Proteomics of the Dark-Ventral-Patch Sexual Signal in Male Red Deer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
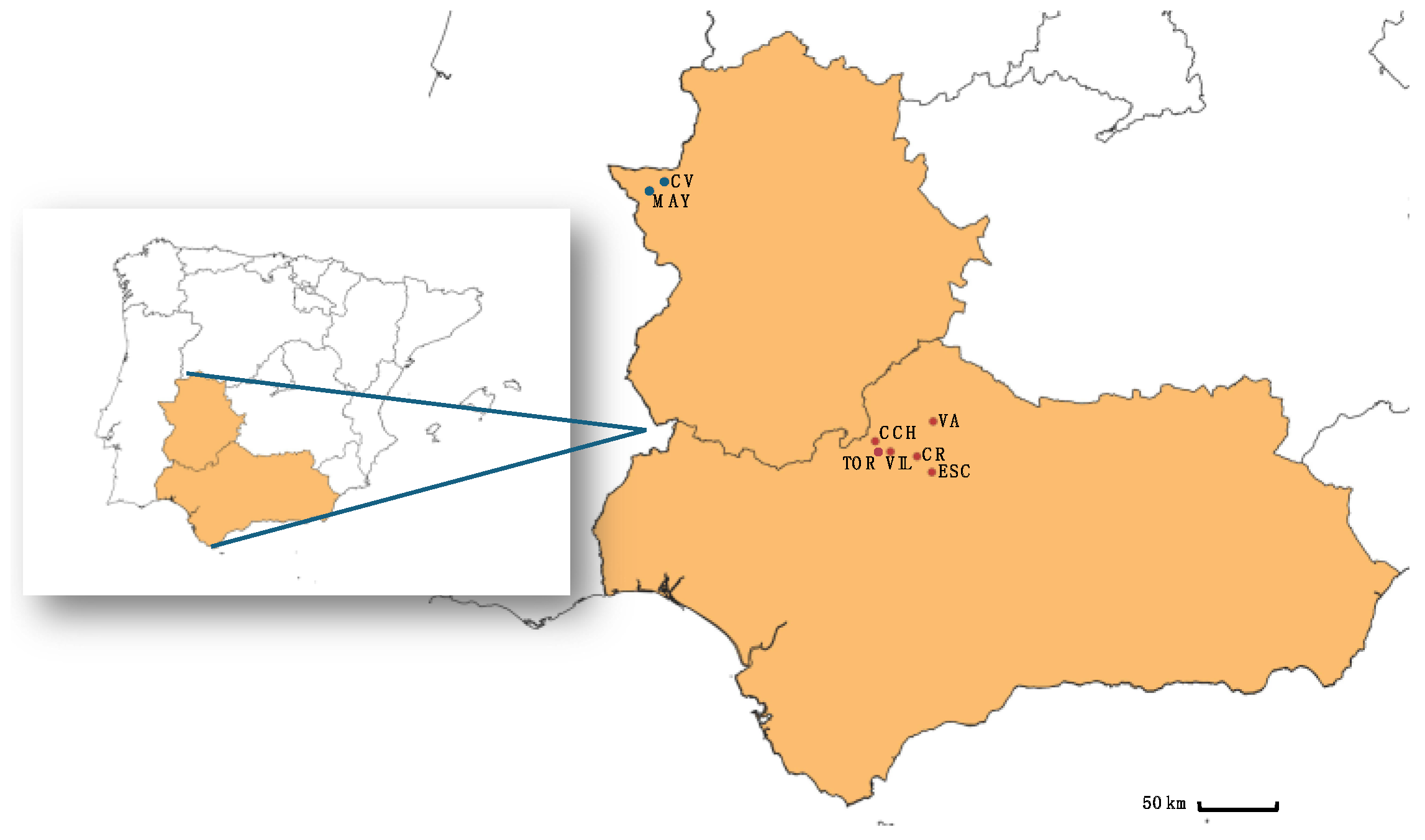
2.1. Study Site and Sample Collection
2.2. Hair Protein Isolation
2.3. Gel-Free LC-MS/MS Analysis
2.4. Label-Free Protein Quantification
2.5. Statistical Analyses
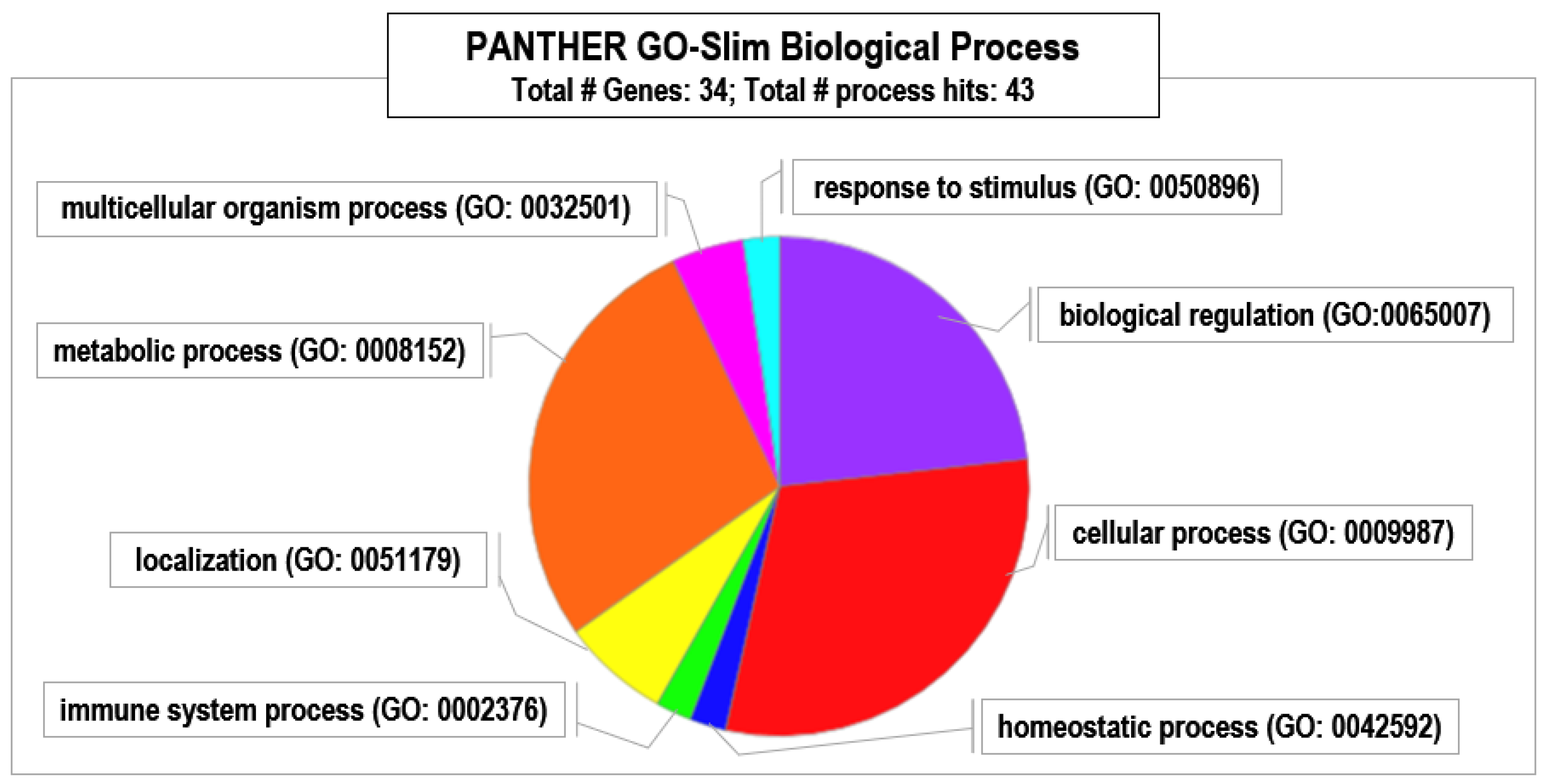
3. Results and Discussion
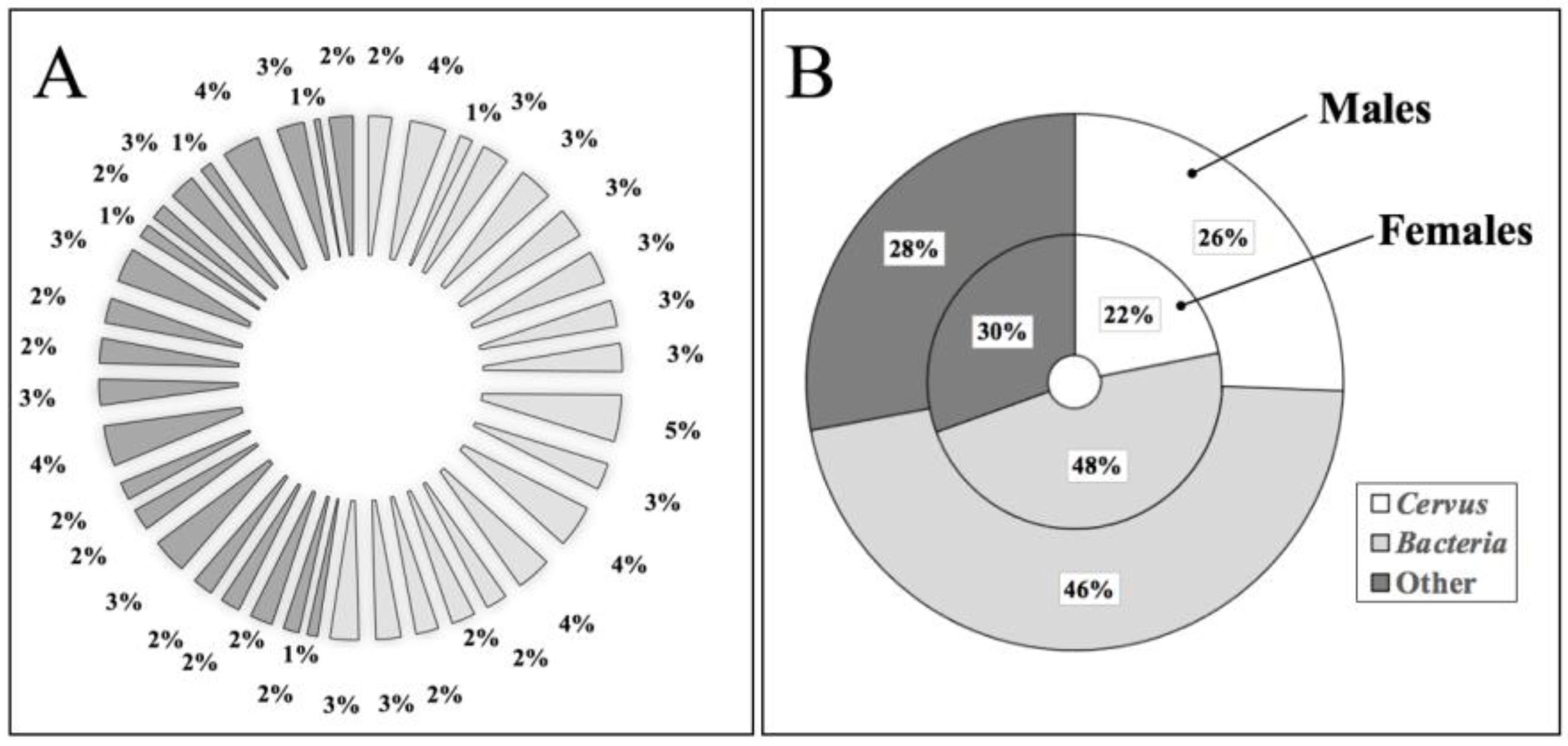
3.1. Cervus Proteins Exclusive to Male Red Deer Dark Ventral Patch
3.2. Possible Effect of the Level of Male–Male Competition
3.3. Possible Redundancy of Sex Traits: Proteins Related to Antler Development
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andersson, M. Sexual Selection; Princeton University Press: Princeton, NJ, USA, 1994. [Google Scholar]
- Smith, J.M.; Harper, D. Animal Signals; OUP Oxford: Oxford, UK, 2003. [Google Scholar]
- Dawkins, R.; Krebs, J.R. Animal Signals: Information or Manipulation? In Behavioural Ecology: An Evolutionary Approach; Krebs, J.R., Davies, N.B., Eds.; Blackwell Scientific: Hoboken, NJ, USA, 1978; pp. 282–309. [Google Scholar]
- Garnham, L.; Løvlie, H. Sophisticated fowl: The complex behaviour and cognitive skills of chickens and red junglefowl. Behav. Sci. 2018, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Lapshin, A.S.; Andreychev, A.V.; Kuznetcov, V.A. Daily and seasonal dynamics of the vocalization of the Eagle Owl (Bubo bubo, Strigiformes, Strigidae) in the central Volga region. Zool. Zhurnal 2018, 97, 77–88. [Google Scholar] [CrossRef]
- Andersson, M. Female choice selects for extreme tail length in a widowbird. Nature 1982, 299, 818–820. [Google Scholar] [CrossRef]
- Arak, A. Sexual selection by male–male competition in natterjack toad choruses. Nature 1983, 306, 261–262. [Google Scholar] [CrossRef]
- Borgia, G. Bower quality, number of decorations and mating success of male satin bowerbirds (Ptilonorhynchus violaceus): An experimental analysis. Anim. Behav. 1985, 33, 266–271. [Google Scholar] [CrossRef]
- Brückner, A.; Parker, J. Molecular evolution of gland cell types and chemical interactions in animals. J. Exp. Biol. 2020, 223, jeb211938. [Google Scholar] [CrossRef] [PubMed]
