A New Species of Pachytriton (Amphibia: Caudata: Salamandridae) from Anhui, China
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Molecular Data and Phylogenetic Analysis
2.3. Morphological Comparison and Analyses
3. Results
3.1. Molecular Phylogenetic and Species Delimitation Analyses
3.2. Morphological Comparisons
3.3. Taxonomic Accounts
- Holotype
- Paratype
- Etymology
- Diagnosis
- Description of the holotype
- Coloration
- Variations
- Distribution and habitat
- Comparisons
- Conservation recommendation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luedtke, J.A.; Chanson, J.; Neam, K.; Hobin, L.; Maciel, A.O.; Catenazzi, A.; Borzée, A.; Hamidy, A.; Aowphol, A.; Jean, A.; et al. Ongoing declines for the world’s amphibians in the face of emerging threats. Nature 2023, 622, 308–314. [Google Scholar] [CrossRef]
- Nishikawa, K.; Matsui, M.; Wang, B.; Yoshikawa, N.; Jiang, J.P. Taxonomic relationship of two newt species of Pachytriton recently described from China (Amphibia: Urodela: Salamandridae). Curr. Herpetol. 2013, 32, 150–158. [Google Scholar] [CrossRef]
- Jiang, J.P.; Cai, B.; Wang, B.; Chen, W.T.; Wen, Z.X.; Zhang, D.Z.; Sui, L.L.; Ma, S. New vertebrate species discovered in China in 2022. Biodivers. Sci. 2023, 31, 23241. [Google Scholar] [CrossRef]
- Jiang, J.P.; Cai, B.; Wang, B.; Chen, W.T.; Wen, Z.X.; Zhang, D.Z.; Sui, L.L.; Ma, S. New vertebrate species discovered in China in 2023. Biodivers. Sci. 2024, 32, 24257. [Google Scholar] [CrossRef]
- Huang, P.Q.; Wang, F.; Lu, H.; Rong, P.; Xiao, X.Y.; Zhang, F.; Huang, S. Pachytriton granulosus (Caudata: Salamandridae): A New Amphibian Record in Anhui Province, China. Sichuan J. Zool. 2023, 42, 420–427, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- AmphibiaChina. AmphibiaChina Information System. 2025. Available online: http://www.amphibiachina.org/ (accessed on 1 August 2025).
- Frost, D.R. Amphibian Species of the World: An Online Reference, Version 6.2; American Museum of Natural History: New York, NY, USA, 2025; Available online: http://research.amnh.org/herpetology/amphibia/index.php/ (accessed on 8 October 2025).
- Li, C.; Yuan, Z.; Li, H.; Wu, Y. The tenth member of stout newt (Amphibia: Salamandridae: Pachytriton): Description of a new species from Guangdong, southern China. Zootaxa 2018, 4399, 207–219. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Jiang, K.; Chen, X.; Hanken, J. Homoplastic evolution of external coloration in Asian stout newts (Pachytriton) inferred from molecular phylogeny. Zool. Scr. 2010, 39, 9–22. [Google Scholar] [CrossRef]
- Yuan, Z.Y.; Zhang, B.L.; Che, J. A new species of the genus Pachytriton (Caudata: Salamandridae) from Hunan and Guangxi, southeastern China. Zootaxa 2016, 4085, 219–232. [Google Scholar] [CrossRef]
- Nishikawa, K.; Jiang, J.P.; Matsui, M. Two new species of Pachytriton from Anhui and Guangxi, China (Amphibia: Urodela: Salamandridae). Curr. Herpetol. 2011, 30, 15–31. [Google Scholar] [CrossRef][Green Version]
- Shen, Y.-P.; Nishikawa, K.; Jiang, J.-P.; Matsui, M.; Rao, D.-Q.; Yoshikawa, N.; Tominaga, A.; Sanamxay, D. Lotic specialization in modern Asian newts (Caudata: Salamandridae): Phylogeny, historical biogeography, and ancestral traits based on combined DNA data. Zool. Res. Divers. Conserv. 2024, 1, 191–200. [Google Scholar] [CrossRef]
- Lyu, Z.T.; Wang, J.; Zeng, Z.C.; Zhou, J.J.; Qi, S.; Wan, H.; Li, Y.Y.; Wang, Y.Y. A new species of the genus Tylototriton (Caudata, Salamandridae) from Guangdong, southern China, with discussion on the subgenera and species groups within the genus. Vertebr. Zool. 2021, 71, 697–710. [Google Scholar] [CrossRef]
- Vieites, D.R.; Min, M.S.; Wake, D.B. Rapid diversification and dispersal during periods of global warming by plethodontid salamanders. Proc. Natl. Acad. Sci. USA 2007, 104, 19903–19907. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, X.; Jin, E.; Wang, A.; Chen, T.; Zhang, X.; Zhu, J.; Dong, L.; Sun, Y.; Yu, C.; et al. The GSA Family in 2025: A Broadened Sharing Platform for Multi-omics and Multimodal Data. Genom. Proteom. Bioinform. 2025, 23, qzaf072. [Google Scholar] [CrossRef]
- CNCB-NGDC Members and Partners. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2025. Nucleic Acids Res. 2025, 53, D30–D44. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A. Tracer v1.4. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 19 January 2016).
