genes 2024, 15, 351. [Google Scholar] [CrossRef] [PubMed]
- Hunter, P. Genetics against extinction: New conservation strategies consider genetic diversity and habitat loss. EMBO Rep. 2023, 24, e57521. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, F.; Di Menna, L.; Iacovelli, L.; Orlando, R.; Zuena, A.R.; Conn, P.J.; Dogra, S.; Joffe, M.E. GPCR interactions involving metabotropic glutamate receptors and their relevance to the pathophysiology and treatment of CNS disorders. Neuropharmacology 2023, 235, 109569. [Google Scholar] [CrossRef]
- Arundine, M.; Tymianski, M. Molecular mechanisms of glutamate-dependent neurodegeneration in ischemia and traumatic brain injury. Cell Mol. Life Sci. 2004, 61, 657–668. [Google Scholar] [CrossRef]
- Mangan, R.J.; Alsina, F.C.; Mosti, F.; Sotelo-Fonseca, J.E.; Snellings, D.A.; Au, E.H.; Carvalho, J.; Sathyan, L.; Johnson, G.D.; Reddy, T.E.; et al. Adaptive sequence divergence forged new neurodevelopmental enhancers in humans. Cell 2022, 185, 4587–4603.e4523. [Google Scholar] [CrossRef]
- Schumacher, T.; Reyer, H.; Maak, S.; Röntgen, M. Homer 1 genotype AA variant relates to congenital splay leg syndrome in piglets by repressing Pax7 in myogenic progenitors. Front. Vet. Sci. 2023, 10, 1028879. [Google Scholar] [CrossRef]
- Blottner, D.; Trautmann, G.; Furlan, S.; Gambara, G.; Block, K.; Gutsmann, M.; Sun, L.W.; Worley, P.F.; Gorza, L.; Scano, M.; et al. Reciprocal Homer1a and Homer2 Isoform Expression Is a Key Mechanism for Muscle Soleus Atrophy in Spaceflown Mice. Int. J. Mol. Sci. 2021, 23, 75. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, P.; Zhang, C.; Zuo, Y.; Zhou, Y.; Han, R. Defective BVES-mediated feedback control of cAMP in muscular dystrophy. Nat. Commun. 2023, 14, 1785. [Google Scholar] [CrossRef]
- Zheng, H.; An, M.; Luo, Y.; Diao, X.; Zhong, W.; Pang, M.; Lin, Y.; Chen, J.; Li, Y.; Kong, Y.; et al. PDGFRα(+)ITGA11(+) fibroblasts foster early-stage cancer lymphovascular invasion and lymphatic metastasis via ITGA11-SELE interplay. Cancer Cell 2024, 42, 682–700.e612. [Google Scholar] [CrossRef]
- Song, J.; Li, Y.; Wu, K.; Hu, Y.; Fang, L. MyD88 and Its Inhibitors in Cancer: Prospects and Challenges. Biomolecules 2024, 14, 562. [Google Scholar] [CrossRef] [PubMed]
- Michell, R.H.; Allan, D. Inositol cyclis phosphate as a product of phosphatidylinositol breakdown by phospholipase C (Bacillus cereus). FEBS Lett. 1975, 53, 302–304. [Google Scholar] [CrossRef]
- Streb, H.; Irvine, R.F.; Berridge, M.J.; Schulz, I. Release of Ca2+ from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1,4,5-trisphosphate. Nature 1983, 306, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.K. How to choose a sampling technique and determine sample size for research: A simplified guide for researchers. Oral Oncol. Rep. 2024, 12, 100662. [Google Scholar] [CrossRef]
- Sham, P.C.; Purcell, S.M. Statistical power and significance testing in large-scale genetic studies. Nat. Rev. Genet. 2014, 15, 335–346. [Google Scholar] [CrossRef]
- Xu, Y.; Jiang, Y.; Shi, T.; Cai, H.; Lan, X.; Zhao, X.; Plath, M.; Chen, H. Whole-genome sequencing reveals mutational landscape underlying phenotypic differences between two widespread Chinese cattle breeds. PLoS ONE 2017, 12, e0183921. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Liu, X.; Liu, J.; Fu, T.; Huang, H.; Han, M.; Liang, D.; Gao, T. Genomic Insights into Genetic Diversity and Adaptation of Nanyang Cattle: Implications for Conservation and Breeding. Animals 2025, 15, 3033. https://doi.org/10.3390/ani15203033
Zhang Y, Liu X, Liu J, Fu T, Huang H, Han M, Liang D, Gao T. Genomic Insights into Genetic Diversity and Adaptation of Nanyang Cattle: Implications for Conservation and Breeding. Animals. 2025; 15(20):3033. https://doi.org/10.3390/ani15203033
Chicago/Turabian StyleZhang, Yan, Xian Liu, Jiakun Liu, Tong Fu, Hetian Huang, Mingpeng Han, Dong Liang, and Tengyun Gao. 2025. "Genomic Insights into Genetic Diversity and Adaptation of Nanyang Cattle: Implications for Conservation and Breeding" Animals 15, no. 20: 3033. https://doi.org/10.3390/ani15203033
APA StyleZhang, Y., Liu, X., Liu, J., Fu, T., Huang, H., Han, M., Liang, D., & Gao, T. (2025). Genomic Insights into Genetic Diversity and Adaptation of Nanyang Cattle: Implications for Conservation and Breeding. Animals, 15(20), 3033. https://doi.org/10.3390/ani15203033