- Buchinger, T.J.; Li, W. Chemical communication and its role in sexual selection across Animalia. Commun. Biol. 2023, 6, 1178. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, T.D. Pheromones and Animal Behavior: Chemical Signals and Signatures; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar] [CrossRef]
- Steiger, S.; Schmitt, T.; Schaefer, H.M. The origin and dynamic evolution of chemical information transfer. Proc. R. Soc. B 2011, 278, 970–979. [Google Scholar] [CrossRef]
- Carranza, J.; Alvarez, F.; Redondo, T. Territoriality as a mating strategy in red deer. Anim. Behav. 1990, 40, 79–88. [Google Scholar] [CrossRef]
- Carranza, J.; Valencia, J. Red deer females collect on male clumps at mating areas. Behav. Ecol. 1999, 10, 525–532. [Google Scholar] [CrossRef]
- Geist, V. Mountain Sheep; University of Chicago Press: Chicago, IL, USA, 1974. [Google Scholar]
- Leboeuf, B.J. Sexual Behavior in the Northern Elephant Seal Mirounga Angustirostris. Behaviour 1972, 41, 1–26. [Google Scholar] [CrossRef]
- Hausfater, G. Dominance and reproduction in Baboons (Papio cynocephalus). Contrib. Primatol. 1975, 7, 1–150. [Google Scholar] [PubMed]
- Clutton-Brock, T.H.; Guinness, F.E.; Albon, S.D.; Barrett, P. Red Deer: Behavior and Ecology of Two Sexes; University of Chicago Press: Chicago, IL, USA, 1982. [Google Scholar]
- Martín, J.; Carranza, J.; López, P.; Alarcos, S.; Pérez-González, J. A new sexual signal in rutting male red deer: Age related chemical scent constituents in the belly black spot. Mamm. Biol. 2014, 79, 362–368. [Google Scholar] [CrossRef]
- Galván, I.; Solano, F.; Zougagh, M.; de Andrés, F.; Murtada, K.; Ríos, A.; de la Peña, E.; Carranza, J. Unprecedented high catecholamine production causing hair pigmentation after urinary excretion in red deer. Cell. Mol. Life Sci. 2019, 76, 397–404. [Google Scholar] [CrossRef] [PubMed]
- de la Peña, E.; Martín, J.; Carranza, J. The intensity of male-male competition may affect chemical scent constituents in the dark ventral patch of male Iberian red deer. PLoS ONE 2019, 14, e0221980. [Google Scholar] [CrossRef]