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D.; Nakagawa, S. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Wu, Y.; Murphy, R.W. Concordant species delimitation from multiple independent evidence: A case study with the Pachytriton brevipes complex (Caudata: Salamandridae). Mol. Phylogenet. Evol. 2015, 92, 108–117. [Google Scholar] [CrossRef]
- Heled, J.; Drummond, A.J. Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 2010, 27, 570–580. [Google Scholar] [CrossRef]
- Edwards, S.V.; Xi, Z.; Janke, A.; Faircloth, B.C.; McCormack, J.E.; Glenn, T.C.; Zhong, B.; Wu, S.; Lemmon, E.M.; Lemmon, A.R.; et al. Implementing and testing the multispecies coalescent model: A valuable paradigm for phylogenomics. Mol. Phylogenet. Evol. 2016, 94, 447–462. [Google Scholar] [CrossRef] [PubMed]
- Solís-Lemus, C.; Knowles, L.L.; Ané, C. Bayesian species delimitation combining multiple genes and traits in a unified framework. Evolution 2015, 69, 492–507. [Google Scholar] [CrossRef] [PubMed]
- Kornai, D.; Jiao, X.; Ji, J.; Flouri, T.; Yang, Z. Hierarchical heuristic species delimitation under the multispecies coalescent model with migration. Syst. Biol. 2024, 73, 1015–1037. [Google Scholar] [CrossRef] [PubMed]
- Villamil, J.; Morando, M.; Avila, L.J.; Lanna, F.M.; Fonseca, E.M.; Sites, J.W., Jr.; Camargo, A. Revisiting the Multispecies Coalescent Model fit with an example from a complete molecular phylogeny of the Liolaemus wiegmannii species group (Squamata: Liolaemidae). Syst. Biol. 2025, 74, syaf048. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Baele, G.; Lemey, P.; Bedford, T.; Rambaut, A.; Suchard, M.A.; Alekeyenko, A.V. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. 2012, 29, 2157–2167. [Google Scholar] [CrossRef]
- Xie, W.; Lewis, P.O.; Fan, Y.; Kuo, L.; Chen, M.H. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst. Biol. 2011, 60, 150–160. [Google Scholar] [CrossRef]
- Kass, R.E.; Raftery, A.E. Bayes factors. J. Am. Stat. Assoc. 1995, 90, 773–795. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Jiang, K.; Hanken, J. Significance of pre-Quaternary climate change for montane species diversity: Insights from Asian salamanders (Salamandridae: Pachytriton). Mol. Phylogenet. Evol. 2013, 66, 380–390. [Google Scholar] [CrossRef]
- Nishikawa, K.; Jiang, J.P.; Matsui, M.; Mo, Y.M. Unmasking Pachytriton labiatus (Amphibia: Urodela: Salamandridae), with description of a new species of Pachytriton from Guangxi, China. Zool. Sci. 2011, 28, 453–461. [Google Scholar] [CrossRef]
- Nishikawa, K.; Matsui, M.; Jiang, J.P. A new species of Pachytriton from China (Amphibia: Urodela: Salamandridae). Curr. Herpetol. 2012, 31, 21–27. [Google Scholar] [CrossRef]
- Thorpe, R.S. Quantitative handling of characters useful in snake systematics with particular reference to intraspecific variation in the Ringed Snake Natrix natrix. Biol. J. Linn. Soc. 1975, 7, 27–43. [Google Scholar] [CrossRef]
- Lyu, Z.T.; Chen, Y.; Yang, J.H.; Zeng, Z.C.; Wang, J.; Zhao, J.; Wan, H.; Pang, H.; Wang, Y.Y. A new species of Nidirana from the N. pleuraden group (Anura, Ranidae) from western Yunnan, China. Zootaxa 2020, 4861, 43–62. [Google Scholar] [CrossRef]
- Fei, L.; Hu, S.Q.; Ye, C.Y.; Huang, Y.Z. Fauna Sinica, Amphibia: General Accounts of Gymnophiona and Urodela; Science Press: Beijing, China, 2006; Volume 1. (In Chinese) [Google Scholar]
