Genetic Analysis of the Conserved Population of Dengchuan Cattle Based on High Concordance SNP loci
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Experimental Methods
2.2.1. Genomic DNA Extraction and Quality Testing of Blood Samples
2.2.2. SNP Typing and Quality Control
2.2.3. Purity Identification and Screening
2.2.4. Genetic Diversity Analysis of Dengchuan Cattle Conservation Populations
2.2.5. Kinship Analysis of Dengchuan Cattle Conservation Groups
2.2.6. Family Structure Analysis of Dengchuan Cattle Breeding Groups
3. Results
3.1. DNA Detection
3.2. SNP Typing and Quality Control of 74 Dengchuan Cattle
3.3. Results of Purity Identification and Screening
3.4. SNP Typing and Quality Control of 60 Dengchuan Cattle
3.5. Genetic Diversity Analysis of Dengchuan Cattle Breeding Population
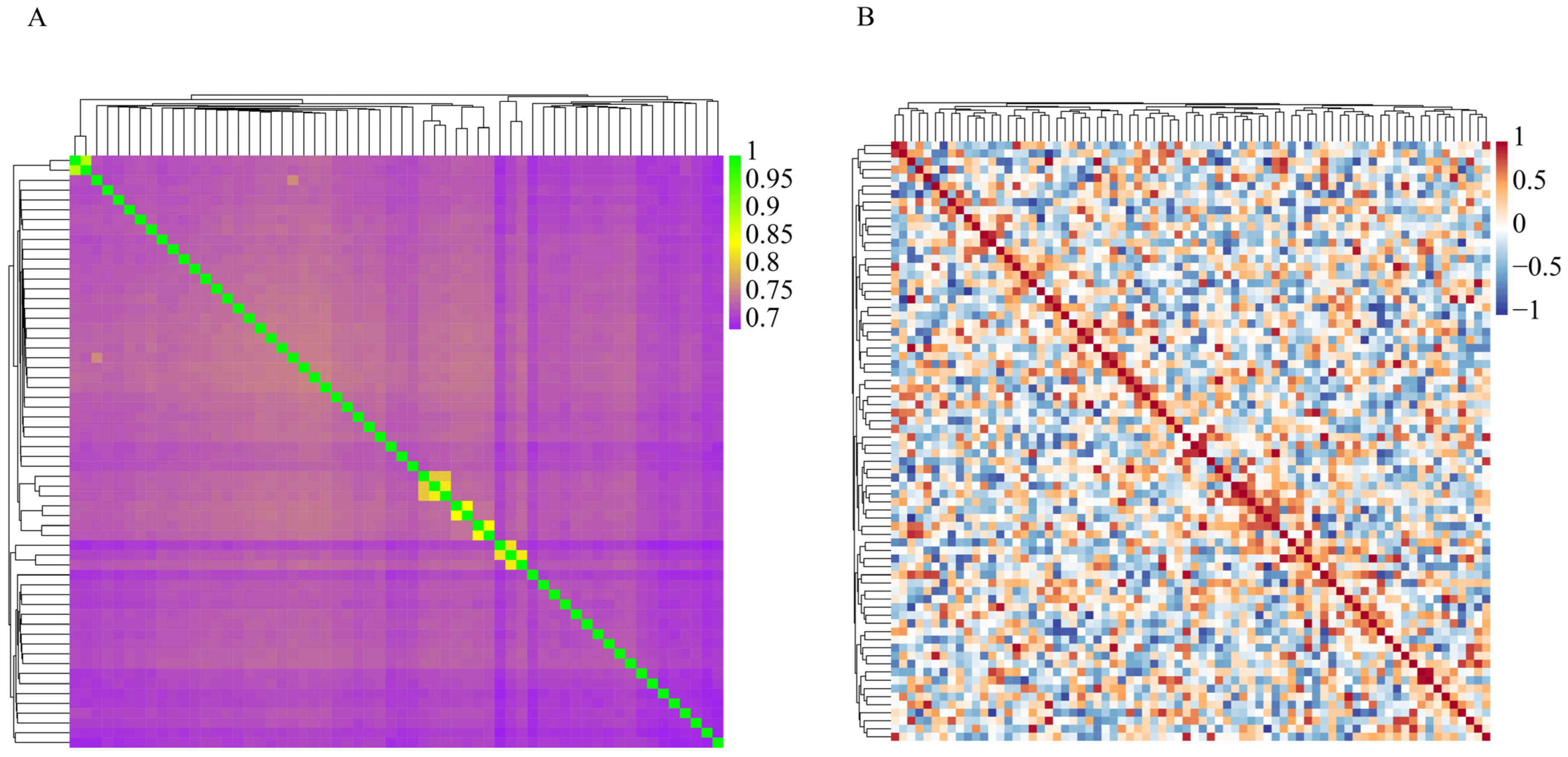
3.6. IBS and Genomic Relationship (G) Matrix Analyses of the Dengchuan Conservation Population
3.7. ROH-Based Inbreeding Coefficient Analysis of Dengchuan Cattle Conservation Population
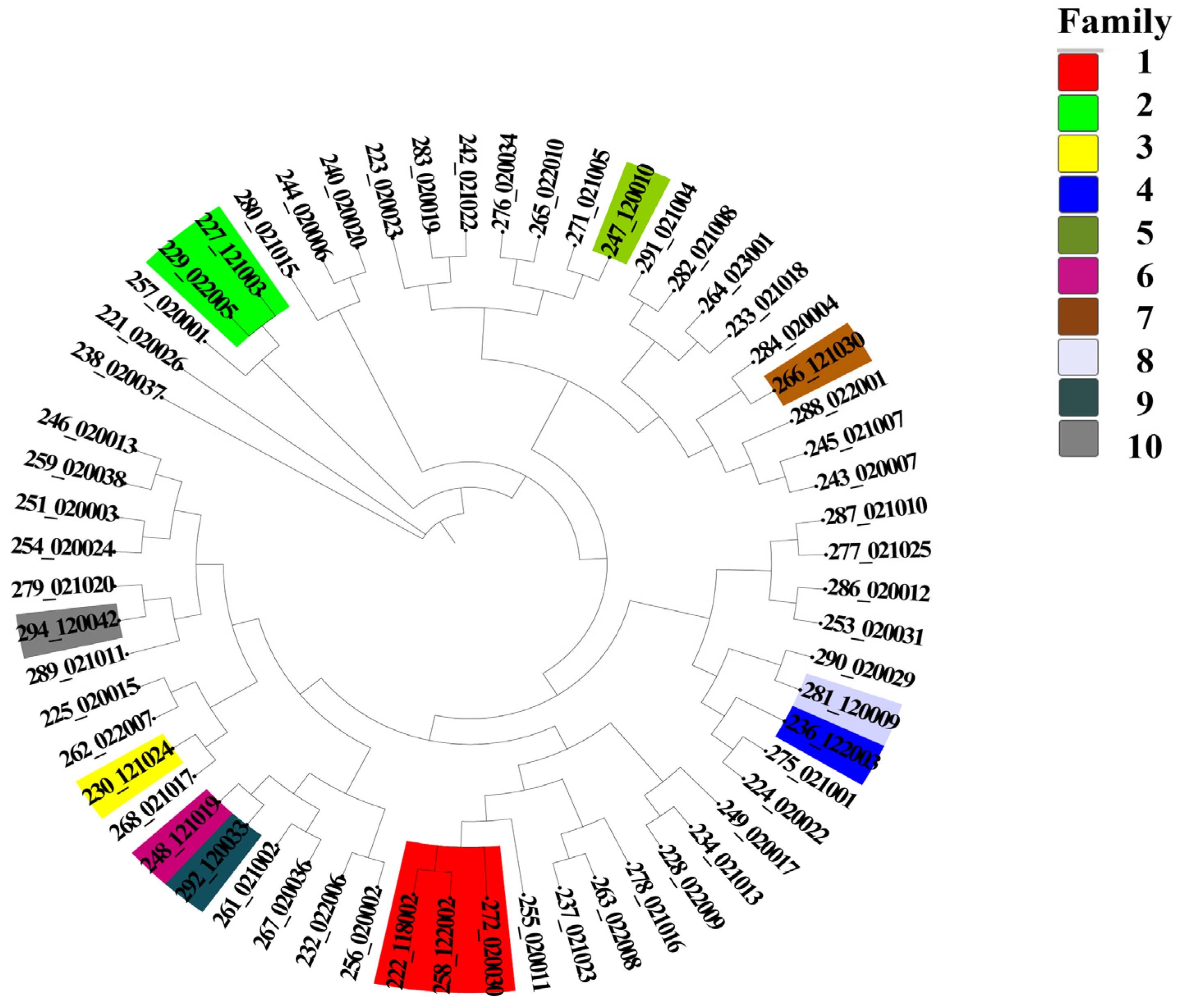
3.8. Analysis of the Family Structure of the Dengchuan Cattle Conservation Group
4. Discussion
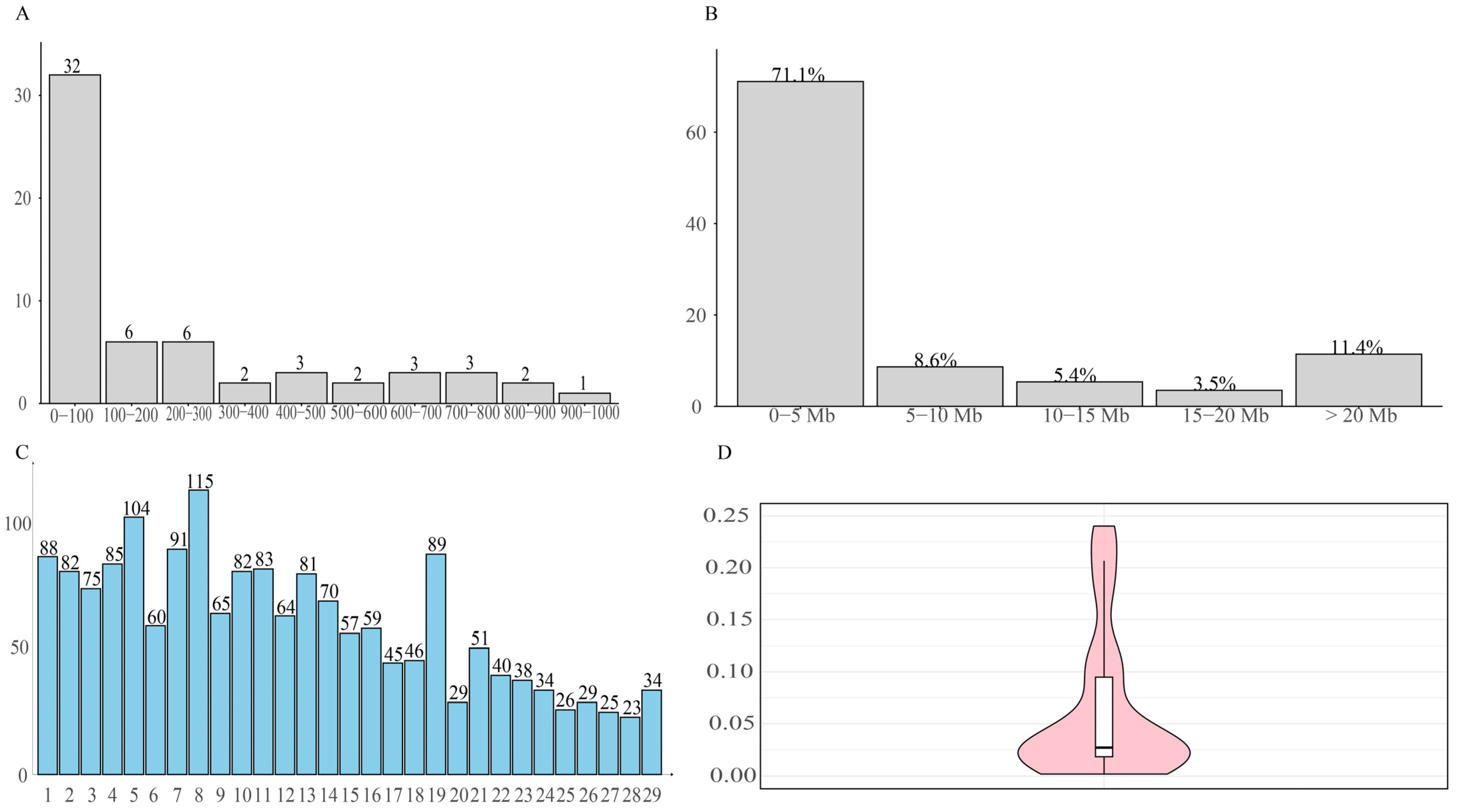
- Overall, a total of 1770 ROHs were detected in 74 Dengchuan cattle, yielding an average of 29.50 ± 12.43(SD) ROHs per individual. This finding indicates significant variability in the presence of pure fragments among individuals within the population.