- de La Peña, E.; Martín, J.; Barja, I.; Carranza, J. Testosterone and the dark ventral patch of male red deer: The role of the social environment. Sci. Nat. 2020, 107, 18. [Google Scholar] [CrossRef]
- Carranza, J.; de la Peña, E.; Mateos, C.; Pérez-González, J.; Alarcos, S.; Torres-Porras, J.; Valencia, J.; Sánchez-Prieto, C.; Castillo, L. The dark ventral patch: A bimodal flexible trait related to male competition in red deer. PLoS ONE 2020, 15, e0241374. [Google Scholar] [CrossRef] [PubMed]
- De La Peña, E.; Pérez-González, J.; Martín, J.; Vedel, G.; Carranza, J. The dark-ventral-patch of male red deer, a sexual signal that conveys the degree of involvement in rutting behavior. BMC Zool. 2021, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Touhara, K. Sexual communication via peptide and protein pheromones. Curr. Opin. Pharmacol. 2008, 8, 759–764. [Google Scholar] [CrossRef] [PubMed]
- De La Peña, E.; Martín, J.; Carranza, J. Ultrastructural morphological features of the hair in a sexual signal: The dark ventral patch of male red deer. J. Zool. 2021, 313, 66–75. [Google Scholar] [CrossRef]
- Li, Y.F.; Radivojac, P. Computational approaches to protein inference in shotgun proteomics. BMC Bioinform. 2012, 13, S4. [Google Scholar] [CrossRef]
- Parker, G.J.; Leppert, T.; Anex, D.S.; Hilmer, J.K.; Matsunami, N.; Baird, L.; Stevens, J.; Parsawar, K.; Durbin-Johnson, B.P.; Rocke, D.M.; et al. Demonstration of Protein-Based Human Identification Using the Hair Shaft Proteome. PLoS ONE 2016, 11, e0160653. [Google Scholar] [CrossRef] [PubMed]
- Belant, J.L.; Seamans, T.W.; Paetkau, D. Genetic tagging free-ranging white-tailed deer using hair snares. Ohio J. Sci. 2007, 107, 50–56. [Google Scholar]
- Torres-Porras, J.; Carranza, J.; Pérez-González, J.; Mateos, C.; Alarcos, S. The tragedy of the commons: Unsustainable population structure of Iberian red deer in hunting estates. Eur. J. Wildl. Res. 2014, 60, 351–357. [Google Scholar] [CrossRef]