- Chen, B.H. Anhui Amphibians and Reptiles Fauna; Anhui Science and Technology Press: Hefei, China, 1991. (In Chinese) [Google Scholar]
- Longcore, J.E.; Pessier, A.P.; Nichols, D.K. Batrachochytrium dendrobatidis gen. et sp. nov., a chytrid pathogenic to amphibians. Mycologia 1999, 91, 219–227. [Google Scholar] [CrossRef]
- Martel, A.; Spitzen-van der Sluijs, A.; Blooi, M.; Bert, W.; Ducatelle, R.; Fisher, M.C.; Woeltjes, A.; Bosman, W.; Chiers, K.; Bossuyt, F.; et al. Batrachochytrium salamandrivorans sp. nov. causes lethal chytridiomycosis in amphibians. Proc. Natl. Acad. Sci. USA 2013, 110, 15325–15329. [Google Scholar] [CrossRef]
- Wang, S.P.; Zhu, W.; Fan, L.Q.; Li, J.Q.; Li, Y.M. Amphibians testing negative for Batrachochytrium dendrobatidis and Batrachochytrium salamandrivorans on the Qinghai-Tibetan Plateau, China. Asian Herpetol. Res. 2017, 8, 190–198. [Google Scholar] [CrossRef]
- He, Z.R.; Wu, S.Y.; Shi, Y.Y.; Wang, Y.T.; Jiang, Y.X.; Zhang, C.N.; Zhao, N.; Wang, S.P. Current status and challenges on the effects of chytrid infection on amphibian populations. Biodivers. Sci. 2024, 32, 23274. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Hanken, J. New species of Pachytriton (Caudata: Salamandridae) from the Nanling mountain range, southeastern China. Zootaxa 2012, 3388, 1–16. [Google Scholar] [CrossRef]
- Xu, W.; Wu, Y.H.; Zhou, W.W.; Chen, H.M.; Zhang, B.L.; Chen, J.M.; Xu, W.; Rao, D.-Q.; Zhao, H.; Yan, F. Hidden hotspots of amphibian biodiversity in China. Proc. Natl. Acad. Sci. USA 2024, 121, e2320674121. [Google Scholar] [CrossRef]
- Fei, L.; Ye, C.; Jiang, J. Colored Atlas of Chinese Amphibians and Their Distributions; Chongqing Publishing House: Chongqing, China, 2012. (In Chinese) [Google Scholar]
- Brown, R.M.; Siler, C.D.; Oliveros, C.H.; Welton, L.J.; Rock, A.; Swab, J.; Van Weerd, M.; van Beijnen, J.; Jose, E.; Rodriguez, D.; et al. The amphibians and reptiles of Luzon Island, Philippines, VIII: The herpetofauna of Cagayan and Isabela Provinces, northern Sierra Madre Mountain Range. ZooKeys 2013, 266, 1–120. [Google Scholar] [CrossRef]
- Wake, D.B.; Deban, S.M. Terrestrial feeding in salamanders. In Functional Vertebrate Morphology; Hildebrand, M., Bramble, D.M., Liem, K.F., Wake, D.B., Eds.; Harvard University Press: Cambridge, MA, USA, 1985; pp. 95–116. [Google Scholar]
- Blake, R.W. Fish functional design and swimming performance. J. Fish Biol. 2004, 65, 1193–1222. [Google Scholar] [CrossRef]
- Ashley-Ross, M.A.; Bechtel, B.F. Kinematics of the transition between aquatic and terrestrial locomotion in the newt Taricha torosa. J. Exp. Biol. 2004, 207, 461–474. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guo, P.; Liu, Q.; Xu, Y.; Jiang, K.; Hou, M.; Ding, L.; Pyron, R.A.; Burbrink, F.T. Out of Asia: Natricine snakes support the Cenozoic Beringian Dispersal Hypothesis. Mol. Phylogenet. Evol. 2016, 102, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Freeman, B.G.; Scholer, M.N.; Ruiz-Gutierrez, V.; Fitzpatrick, J.W. Climate change causes upslope shifts and mountaintop extirpations in a tropical bird community. Science 2018, 362, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, J.; Gao, J.; Wei, Z.; Jin, Y. The impact of elevation and prediction of climate change on an ultra—high-elevation ectotherm. Ecol. Indic. 2025, 169, 106644. [Google Scholar] [CrossRef]