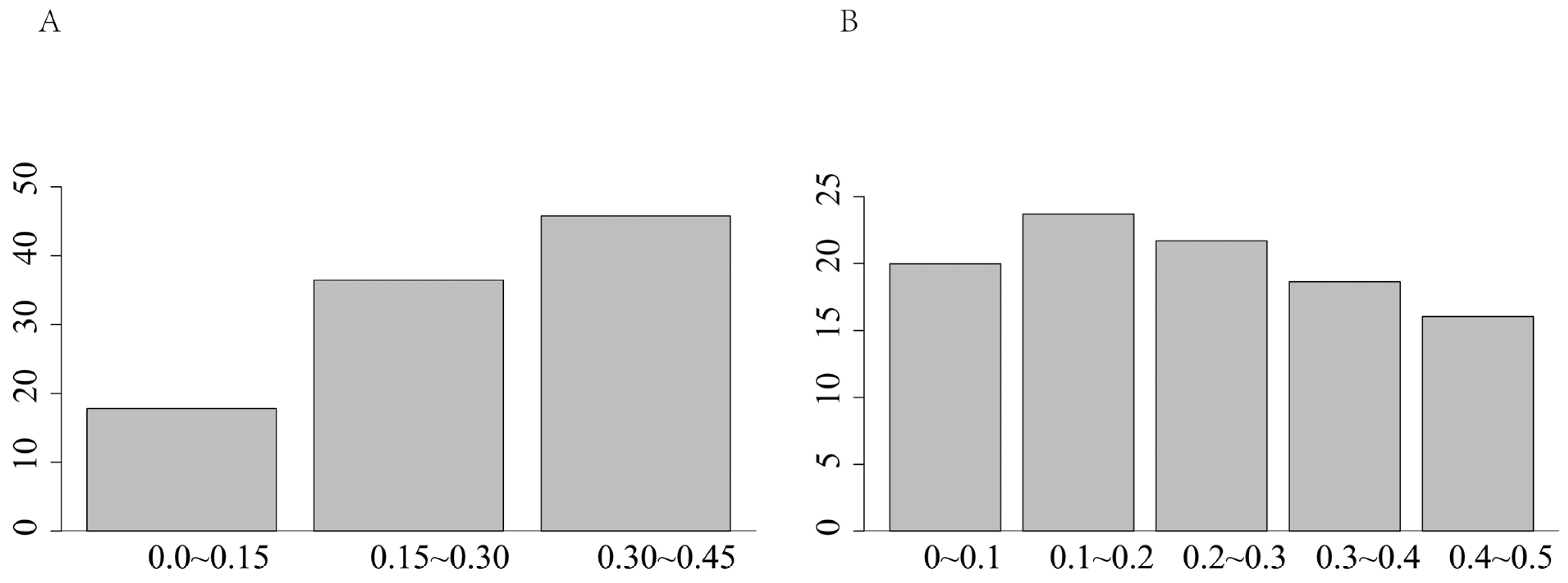
- The study demonstrated that inbreeding leads to an increase in the length of ROH fragments [35]. The length of single ROH in the population predominantly ranged from 0 to 5 Mb, accounting for 71.1% of the total. This finding aligns with the results reported by Xu et al. [36]. which indicated that most ROH lengths among eight cattle breeds in China were also within 5 Mb. Conversely, the occurrence of single ROH lengths ranging from 15 to 20 Mb was minimal, constituting only 3.50% of the total. This suggests that the population is primarily characterized by short fragments resulting from distant inbreeding, with relatively few recent inbreeding events.