- Pérez-Barbería, F.; Duff, E.; Brewer, M.; Guinness, F. Evaluation of methods to age Scottish red deer: The balance between accuracy and practicality. J. Zool. 2014, 294, 180–189. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Molina, A.M.; Abril, N.; Lora, A.J.; Huertas-Abril, P.V.; Ayala, N.; Blanco, C.; Moyano, M. Proteomic profile of the effects of low-dose bisphenol A on zebrafish ovaries. Food Chem. Toxicol. 2021, 156, 112435. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Molecular. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, R.; Cheung, A.S.; Pang, K.; Saffery, R.; Novakovic, B. Sexual Dimorphism in Innate Immunity: The Role of Sex Hormones and Epigenetics. Front. Immunol. 2021, 11, 604000. [Google Scholar] [CrossRef] [PubMed]
- Kierdorf, U.; Kierdorf, H.; Schultz, M.; Rolf, H.J. Histological structure of antlers in castrated male fallow deer (Dama dama). Anat. Rec. 2004, 281A, 1352–1362. [Google Scholar] [CrossRef]
- De La Peña, E.; Martin, J.; Barja, I.; Pérez-Caballero, R.; Acosta, I.; Carranza, J. Immune challenge of mating effort: Steroid hormone profile, dark ventral patch and parasite burden in relation to intrasexual competition in male Iberian red deer. Integr. Zool. 2020, 15, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Viney, M.E.; Riley, E.M.; Buchanan, K.L. Optimal immune responses: Immunocompetence revisited. Trends Ecol. Evol. 2005, 20, 665–669. [Google Scholar] [CrossRef] [PubMed]
- Malo, A.F.; Roldan, E.R.S.; Garde, J.J.; Soler, A.J.; Vicente, J.; Gortazar, C.; Gomendio, M. What does testosterone do for red deer males? Proc. R. Soc. B. 2009, 276, 971–980. [Google Scholar] [CrossRef]
- Choi, G.; Levi, M.; Van, D. The relationship between inflammation and the coagulation system. Swiss Med. Wkly. 2006, 136, 139–144. [Google Scholar] [CrossRef]
- Lidington, E.A.; Haskard, D.O.; Mason, J.C. Induction of decay-accelerating factor by thrombin through a protease-activated receptor 1 and protein kinase C-dependent pathway protects vascular endothelial cells from complement-mediated injury. Blood 2000, 96, 2784–2792. [Google Scholar] [CrossRef]
- Ikeda, K.; Nagasawa, K.; Horiuchi, T.; Tsuru, T.; Nishizaka, H.; Niho, Y. C5a induces tissue factor activity on endothelial cells. Thromb. Haemost. 1997, 77, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Molnár, A.; Gyurján, I.; Korpos, É.; Borsy, A.; Stéger, V.; Buzás, Z.; Kiss, I.; Zomborszky, Z.; Papp, P.; Deák, F.; et al. Identification of differentially expressed genes in the developing antler of red deer Cervus elaphus. Mol. Genet. Genom. 2007, 277, 237–248. [Google Scholar] [CrossRef] [PubMed]