- Imperio, S.; Bionda, R.; Viterbi, R.; Provenzale, A. Climate change and human disturbance can lead to local extinction of Alpine rock ptarmigan: New insight from the western Italian Alps. Glob. Change Biol. 2014, 20, 1217–1227. [Google Scholar] [CrossRef]
- Ferrarini, A.; Alatalo, J.M.; Gustin, M. Climate change will seriously impact bird species dwelling above the treeline: A prospective study for the Italian Alps. Sci. Total Environ. 2017, 586, 960–968. [Google Scholar] [CrossRef]
- Hebblewhite, M.; Merrill, E.H.; McDonald, T.L.; Musiani, M.; Paquet, P.C.; Mattson, D.J. Projecting the future of an alpine ungulate under climate change scenarios. Conserv. Biol. 2017, 31, 1347–1358. [Google Scholar] [CrossRef]
- Rangel-Chamorro, C.; Renjifo, L.M. Elevational ranges of montane birds and deforestation in the Western Andes of Colombia. PLoS ONE 2015, 10, e0143311. [Google Scholar] [CrossRef]
- Li, B.; Liang, C.; Song, P.; Liu, D.; Qin, W.; Jiang, F.; Gu, H.; Gao, H.; Zhang, T. Threatened birds face new distribution under future climate change on the Qinghai-Tibet Plateau (QTP). Ecol. Indic. 2023, 150, 110217. [Google Scholar] [CrossRef]
- Fisher, M.C.; Garner, T.W.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef]

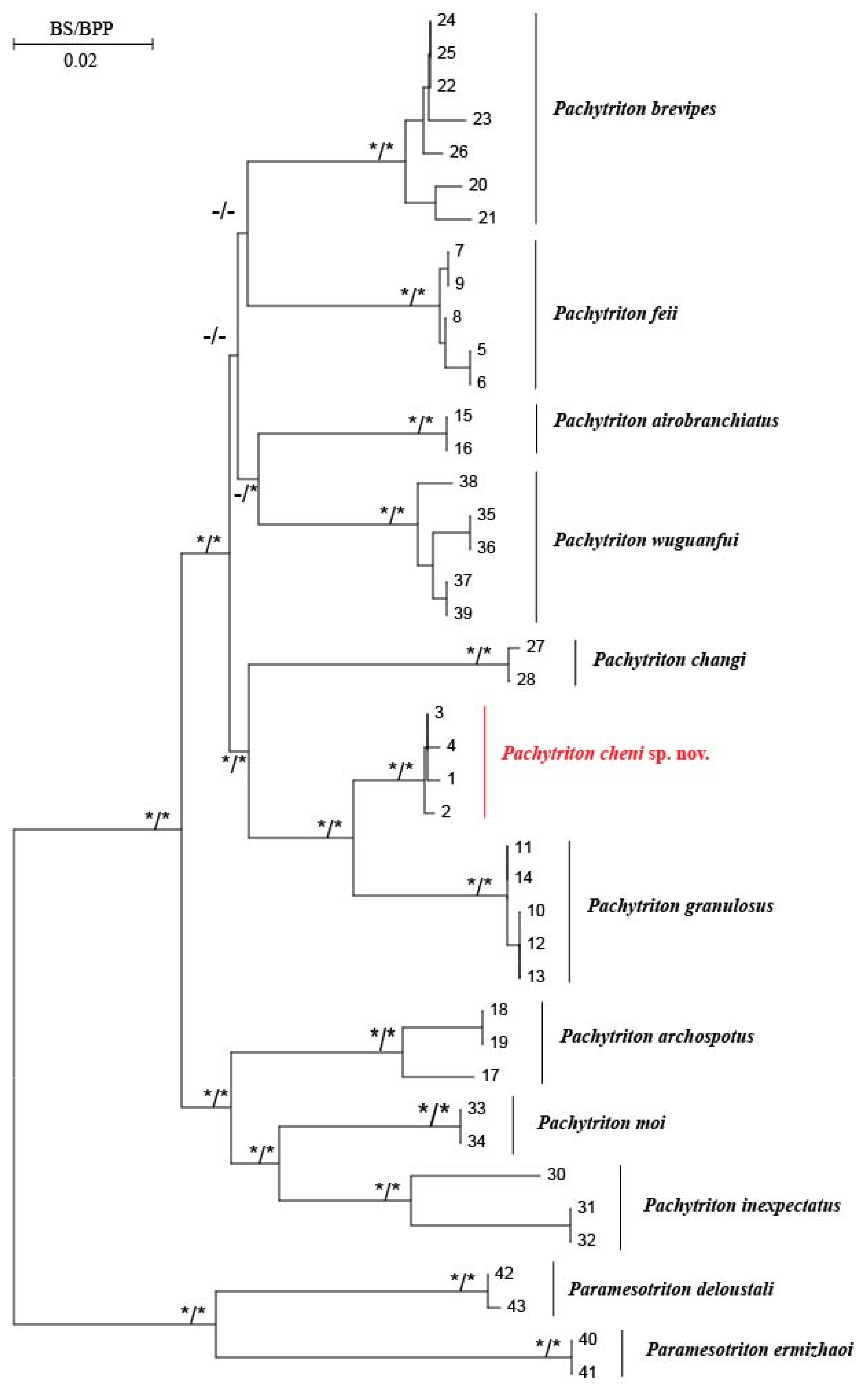

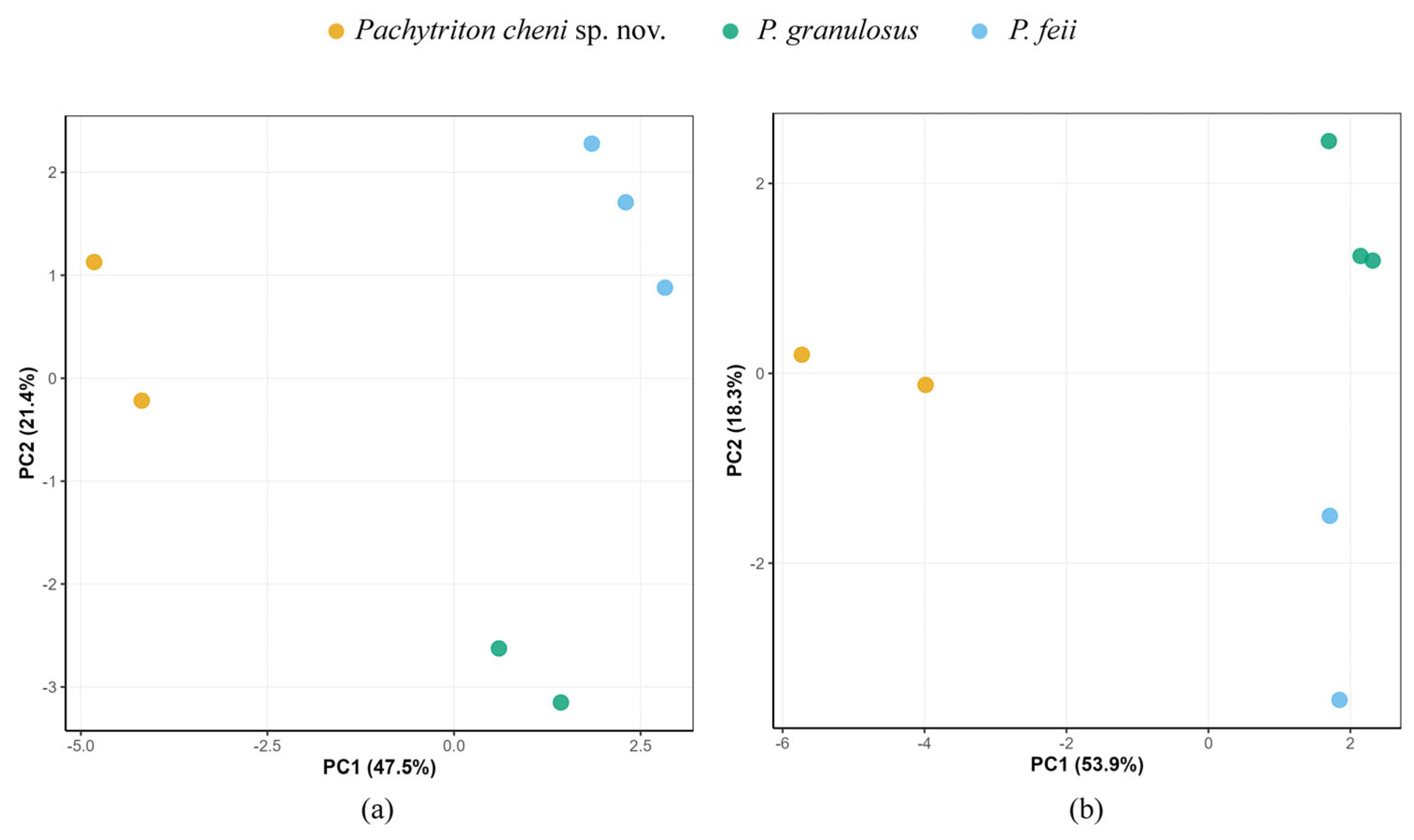
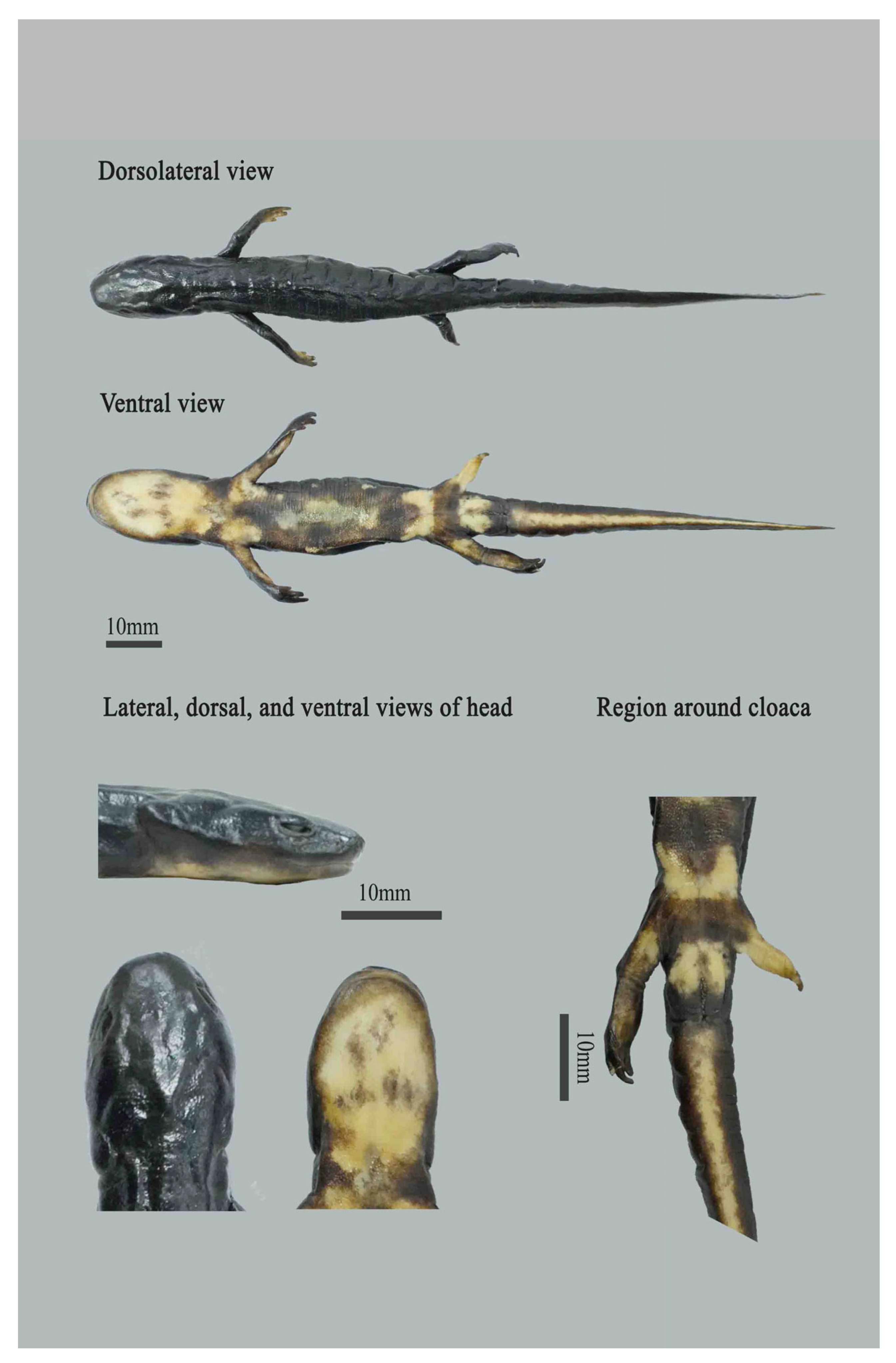


| ID No. | Species Name | Voucher | Localities | GenBank No. | Source | |||
|---|---|---|---|---|---|---|---|---|
| ND2 | cytb | RAG1 | POMC | |||||
| 1 | Pachytriton cheni sp. nov. | ANU20030001 | China: Anhui: Huangshan: She | N/A | N/A | N/A | N/A | This study |
| 2 | Pachytriton cheni sp. nov. | ANU20030002 | China: Anhui: Huangshan: She | N/A | N/A | N/A | N/A | This study |
| 3 | Pachytriton cheni sp. nov. | ANU20030007 | China: Anhui: Huangshan: She | N/A | N/A | N/A | N/A | This study |
| 4 | Pachytriton cheni sp. nov. | ANU20030009 | China: Anhui: Huangshan: She | N/A | N/A | N/A | N/A | This study |
| 5 | Pachytriton feii | ANU20030015 | China: Anhui: Chizhou: Shitai | N/A | N/A | N/A | N/A | This study |
| 6 | Pachytriton feii | ANU20030016 | China: Anhui: Chizhou: Shitai | N/A | N/A | N/A | N/A | This study |
| 7 | Pachytriton feii | ANU20030017 | China: Anhui: Chizhou: Shitai | N/A | N/A | N/A | N/A | This study |
| 8 | Pachytriton feii | ANU20030018 | China: Anhui: Chizhou: Shitai | N/A | N/A | N/A | N/A | This study |
| 9 | Pachytriton feii | ANU20030019 | China: Anhui: Chizhou: Shitai | N/A | N/A | N/A | N/A | This study |
| 10 | Pachytriton granulosus | ANU20030022 | China: Zhejiang: Shaoxing: Xingchang | N/A | N/A | N/A | N/A | This study |
| 11 | Pachytriton granulosus | ANU20030023 | China: Zhejiang: Ningbo: Fenghua | N/A | N/A | N/A | N/A | This study |
| 12 | Pachytriton granulosus | ANU20030024 | China: Zhejiang: Taizhou: Tiantai | N/A | N/A | N/A | N/A | This study |
| 13 | Pachytriton granulosus | ANU20030025 | China: Zhejiang: Lishui: Jinyun | N/A | N/A | N/A | N/A | This study |
| 14 | Pachytriton granulosus | ANU20030026 | China: Zhejiang: Lishui: Jinyun | N/A | N/A | N/A | N/A | This study |