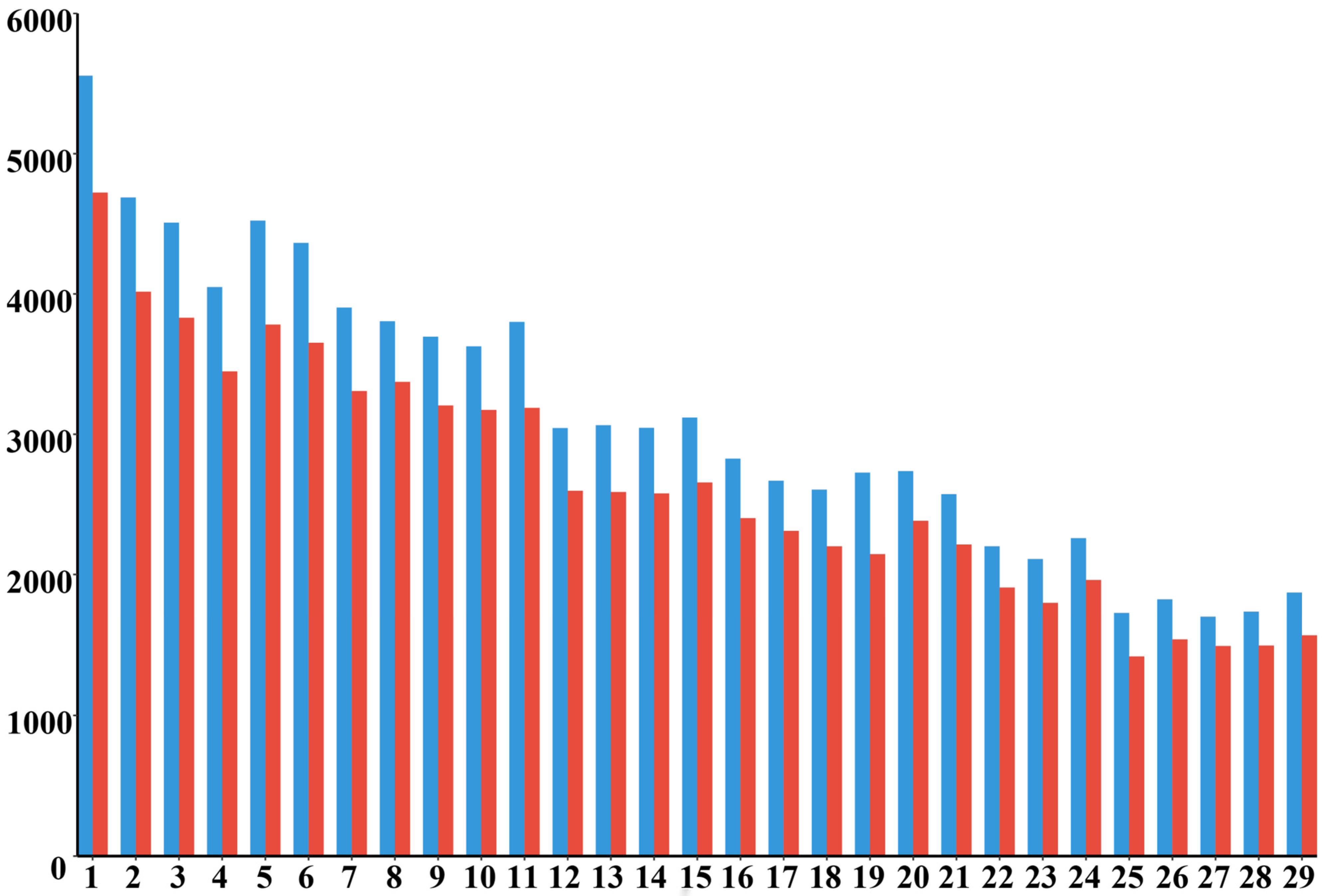
- At the chromosome level, the distribution of ROH across the 29 autosomes was uneven. Chromosome 8 exhibited the highest number of ROH fragments, totaling 115, while chromosome 27 had the lowest, with only 23 fragments. This disparity in the accumulation of pure fragments across different chromosomes may be attributed to genetic drift or selective pressures acting on specific chromosomes, such as those associated with quantitative trait loci (QTL) related to meat quality, body size, and reproduction. These factors are likely intensified through repeated selection during conservation or breeding processes.