- López-Pedrouso, M.; Lorenzo, J.M.; Landete-Castillejos, T.; Chonco, L.; Pérez-Barbería, F.J.; García, A.; López-Garrido, M.-P.; Franco, D. SWATH-MS Quantitative Proteomic Analysis of Deer Antler from Two Regenerating and Mineralizing Sections. Biology 2021, 10, 679. [Google Scholar] [CrossRef] [PubMed]
- Candolin, U. The use of multiple cues in mate choice. Biol. Rev. 2003, 78, 575–595. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, R.A. Multiple displays in animal communication: ‘backup signals’ and ‘multiple messages’. Philos. Trans. R. Soc. Lond. B 1996, 351, 329–338. [Google Scholar]
- Moller, A.P.; Pomiankowski, A. Why have birds got multiple sexual ornaments? Behav. Ecol. Sociobiol. 1993, 32, 167–176. [Google Scholar] [CrossRef]
- Searcy, W.A.; Nowicki, S. The Evolution of Animal Communication: Reliability and Deception in Signaling Systems; Princeton University Press: Princeton, NJ, USA, 2005. [Google Scholar]
- Senar, J.C.; Negro, J.; Quesada, J.; Ruiz, I.; Garrido, J. Two pieces of information in a single trait? The yellow breast of the Great Tit (Parus major) reflects both pigment acquisition and body condition. Behaviour 2008, 145, 1195–1210. [Google Scholar]
- Zuk, M.; Ligon, J.D.; Thornhill, R. Effects of experimental manipulation of male secondary sex characters on female mate preference in red jungle fowl. Anim. Behav. 1992, 44, 999–1006. [Google Scholar] [CrossRef]

| GO-Term | Description | Count in Network a | Strength b | FDR c |
|---|---|---|---|---|
| Catabolism of aromatic amino acids | ||||
| GO:0006572 | Tyrosine catabolic process | 3/5 | 1.74 | 9.50 × 10−03 |
| GO:0006559 | L-phenylalanine catabolic process | 4/8 | 1.66 | 1.40 × 10−03 |
| GO:0009074 | Aromatic amino acid family catabolic process | 5/15 | 1.49 | 6.20 × 10−04 |
| GO:0006570 | Tyrosine metabolic process | 4/13 | 1.45 | 4.60 × 10−03 |
| GO:0009072 | Aromatic amino acid family metabolic process | 6/25 | 1.34 | 3.20 × 10−04 |