| 15 | Pachytriton airobranchiatus | SWUFYZY0301 | China: Guangdong: Huidong: Mt. Lianhua | MG732934 | MG732932 | [8] | ||
| 16 | Pachytriton airobranchiatus | SWUFYZY0213 | China: Guangdong: Huidong: Mt. Lianhua | MG732933 | MG732931 | [8] | ||
| 17 | Pachytriton archospotus | CIB95953 | China: Hunan: Guidong: Mt. Qiyun | GQ303628 | GQ303665 | GQ303706 | [9] | |
| 18 | Pachytriton archospotus | KIZ04564 | China: Hunan: Guidong | KU375008 | KU374979 | KU375036 | [9] | |
| 19 | Pachytriton archospotus | CIB95949 | China: Hunan: Guidong: Mt. Qiyun | GQ303630 | GQ303667 | GQ303708 | [9] | |
| 20 | Pachytriton brevipes | CIB95926 | China: Jiangxi: Nanfeng: Mt. Junfeng | GQ303626 | GQ303663 | GQ303704 | [9] | |
| 21 | Pachytriton brevipes | CIB95930 | China: Jiangxi: Nanfeng: Mt. Junfeng | GQ303627 | GQ303664 | GQ303705 | [9] | |
| 22 | Pachytriton brevipes | CIB88221 | China: Fujian: Wuyi Shan: Mt. Wuyi | GQ303615 | GQ303652 | GQ303693 | [9] | |
| 23 | Pachytriton brevipes | CIB88194 | China: Fujian: Wuyi Shan: Mt. Wuyi | GQ303616 | GQ303653 | GQ303694 | [9] | |
| 24 | Pachytriton brevipes | CIB88188 | China: Fujian: Wuyi Shan: Mt. Wuyi | GQ303617 | GQ303654 | GQ303695 | [9] | |
| 25 | Pachytriton brevipes | CIB88197 | China: Fujian: Wuyi Shan: Mt. Wuyi | GQ303618 | GQ303655 | GQ303696 | [9] | |
| 26 | Pachytriton brevipes | KIZ08928 | China: Fujian: Wuyi Shan: Mt. Wuyi | KU375010 | KU374981 | KU375037 | [10] | |
| 27 | Pachytriton changi | KUHE 39832 | Unknown locality | AB638711 | [8] | |||
| 28 | Pachytriton changi | KUHE 39763 | Unknown locality | AB638709 | [11] | |||
| 29 | Pachytriton changi | KUHE:44985 | China: Hunan: Nanling | LC746909 | [12] | |||
| 30 | Pachytriton inexpectatus | KIZ08711 | China: Hunan: Jiangyong | KU375031 | KU375002 | KU375044 | [8] | |
| 31 | Pachytriton inexpectatus | KIZ05203 | China: Guangxi: Dayaoshan | KU375029 | KU375000 | KU375045 | [8] | |
| 32 | Pachytriton inexpectatus | KIZ05204 | China: Guangxi: Dayaoshan | KU375030 | KU375001 | [8] | ||
| 33 | Pachytriton moi | KIZ07767 | China: Guangxi: Maoershan | KU375032 | KU375003 | [8] | ||
| 34 | Pachytriton moi | KIZ07768 | China: Guangxi: Maoershan | KU375033 | KU375004 | KU375046 | [8] | |
| 35 | Pachytriton wuguanfui | KIZ08756 | China: Guangxi: Guposhan | KU375012 | KU374983 | [10] | ||
| 36 | Pachytriton wuguanfui | KIZ08761 | China: Guangxi: Guposhan | KU375013 | KU374984 | KU375040 | [10] | |
| 37 | Pachytriton wuguanfui | KIZ021705 | China: Hunan: Jiuweishan | KU375014 | KU374985 | [10] | ||
| 38 | Pachytriton wuguanfui | KIZ021706 | China: Hunan: Jiuweishan | KU375015 | KU374986 | KU375041 | [10] | |
| 39 | Pachytriton wuguanfui | KIZ021707 | China: Hunan: Jiuweishan | KU375016 | KU374987 | [10] | ||
| 40 | Paramesotriton ermizhaoi | CIB88141 | China: Guangxi: Jin Xiu: Mt. Dayao | FJ744601 | GQ303670 | [9] | ||
| 41 | Paramesotriton ermizhaoi | CIB88140 | China: Guangxi: Jin Xiu: Mt. Dayao | FJ744602 | GQ303671 | [9] | ||
| 42 | Paramesotriton deloustali | MVZ223628 | Vietnam: Vinh Phu Province: Tam Dao: Vinh Yen District | FJ744599 | GQ303668 | [9] | ||
| 43 | Paramesotriton deloustali | MVZ223629 | Vietnam: Vinh Phu Province: Tam Dao: Vinh Yen District | FJ744600 | GQ303669 | [9] | ||
| No. | Morphological Character | Abbreviation | Description |