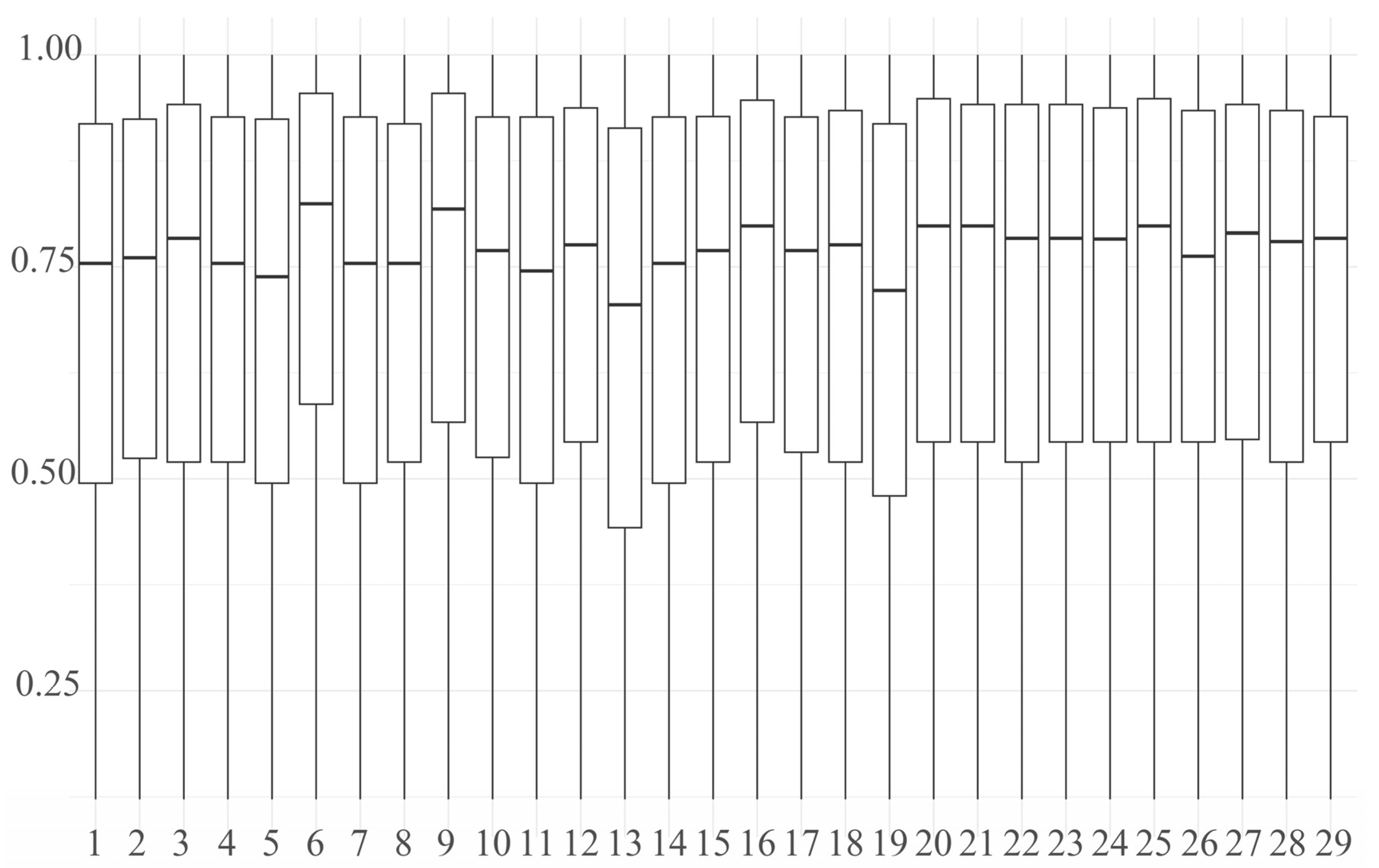
- The mean value of the FROH group was 0.0928 ± 0.1054, which is higher than the inbreeding coefficients reported for 11 Polish cattle breeds studied by Szmatoła [37]. Specifically, the coefficients for Polish Red and White Friesian cattle, Limousin cattle, Charolais cattle, Simmental cattle, Polish Red cattle, Polish Red and White cattle, and Polish Black and White cattle were 0.042, 0.059, 0.065, 0.068, 0.053, 0.042, and 0.054, respectively. Additionally, Holstein-Friesian Red and White cattle had an inbreeding coefficient of 0.087, while Hereford cattle, Holstein-Friesian Black and White cattle, and Montbéliard cattle had coefficients of 0.151, 0.118, and 0.108, respectively. These findings indicate that the inbreeding level of the Dengchuan cattle population is low; however, it is essential to introduce new purebred bloodlines to enhance population size and genetic diversity. Consistently, the mean F based on Hardy–Weinberg equilibrium was 0.071 ± 0.122 (SD), also significantly greater than zero (p < 0.01). The similarity of FROH and F results provides complementary evidence for the existence of a low but significant level of inbreeding in the conservation population. The highest percentage of individuals with FROH values ranging from 0 to 0.06 reflects a generally low overall inbreeding level within the population, although significant variation exists among individuals. Further analysis of sex differences revealed that the FROH was 0.119 ± 0.107(SD) for bulls and 0.087 ± 0.105(SD) for females, indicating a slightly higher level of inbreeding among bulls. This may be attributed to their more frequent use as primary breeding individuals and the concentration of breeding sources. The results of the NJ cluster analysis indicated that the 11 bulls could be classified into 10 lineages, with 9 lineages (lineages 2 to 10) comprising a small number of bulls, leading to an unbalanced structure. Therefore, careful attention should be paid to the selection and breeding of progeny in the future to prevent loss of pedigree. The 46 females classified under the category of ‘Others’ exhibited a kinship coefficient of less than 0.1 with all the bulls, allowing them to be mated with any bulls within the group.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Djedovic, R.; Radojkovic, D.; Stanojevic, D.; Savic, R.; Vukasinovic, N.; Popovac, M.; Bogdanovic, V.; Radovic, C.; Gogic, M.; Gligovic, N.; et al. Base characteristics, preservation methods, and assessment of the genetic diversity of autochthonous breeds of cattle, sheep and pigs in Serbia: A review. Animals 2024, 14, 1894. [Google Scholar] [CrossRef]
- Shaw, R.E.; Farquharson, K.A.; Bruford, M.W.; Coates, D.J.; Elliott, C.P.; Mergeay, J.; Ottewell, K.M.; Segelbacher, G.; Hoban, S.; Hvilsom, C.; et al. Global meta-analysis shows action is needed to halt genetic diversity loss. Nature 2025, 638, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Xiang, W.; Qu, K.; Ahmed, Z.; Liu, J.; Cai, M.; Zhang, J.; Chen, N.; Lei, C.; Huang, B. Whole genome insights into genetic diversity, introgression, and adaptation of Yunnan indigenous cattle of Southwestern China. BMC Genom. 2025, 26, 216. [Google Scholar] [CrossRef]