| GO:0009063 | Cellular amino acid catabolic process | 10/101 | 0.96 | 1.50 × 10−04 |
| GO:1901606 | Alpha-amino acid catabolic process | 10/86 | 1.03 | 4.32 × 10−05 |
| GO:0008652 | Cellular amino acid biosynthetic process | 6/72 | 0.88 | 2.22 × 10−02 |
| GO:1901605 | Alpha-amino acid metabolic process | 14/178 | 0.86 | 1.42 × 10−05 |
| Catabolism of lipids | ||||
| GO:0034440 | Lipid oxidation | 6/78 | 0.85 | 3.00 × 10−02 |
| GO:0060192 | Negative regulation of lipase activity | 3/9 | 1.49 | 2.62 × 10−02 |
| GO:0006635 | Fatty acid beta-oxidation | 5/56 | 0.91 | 4.82 × 10−02 |
| GO:0034374 | Low-density lipoprotein particle remodeling | 3/10 | 1.44 | 3.25 × 10−02 |
| Catabolism of carbohydrates | ||||
| GO:0016052 | Carbohydrate catabolic process | 7/88 | 0.86 | 1.04 × 10−02 |
| GO:0046365 | Monosaccharide catabolic process | 4/24 | 1.18 | 2.39 × 10−02 |
| GO:0006090 | Pyruvate metabolic process | 5/57 | 0.91 | 4.94 × 10−02 |
| GO:0046395 | Carboxylic acid catabolic process | 15/199 | 0.84 | 9.29 × 10−06 |
| GO:0006026 | Aminoglycan catabolic process | 4/28 | 1.12 | 3.66 × 10−02 |
| Response to stress | ||||
| GO:1990748 | Cellular detoxification | 9/107 | 0.89 | 1.30 × 10−03 |
| GO:0098869 | Cellular oxidant detoxification | 8/94 | 0.89 | 2.80 × 10−03 |
| GO:0042743 | Hydrogen peroxide metabolic process | 5/38 | 1.08 | 1.26 × 10−02 |
| GO:0071456 | Cellular response to hypoxia | 6/73 | 0.88 | 2.33 × 10−02 |
| GO:0051085 | Chaperone cofactor-dependent protein refolding | 4/30 | 1.09 | 4.53 × 10−02 |
| GO:0006749 | Glutathione metabolic process | 5/56 | 0.91 | 4.82 × 10−02 |
| GO:0019835 | Cytolysis | 5/25 | 1.26 | 3.00 × 10−03 |
| Metabolism of structural proteins | ||||
| GO:0048251 | Elastic fiber assembly | 4/11 | 1.52 | 3.00 × 10−03 |
| GO:0045104 | Intermediate filament cytoskeleton organization | 8/91 | 0.91 | 2.50 × 10−03 |
| GO:0045109 | Intermediate filament organization | 6/75 | 0.87 | 2.60 × 10−02 |
| GO:0010712 | Regulation of collagen metabolic process | 4/19 | 1.29 | 1.28 × 10−02 |
| Complement activation | ||||
| GO:0006956 | Complement activation | 10/62 | 1.17 | 4.78 × 10−06 |
| GO:0006958 | Complement activation, classical pathway | 8/43 | 1.23 | 3.54 × 10−05 |