|---|---|---|---|
| 1 | Snout-vent length | SVL | From tip of snout to anterior tip of vent |
| 2 | Head length | HL | From tip of snout to wrinkle of throat |
| 3 | Head width | HW | Measured at angle anterior to parotid grand |
| 4 | Maximum head width | MXHW | Measured at widest point |
| 5 | Snout length | SL | From tip of snout to anterior tip of upper eyelid |
| 6 | Eyelid-nostril length | ENL | Minimum distance between eyelid and nostril |
| 7 | Internarial distance | IND | Minimum distance between the external nares |
| 8 | Interorbital distance | IOD | Minimum distance between upper eyelids |
| 9 | Upper eyelid width | UEW | Greatest width of upper eyelid |
| 10 | Upper eyelid length | UEL | Greatest length of upper eyelid |
| 11 | Orbit length | OL | Maximum length of orbit |
| 12 | Axilla-groin distance | AGD | Minimum distance between axilla and groin |
| 13 | Trunk length | TRL | From wrinkle of throat to anterior tip of vent |
| 14 | Tail length | TAL | From anterior tip of vent to tail tip |
| 15 | Vent length | VL | From anterior to posterior tip of vent |
| 16 | Basal tail width | BTAW | Tail width measured at root of tail |
| 17 | Medial tail width | MTAW | Tail width measured at middle |
| 18 | Basal tail height | BTAH | Tail height measured at base of tail |
| 19 | Maximum tail height | MXTAH | Tail height measured at highest point |
| 20 | Forelimb length | FLL | Distance from axilla to tip of longest finger |
| 21 | Hindlimb length | HLL | Distance from groin to tip of longest toe |
| ID | Species | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Pachytriton cheni sp. nov. | 0.39 | |||||||||
| 2 | Pachytriton granulosus | 4.39 | 0.12 | ||||||||
| 3 | Pachytriton feii | 6.48 | 7.55 | 0.35 | |||||||
| 4 | Pachytriton archospotus | 8.11 | 9.93 | 7.25 | 1.50 | ||||||
| 5 | Pachytriton brevipes | 6.54 | 7.52 | 5.30 | 8.41 | 0.76 | |||||
| 6 | Pachytriton wuguanfui | 6.31 | 8.36 | 7.14 | 7.84 | 6.59 | 0.51 | ||||
| 7 | Pachytriton airobranchiatus | 6.63 | 8.17 | 5.98 | 8.37 | 6.27 | 6.32 | - | |||
| 8 | Pachytriton changi | 6.77 | 7.77 | 7.85 | 9.46 | 7.22 | 7.08 | 7.48 | 0.18 | ||
| 9 | Pachytriton inexpectatus | 10.22 | 11.70 | 10.34 | 10.75 | 10.87 | 11.45 | 10.27 | 9.76 | 3.07 | |
| 10 | Pachytriton moi | 7.56 | 8.85 | 8.10 | 7.92 | 8.26 | 9.30 | 8.54 | 9.64 | 8.64 | - |
| Competing Delimitations | Path Sampling |
|---|---|
| P. sp. (Qingliangfeng) conspecific to P. granulosus | −7335.6368 |
| P. sp. (Qingliangfeng) independent from P. granulosus | −7323.3744 |
| 2lnBf | 24.52 |
| Species | Pachytriton cheni sp. nov. | P. granulosus | P. feii | P. changi | P. wuguanfui | P. airobranchiatus | P. brevipes | P. archospotus | P. moi | P. inexpectatus | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender | Male (n = 2) | Female (n = 2) | Male (n = 2) | Female (n = 3) | Male (n = 3) | Female (n = 2) | Male (n = 2) | Female (n = 1) | Male (n = 4) | Female (n = 2) | Male (n = 2) | Male (n = 3) | Male (n = 1) | Male (n = 14) |
| SVL | 63.1 ± 8.4 (57.1–69.1) | 66.3 ± 3.4 (63.9–68.7) | 74.0 ± 1.6 (72.9–75.2) | 60.6 ± 4.8 (55.1–63.6) | 84.0 ± 4.6 (78.9–87.7) | 85.8 ± 9.0 (79.4–92.1) | 83.0 ± 2.9 (81.8–84.2) | 83.5 | 73.1 ± 30.8 (63.3–76.8) | 66.1 ± 5.4 (64.4–67.7) | 80.4 ± 105.1 (73.1–87.6) | 86.8 ± 5.9 (81.5–93.1) | 100.2 | 87.8 ± 10.1 (68.6–102.1) |