- Yang, L.; Su, Z.; Li, H.; Zhao, T. Investigation and analysis of the current status of Dengchuan cattle breed resources. Heilongjiang Anim. Sci. Vet. Med. 2021, 48, 58–61, 67, 146–147. [Google Scholar]
- Wang, Q.; Lu, Y.; Li, M.; Gao, Z.; Li, D.; Gao, Y.; Deng, W.; Wu, J. Leveraging whole-genome resequencing to uncover genetic diversity and promote conservation strategies for ruminants in Asia. Animals 2025, 15, 831. [Google Scholar] [CrossRef] [PubMed]
- Hayah, I.; Talbi, C.; Chafai, N.; Houaga, I.; Botti, S.; Badaoui, B. Genetic diversity and breed-informative SNPs identification in domestic pig populations using coding SNPs. Front. Genet. 2023, 14, 1229741. [Google Scholar] [CrossRef] [PubMed]
- McVean, G. A genealogical interpretation of principal components analysis. PLoS Genet. 2009, 5, e1000686. [Google Scholar] [CrossRef]
- Burgos-Paz, W.; Gómez-Vargas, Y.; Ramírez-Toro, E.J. Performance of the Illumina GGP Bovine 100K SNP array for buffalo populations genotyping in Colombia. Rev. Cient. 2020, 30, 120. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Corrada Bravo, H.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, H.; Zhang, W.; Jiang, J.; Qian, H.; Man, C.; Gao, H.; Chen, Q.; Du, L.; Chen, S.; et al. Development and validation of a 5K low-density SNP chip for Hainan cattle. BMC Genom. 2024, 25, 873. [Google Scholar] [CrossRef]
- Flury, C.; Tapio, M.; Sonstegard, T.; Drögemüller, C.; Leeb, T.; Simianer, H.; Hanotte, O.; Rieder, S. Effective population size of an indigenous Swiss cattle breed estimated from linkage disequilibrium. J. Anim. Breed. Genet. 2010, 127, 339–347. [Google Scholar] [CrossRef]
- Fernández, J.; Meuwissen, T.H.E.; Toro, M.A.; Mäki-Tanila, A. Management of genetic diversity in small farm animal populations. Animal 2011, 5, 1684–1698. [Google Scholar] [CrossRef]
- Taberlet, P.; Valentini, A.; Rezaei, H.R.; Naderi, S.; Pompanon, F.; Negrini, R.; Ajmone-Marsan, P. Are cattle, sheep, and goats endangered species? Mol. Ecol. 2008, 17, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Sudrajad, P.; Kusminanto, R.Y.; Volkandari, S.D.; Cahyadi, M. Genomic structure of Bali cattle based on linkage disequilibrium and effective population size analyses using 50K single nucleotide polymorphisms data. Vet. World 2022, 15, 449–454. [Google Scholar]
- Wang, P.; Ou, G.; Li, G.; Li, H.; Zhao, T. Analysis of genetic diversity and structure of endangered Dengchuan cattle population using a single-nucleotide polymorphism chip. Anim. Biotechnol. 2024, 35, 2349625. [Google Scholar] [CrossRef]
- Wang, H.L.; Wang, Q.; Xing, S.Y.; Wang, J.; Li, Q.H.; Zheng, K.Q.; Cui, H.X.; Liu, R.R.; Zhao, G.L.; Wen, J. Evaluation of conservation status of Beijing You chicken conservation population based on phenotypic and genomic information. Acta Vet. Zootech. Sin. 2021, 52, 2406–2415. [Google Scholar]
- Sun, H.; Wang, Z.; Zhang, Z.; Xiao, N.; Xu, Z.; Zhang, X.Z.; Yang, H.J.; Zhu, M.X.; Xiao, M.; Liu, X.H.; et al. Analysis of the conservation status of Meishan pigs based on genomic sequencing data. J. Shanghai Jiaotong Univ. (Agric. Sci.) 2017, 35, 65–70. [Google Scholar]