| GO:0006957 | Complement activation, alternative pathway | 4/11 | 1.52 | 3.00 × 10−03 |
| Blood coagulation | ||||
| GO:0030194 | Positive regulation of blood coagulation | 4/15 | 1.39 | 6.70 × 10−03 |
| GO:0050819 | Negative regulation of coagulation | 5/36 | 1.11 | 1.04 × 10−02 |
| GO:0030193 | Regulation of blood coagulation | 6/50 | 1.04 | 4.80 × 10−03 |
| GO:0050818 | Regulation of coagulation | 7/55 | 1.07 | 1.30 × 10−03 |
| GO:0007596 | Blood coagulation | 9/100 | 0.92 | 8.60 × 10−04 |
| GO:0090303 | Positive regulation of wound healing | 5/39 | 1.07 | 1.33 × 10−02 |
| Immune response | ||||
| GO:0006953 | Acute-phase response | 4/21 | 1.24 | 1.64 × 10−02 |
| GO:0002888 | Positive regulation of myeloid leukocyte-mediated immunity | 3/12 | 1.36 | 4.82 × 10−02 |
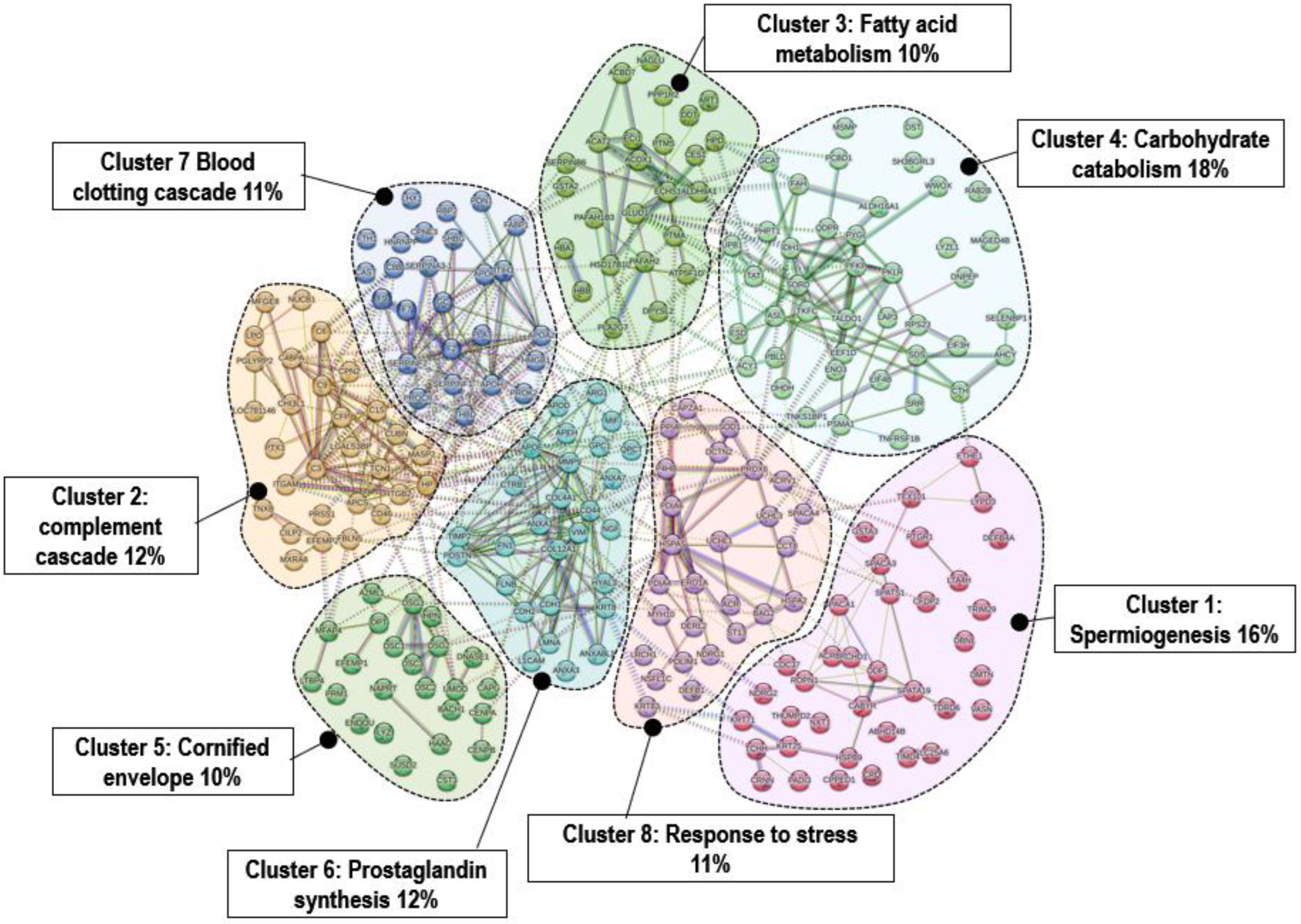
| Cluster ID | Gene Count | Network Stats (Number of Nodes; Average Node Degree; Avg. Local Clustering Coefficient; Expected Number of Edges) and PPI Enrichment p-Value | Biological Process | Protein Names |
|---|---|---|---|---|
| Cluster 1 | 37 | 37; 29; 1.57; 0.362; 1 <1.0 × 10−16 | Spermiogenesis. Acrosome biogenesis | ABHD14B, ACRBP, CABYR, CACHD1, CDC37, CFDP2, CPPED1, CPQ, CRNN, DBNL, DEFB4A, DMTN, ETHE1, GSTA3, HSPB9, KRT25, KRT71, LTA4H, LYPD3, NDRG2, NXT1, ODF1, PADI3, PTGR1, ROPN1, SLC25A6, SPACA1, SPACA3, SPATA19, SPATS1, TCHH, TDRD6, TEX101, TIMD4, TTHUMPD2, RIM29, VASN |
| Cluster 2 | 29 | 29; 50; 3.45; 0.529; 1 <1.0 × 10−16 | Complement cascades | APCS, C1S, C3, C4BPA, C6, C9, CD46, CFP, CHI3L1, CILP2, CPN2, CUBN, EFEMP2, FBLN5, HP, ITGAM, ITGB2, LGALS3BP, LOC781146, LPO, TNX, MASP2, MFGE8, MXRA8, NUCB1, PGLYRP2, PRSS1, PTX3, TCN1, TNXB |
| Cluster 3 | 25 | 25; 29; 2.9; 0.397; 2 <1.0 × 10−16 | Fatty acid metabolism | ACAT2, ACBD7, ACOX1, ALDH9A1, ART1, ATP5F1D, CES1, DDT, DPYSL2, ECHS1, ECI1, GLUD1, GSTA2, HBA1, HBB, HPD, HSD17B10, NAGLU, PAFAH1B3, PAFAH2, PLA2G7, PPP1R2, PTMA, PTMS, SERPINB6 |
| Cluster 4 | 42 | 42; 61; 2.32; 0.679; 9 <1.0 × 10−16 | Carbohydrate catabolism | ACY1, AHCY, ALDH16A1, ASL, CTH, DHDH, DNPEP, DST, EEF1D, EIF3H, EIF4B, ENO3, ESD, FAH, GCAT, IDH1, LAP3, LYZL1, MAGED4B, MSMP, PBLD, PCBD1, PFKP, PHPT1, PKLR, PSMA1, PYGL, QDPR, RAB2B, RPS23, SDS, SELENBP1, SH3BGRL3, SORD, SRR, TALDO1, TAT, TKFC, TNFRSF1B, TNKS1BP1, UPB1, WWOX |