| RHL | 30.0 ± 5.7 (26.0–34.0) | 29.6 ± 0.8 (29.1–30.1) | 26.8 ± 0.8 (26.2–27.4) | 26.5 ± 1.1 (25.2–27.4) * | 28.1 ± 2.5 (26.2–30.9) | 25.1 ± 0.2 (25.0–25.2) | 25.3 ± 0.02 (25.2–25.4) | 24.2 | 23.0 ± 1.0 (21.3–23.9) | 22.2 ± 0.7 (21.6–22.7) | 24.9 ± 1.1 (24.1–25.6) | 25.5 (23.5–29.4) | 35.9 | 28.7 (24.6–31.6) |
| RHW | 17.2 ± 0.6 (16.8–17.7) | 14.2 ± 0.7 (13.7–14.7) | 18.4 ± 0.2 (18.2–18.6) | 19.0 ± 0.9 (18.0–19.7) * | 19.7 ± 0.2 (19.5–19.8) | 18.7 ± 1.0 (18.0–19.4) * | 18.5 ± 0.2 (18.2–18.8) | 19.5 | 19.7 ± 0.1 (19.4–20.3) | 19.3 ± 0.1 (19.1–19.6) | 19.3 ± 0.2 (19.0–19.6) | 21.7 (21.7–22.2) | 23.2 | 19.8 (18.5–21.7) |
| RMXHW | 19.8 ± 1.4 (18.8–20.8) | 17.9 ± 0.9 (17.3–18.5) | 19.6 ± 0.8 (19.0–20.1) | 19.5 ± 1.0 (18.4–20.1) | 20.7 ± 1.3 (19.9–22.2) | 19.4 ± 0.2 (19.3–19.5) | 19.9 ± 0.7 (19.3–20.5) | - | - | - | 21.2 ± 1.1 (20.4–21.9) | 24.3 (23.8–24.6) | 25.3 | 22.3 (20.7–25.2) |
| RSL | 7.3 ± 0.3 (7.1–7.5) | 6.1 ± 0.7 (5.6–6.6) | 8.6 ± 0.7 (8.1–9.1) | 8.4 ± 0.3 (8.2–8.8) | 8.7 ± 0.2 (8.5–8.8) | 8.4 ± 0.7 (7.9–8.9) | 10.0 ± 0.0 (9.8–10.1) | 9.2 | 10.2 ± 0.5 (9.5–11.1) | 9.6 ± 0.0 (9.5–9.7) | 8.7 ± 0.1 (8.4–8.9) | 8.0 (7.8–8.3) | 11.3 | 9.5 (8.6–10.6) |
| RENL | 6.7 ± 0.1 (6.6–6.8) | 5.6 ± 0.1 (5.5–5.7) | 6.8 ± 0.0 (6.8–6.8) | 6.9 ± 0.4 (6.5–7.3) * | 6.7 ± 0.1 (6.6–6.8) | 6.8 ± 0.0 (6.7–6.8) * | 7.3 ± 0 (7.2–7.3) | - | - | - | 6.6 ± 0.0 (6.6–6.6) | 5.7 (5.4–5.9) | 8.6 | 7.0 (6.4–7.9) |
| RIND | 6.3 ± 0.0 (6.3–6.3) | 7.0 ± 0.0 (7.0–7.0) | 6.0 ± 0.1 (6.0–6.0) | 5.8 ± 0.9 (4.8–6.3) | 6.5 ± 0.1 (6.4–6.6) * | 6.1 ± 0.2 (6.0–6.3) | 7.0 ± 0.04 (6.8–7.1) | 4.6 | 9.1 ± 4.3 (5.6–10.7) | 9.5 ± 0.9 (8.9–10.2) | 6.0 ± 1.0 (5.3–6.7) | 5.0 (4.9–5.8) | 7.5 | 6.5 (5.9–7.4) |
| RIOD | 11.8 ± 0.6 (11.4–12.1) | 11.2 ± 0.0 (11.2–11.3) | 6.9 ± 0.9 (6.3–7.6) | 7.1 ± 0.6 (6.5–7.5) ** | 8.6 ± 0.3 (8.4–8.9) * | 7.1 ± 0.9 (6.5–7.7) | 7.4 ± 0.3 (7.0–7.7) | 9.2 | 10.7 ± 0.2 (10.2–11.4) | 10.4 ± 0.1 (10.1–10.6) | 7.2 ± 4.2 (5.7–8.6) | 9.3 (8.8–9.6) | 8.0 | 6.9 (6.4–7.9) |
| RUEW | 0.9 ± 0.1 (0.8–1.0) | 1.3 ± 0.0 (1.3–1.3) | 3.4 ± 0.4 (3.1–3.6) * | 3.4 ± 0.4 (3.0–3.8) ** | 2.8 ± 0.2 (2.6–3.0) * | 2.6 ± 0.3 (2.4–2.9) | 2.7 ± 0.1 (2.4–2.9) | - | - | - | 2.7 ± 0.1 (2.4–2.9) | 1.5 (1.2–1.6) | 2.7 | 2.5 (2.0–3.2) |
| RUEL | 4.8 ± 1.1 (4.1–5.6) | 4.5 ± 1.0 (3.8–5.3) | 5.9 ± 0.5 (5.5–6.3) | 6.5 ± 0.3 (6.2–6.7) | 5.2 ± 0.4 (4.7–5.5) | 5.0 ± 0.1 (4.9–5.1) | 4.7 ± 0.1 (4.5–4.9) | - | 3.3 ± 0.7 (2.3–4.3) | 3.9 ± 0.2 (3.5–4.2) | 4.8 ± 0.0 (4.7–4.8) | 3.8 (3.7–4.2) | 4.7 | 4.9 (3.9–6.1) |
| ROL | 1.4 ± 0.3 (1.2–1.6) | 1.5 ± 0.1 (1.5–1.6) | 3.1 ± 0.4 (2.9–3.4) | 3.1 ± 0.4 (2.7–3.4) ** | 3.8 ± 0.2 (3.5–4.0) | 3.1 ± 0.6 (2.6–3.5) | 3.4 ± 0.3 (3.0–3.7) | - | - | - | 3.4 ± 0.0 (3.3–3.5) | 2.6 (2.1–2.8) | 3.7 | 2.9 (2.4–3.5) |
| RAGD | 54.6 ± 3.5 (52.2–57.1) | 51.5 ± 6.0 (47.2–55.7) | 51.6 ± 0.6 (51.2–52.0) | 52.8 ± 0.9 (51.9–53.7) | 48.4 ± 1.8 (46.6–50.2) | 49.9 ± 0.0 (49.9–50.0) | 50.1 ± 5.1 (48.5–51.7) | - | - | - | 51.9 ± 0.1 (51.6–52.1) | 50.1 (49.2–50.9) | 52.1 | 49.5 (43.7–53.7) |
| RTRL | 71.9 ± 3.1 (69.7–74.1) | 72.1 ± 0.5 (71.7–72.5) | 73.7 ± 1.5 (72.6–74.7) | 73.4 ± 1.0 (72.3–74.2) | 73.4 ± 1.1 (72.2–74.2) | 74.7 ± 2.1 (73.3–76.2) | 74.7 ± 0.0 (74.5–74.8) | - | - | - | 75.2 ± 1.1 (74.4–75.9) | 74.5 (70.6–76.5) | 64.1 | 71.3 (68.4–75.4) |