- Liu, C.L.; Lu, D.; Zhou, Q.Y.; Wan, M.C.; Hao, X.D.; Zhang, Y.H.; Zheng, R.Q.; Ji, H.Y. Analysis of the population genetic structure of Hang pigs using high-density SNP chips. Acta Vet. Zootech. Sin. 2022, 53, 2502–2513. [Google Scholar]
- Senczuk, G.; Mastrangelo, S.; Ciani, E.; Battaglini, L.; Cendron, F.; Ciampolini, R.; Crepaldi, P.; Mantovani, R.; Bongioni, G.; Pagnacco, G.; et al. The genetic heritage of Alpine local cattle breeds using genomic SNP data. Genet. Sel. Evol. 2020, 52, 40. [Google Scholar] [CrossRef]
- Biscarini, F.; Mastrangelo, S.; Catillo, G.; Senczuk, G.; Ciampolini, R. Insights into genetic diversity, runs of homozygosity and heterozygosity-rich regions in Maremmana semi-feral cattle using pedigree and genomic data. Animals 2020, 10, 2285. [Google Scholar]
- Molina Flores, B.; Camacho Vallejo, M.E.; Delgado Bermejo, J.V.; Navas González, F.J.; Martínez, M.D.A. Do Pharaohs’ cattle still graze the Nile Valley? Genetic characterization of the Egyptian Baladi cattle breed. Anim. Biotechnol. 2023, 34, 645–657. [Google Scholar] [CrossRef]
- Lyu, Y.; Guan, X.; Xu, X.; Wang, P.; Li, Q.; Panigrahi, M.; Zhang, J.; Chen, N.; Huang, B.; Lei, C. A whole genome scan reveals distinct features of selection in Zhaotong cattle of Yunnan province. Anim. Genet. 2023, 54, 731–742. [Google Scholar] [CrossRef]
- Panagiotou, O.A.; Evangelou, E.; Ioannidis, J.P. Genome-wide significant associations for variants with minor allele frequency of 5% or less--an overview: A HuGE review. Am. J. Epidemiol. 2010, 172, 869–889. [Google Scholar]
- Matukumalli, L.K.; Lawley, C.T.; Schnabel, R.D.; Taylor, J.F.; Allan, M.F.; Heaton, M.P.; O’Connell, J.; Moore, S.S.; Smith, T.P.L.; Sonstegard, T.S.; et al. Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE 2009, 4, e5350. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Mao, C.; Zhang, T.; Ren, A.; Jia, X.; Lai, S.-J.; Chen, S.-Y. Genome-wide SNP discovery and genetic diversity evaluation of Liangshan cattle in China. Anim. Biotechnol. 2021, 32, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Gan, J.; Fang, D.; Tang, H.; Wang, H.; Yi, J.; Fu, M. Genome-wide SNP discovery and evaluation of genetic diversity among six Chinese indigenous cattle breeds in Sichuan. PLoS ONE 2018, 13, e0201534. [Google Scholar]
- Konopiński, M.K. Shannon diversity index: A call to replace the original Shannon’s formula with unbiased estimator in the population genetics studies. PeerJ 2020, 8, e9391. [Google Scholar] [CrossRef]
- Wang, R.; Wang, X.; Qi, Y.; Li, Y.; Na, Q.; Yuan, H.; Rong, Y.; Ao, X.; Guo, F.; Zhang, L.; et al. Genetic diversity analysis of Inner Mongolia cashmere goats (Erlangshan subtype) based on whole genome re-sequencing. BMC Genom. 2024, 25, 698. [Google Scholar] [CrossRef]
- Nagylaki, T. Fixation indices in subdivided populations. Genetics 1998, 148, 1325–1332. [Google Scholar] [CrossRef]
- Zhang, Q.; Guldbrandtsen, B.; Bosse, M.; Lund, M.S.; Sahana, G. Runs of homozygosity and distribution of functional variants in the cattle genome. BMC Genom. 2015, 16, 542. [Google Scholar] [CrossRef] [PubMed]