| Cluster 5 | 24 | 24; 15; 1.25; 0.533; 8 <1.0 × 10−16 | Cornified envelope | A2ML1, BACH1, CAPG, CENPA, CENPB, CST3, DNASE1, DPT, DSC1, DSC2, DSC3, DSG2, DSG3, EFEMP1, ENDOU, HAAO, HPN, LTBP4, LYZ, MFAP4, NAPRT, PRM1, SUSD2, UMOD |
| Cluster 6 | 28 | 28; 82; 5.86; 0.714 <1.0 × 10−16 | Signaling and regulation | ANXA1, ANXA3, ANXA7, ANXA8L1, APEH, APOD, APOE, ARG1, CD44, CDH1, CDH2, COL12A1, COL4A1, CTRB1, FLNB, FN1, GPC1, HYAL3, KRT8, L1CAM, LMNA, MIF, MMP9, NGF, POSTN, QPCT, TIMP2, VIM |
| Cluster 7 | 26 | 26; 47; 3.62; 0.636; 1 <1.0 × 10−16 | Blood clotting cascade | APOA2, APOF, APOH, C8B, CAST, CPNE3, F2, F7, F9, FABP1, FTH1, GC, HMGB1, HNRNPF, ITIH3, LHX1, PLAT, PON1, PROCR, PROK2, RBP2, SERPINA3-1, SERPINF1, SERPINF2, SHBG, THBD |
| Cluster 8 | 27 | 27; 35; 2.59; 0.647; 3 <1.0 × 10−16 | Cellular responses to stress | ACR, ACRV1, BAG2, CAPZA1, CCT8, DCTN2, DEFB1, DERL2, ERO1A, HSPA2, HSPA5, KRT83, LRCH1, MYH10, NDRG1, NSFL1C, P4HB, PDIA4, PDIA6, PDLIM1, PPIA, PRDX6, SOD1, SPACA4, ST13, UCHL1, UCHL3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Broggini, C.; Huertas-Abril, P.V.; Membrillo, A.; de la Peña, E.; Abril, N.; Carranza, J. Proteomics of the Dark-Ventral-Patch Sexual Signal in Male Red Deer. Animals 2025, 15, 252. https://doi.org/10.3390/ani15020252
Broggini C, Huertas-Abril PV, Membrillo A, de la Peña E, Abril N, Carranza J. Proteomics of the Dark-Ventral-Patch Sexual Signal in Male Red Deer. Animals. 2025; 15(2):252. https://doi.org/10.3390/ani15020252
Chicago/Turabian StyleBroggini, Camilla, Paula V. Huertas-Abril, Alberto Membrillo, Eva de la Peña, Nieves Abril, and Juan Carranza. 2025. "Proteomics of the Dark-Ventral-Patch Sexual Signal in Male Red Deer" Animals 15, no. 2: 252. https://doi.org/10.3390/ani15020252
APA StyleBroggini, C., Huertas-Abril, P. V., Membrillo, A., de la Peña, E., Abril, N., & Carranza, J. (2025). Proteomics of the Dark-Ventral-Patch Sexual Signal in Male Red Deer. Animals, 15(2), 252. https://doi.org/10.3390/ani15020252