| RTAL | 110.1 ± 2.3 (108.5–111.7) | 113.5 ± 5.9 (109.4–117.7) | 101.7 ± 1.5 (100.6–102.7) | 101.2 ± 2.2 (99.8–103.8) | 102.9 ± 2.9 (99.8–105.6) | 98.3 ± 3.7 (95.7–100.9) | 103.1 ± 8.0 (101.1–105.1) | 86.5 | 92.2 ± 44.3 (84.2–98.8) | 93.4 ± 16.7 (90.5–96.3) | 96.2 ± 49.0 (91.2–101.1) | 95.7 (93.5–95.8) | 90.5 | 90.6 (85.4–98.7) |
| RVL | 11.8 ± 0.7 (11.3–12.2) | 11.2 ± 0.3 (11.0–11.4) | 7.9 ± 0.6 (7.5–8.3) * | 6.7 ± 0.9 (5.7–7.4) * | 7.3 ± 0.6 (6.7–7.8) ** | 4.3 ± 1.1 (3.6–5.1) | 5.9 ± 0.1 (5.7–6.1) | - | - | - | 6.2 ± 2.0 (5.2–7.2) | 5.0 (4.8–5.0) | 5.2 | 5.6 (4.6–7.5) |
| RBTAW | 10.9 ± 0.3 (10.7–11.1) | 10.8 ± 1.2 (9.9–11.6) | 14.7 ± 0.2 (14.6–14.8) * | 14.8 ± 0.6 (14.1–15.3) | 12.3 ± 0.9 (11.4–13.2) | 11.9 ± 1.4 (10.9–12.9) | 12.1 ± 0.0 (12.0–12.2) | 12.1 | 11.2 ± 0.7 (10.1–12.2) | 12.2 ± 2.9 (11–13.4) | 14.7 ± 0.3 (14.3–15.0) | 15.8 (15.7–17.2) | 13.2 | 14.7 (12.3–16.5) |
| RMTAW | 13.2 ± 0.1 (13.2–13.3) | 12.4 ± 0.8 (11.8–12.9) | 11.6 ± 1.1 (10.8–12.3) | 12.0 ± 1.0 (11.0–13.0) | 9.1 ± 0.6 (8.4–9.6) * | 8.7 ± 1.3 (7.8–9.6) | 7.9 ± 0.3 (7.5–8.3) | - | - | - | 10.9 ± 1.6 (10.0–11.8) | 9.8 (8.6–12.5) | 7.8 | 11.2 (8.9–13.5) |
| RBTAH | 12.2 ± 1.4 (11.2–13.2) | 12.2 ± 0.6 (11.7–12.6) | 12.4 ± 1.1 (11.6–13.1) | 12.6 ± 0.8 (11.7–13.3) | 11.6 ± 0.6 (11.0–12.2) | 11.4 ± 2.3 (9.8–13.1) | 10.4 ± 0.3 (10.0–10.8) | 8.4 | 10.4 ± 0.2 (10.2–11.1) | 9.0 ± 1.4 (8.1–9.8) | 13.2 ± 0.2 (12.9–13.5) | 15.9 (14.0–16.0) | 12.8 | 12.2 (10.3–14.2) |
| RMXTAH | 14.1 ± 0.2 (13.9–14.3) | 11.6 ± 0.1 (11.6–11.7) | 16.2 ± 2.2 (14.7–17.8) | 16.1 ± 1.2 (14.7–17.1) * | 15.0 ± 0.5 (14.5–15.4) | 14.9 ± 1.9 (13.6–16.3) | 15.2 ± 0.1 (15.0–15.4) | - | - | - | 20.3 ± 20.5 (17.1–23.5) | 17.5 (16.1–18.0) | 14.4 | 14.9 (12.4–15.7) |
| RFLL | 22.3 ± 0.1 (22.3–22.4) | 25.3 ± 0.6 (24.8–25.7) | 22.5 ± 1.1 (21.8–23.2) | 22.9 ± 0.4 (22.5–23.3) | 26.3 ± 1.5 (24.6–27.4) * | 24.3 ± 3.8 (21.6–27.0) | 23.9 ± 0.7 (23.3–24.5) | 17.6 | 15.3 ± 2.3 (13.8–17.8) | 14.9 ± 9.0 (12.7–17) | 20.7 ± 3.7 (19.3–22.0) | 23.3 (21.7–24.3) | 22.7 | 20.3 (18.5–23.6) |
| RHLL | 24.6 ± 0.8 (24.0–25.1) | 27.4 ± 0.6 (26.9–27.8) | 24.9 ± 1.8 (23.6–26.1) | 25.7 ± 1.4 (24.1–26.8) | 29.2 ± 1.4 (27.7–30.4) * | 29.4 ± 3.5 (26.9–31.9) | 27.2 ± 0.9 (26.5–27.8) | 20.7 | 17.1 ± 3.3 (14.7–19.6) | 18.3 ± 0.2 (18–18.6) | 25.4 ± 0.5 (24.9–25.9) | 29.4 (25.9–29.6) | 24.4 | 24.5 (21.8–35.3) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Z.; Wu, S.; Wang, S.; Ma, L.; Zhao, N.; Wu, X.; Wang, S. A New Species of Pachytriton (Amphibia: Caudata: Salamandridae) from Anhui, China. Animals 2025, 15, 3018. https://doi.org/10.3390/ani15203018
He Z, Wu S, Wang S, Ma L, Zhao N, Wu X, Wang S. A New Species of Pachytriton (Amphibia: Caudata: Salamandridae) from Anhui, China. Animals. 2025; 15(20):3018. https://doi.org/10.3390/ani15203018
Chicago/Turabian StyleHe, Zhirong, Siyu Wu, Shanqing Wang, Li Ma, Na Zhao, Xiaobing Wu, and Supen Wang. 2025. "A New Species of Pachytriton (Amphibia: Caudata: Salamandridae) from Anhui, China" Animals 15, no. 20: 3018. https://doi.org/10.3390/ani15203018
APA StyleHe, Z., Wu, S., Wang, S., Ma, L., Zhao, N., Wu, X., & Wang, S. (2025). A New Species of Pachytriton (Amphibia: Caudata: Salamandridae) from Anhui, China. Animals, 15(20), 3018. https://doi.org/10.3390/ani15203018