- Kirin, M.; McQuillan, R.; Franklin, C.S.; Campbell, H.; McKeigue, P.M.; Wilson, J.F. Genomic runs of homozygosity record population history and consanguinity. PLoS ONE 2010, 5, e13996. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhao, G.; Yang, L.; Zhu, B.; Chen, Y.; Zhang, L.; Gao, X.; Gao, H.; Liu, G.E.; Li, J. Genomic patterns of homozygosity in Chinese local cattle. Sci. Rep. 2019, 9, 16977. [Google Scholar] [CrossRef] [PubMed]
- Szmatoła, T.; Gurgul, A.; Jasielczuk, I.; Ząbek, T.; Ropka-Molik, K.; Litwińczuk, Z.; Bugno-Poniewierska, M. A comprehensive analysis of runs of homozygosity of eleven cattle breeds representing different production types. Animals 2019, 9, 1024. [Google Scholar] [CrossRef]


| Quality Control Criteria | Number of Markers of SNPs |
|---|---|
| Total number of markers | 95,256 |
| Markers on sex chromosomes (X, Y) or with unassigned chromosomal location (0) | 4907 |
| SNP detection rate < 0.90 | 8478 |
| Markers with p < 10−6 in Hardy–Weinberg balance test | 71 |
| Markers with minor allele frequency < 0.01 | 3340 |
| Number of SNPs passing quality control | 78,460 |
| Quality Control Standard | Number of SNPs Markers |
|---|---|
| Total number of markers | 95,256 |
| Markers on sex chromosomes (X, Y) or with unassigned chromosomal location (0) | 4907 |
| SNP detection rate < 0.90 | 8242 |
| Markers with p < 10−6 on Hardy–Weinberg balance test | 50 |
| Markers with minor allele frequency < 0.01 | 5111 |
| Number of SNPs passing quality control | 76,946 |
| Family Name | Sex | Number | Individual Number |
|---|---|---|---|
| Family 1 | Male | 2 | 258_122002 |
| 222_118002 | |||
| Female | 1 | 272_020030 | |
| Family 2 | Male | 1 | 227_121003 |
| Female | 1 | 229_022005 | |
| Family 3 | Male | 1 | 230_121024 |
| Female | |||
| Family 4 | Male | 1 | 236_122003 |
| Female | |||
| Family 5 | Male | 1 | 247_120010 |
| Female | 1 | ||
| Family 6 | Male | 1 | 248_121019 |
| Female | |||
| Family 7 | Male | 1 | 266_121030 |
| Female | |||
| Family 8 | Male | 1 | 281_120009 |
| Female | |||
| Family 9 | Male | 1 | 292_120033 |
| Female | |||
| Family 10 | Male | 1 | 294_120042 |
| Female | |||
| Other | Female | 46 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, J.; Su, J.; Sui, S.; Li, H.; Jiang, R.; Xu, L.; Tan, Y.; Zhang, B. Genetic Analysis of the Conserved Population of Dengchuan Cattle Based on High Concordance SNP loci. Animals 2025, 15, 2937. https://doi.org/10.3390/ani15202937
Long J, Su J, Sui S, Li H, Jiang R, Xu L, Tan Y, Zhang B. Genetic Analysis of the Conserved Population of Dengchuan Cattle Based on High Concordance SNP loci. Animals. 2025; 15(20):2937. https://doi.org/10.3390/ani15202937
Chicago/Turabian StyleLong, Jiangyu, Jingjing Su, Shiyan Sui, Huimin Li, Rong Jiang, Linjie Xu, You Tan, and Birong Zhang. 2025. "Genetic Analysis of the Conserved Population of Dengchuan Cattle Based on High Concordance SNP loci" Animals 15, no. 20: 2937. https://doi.org/10.3390/ani15202937
APA StyleLong, J., Su, J., Sui, S., Li, H., Jiang, R., Xu, L., Tan, Y., & Zhang, B. (2025). Genetic Analysis of the Conserved Population of Dengchuan Cattle Based on High Concordance SNP loci. Animals, 15(20), 2937. https://doi.org/10.3390/ani15202937





