Comprehensive Identification of the Bovine KLF Gene Family and Its Functional Regulation in Muscle Development: Insights from Single-Nuclei Transcriptomics
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of KLF Family
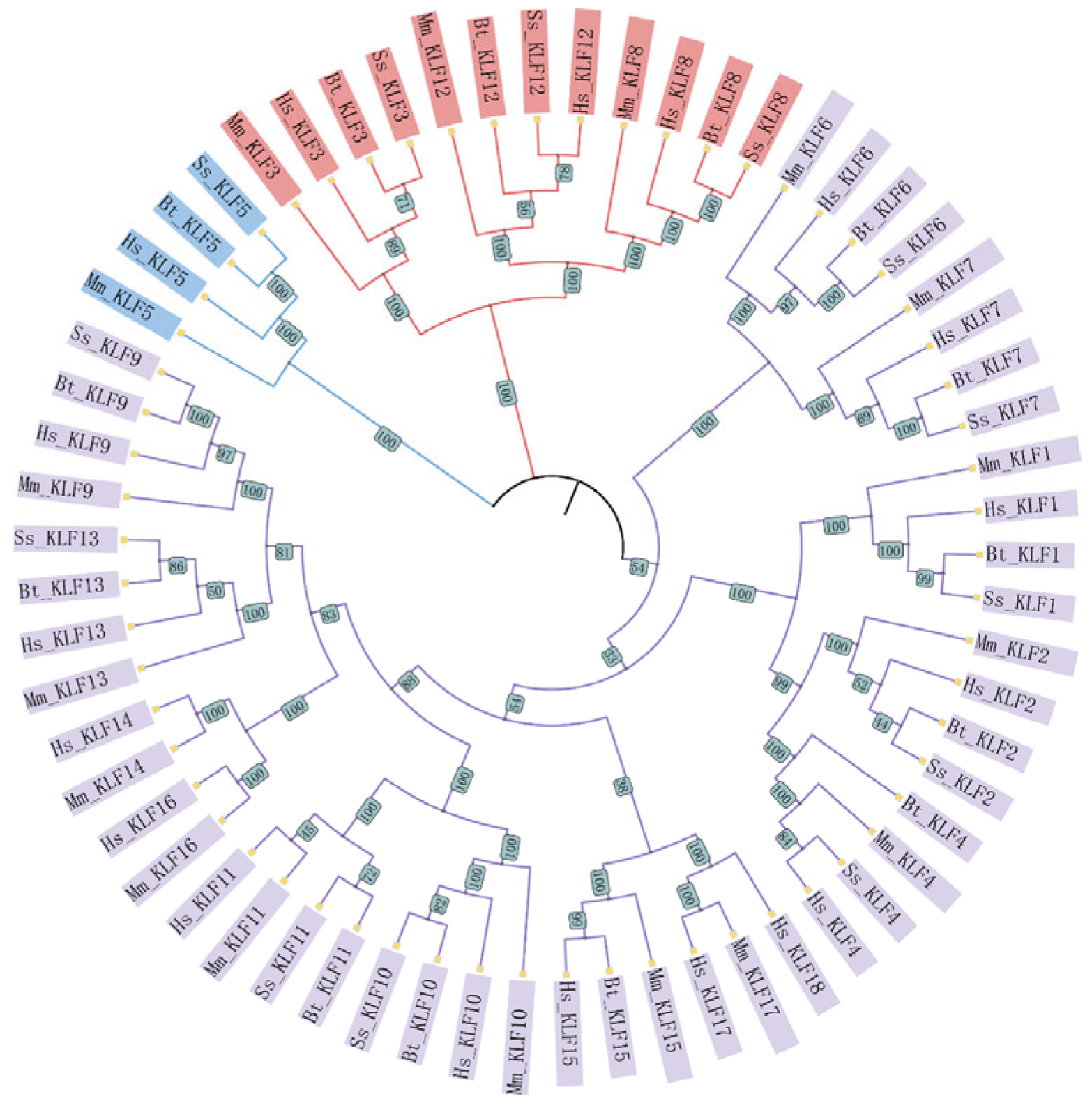
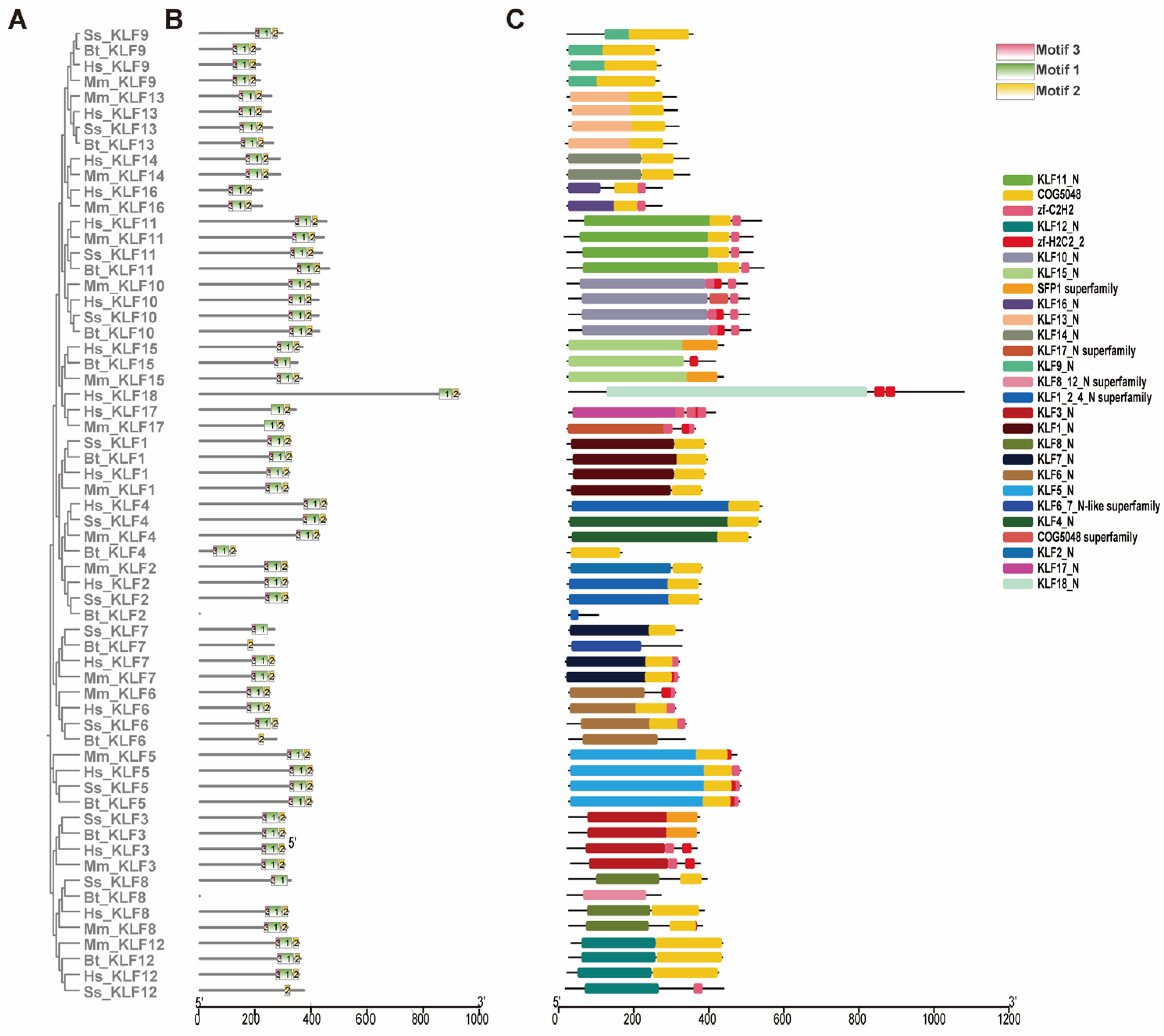
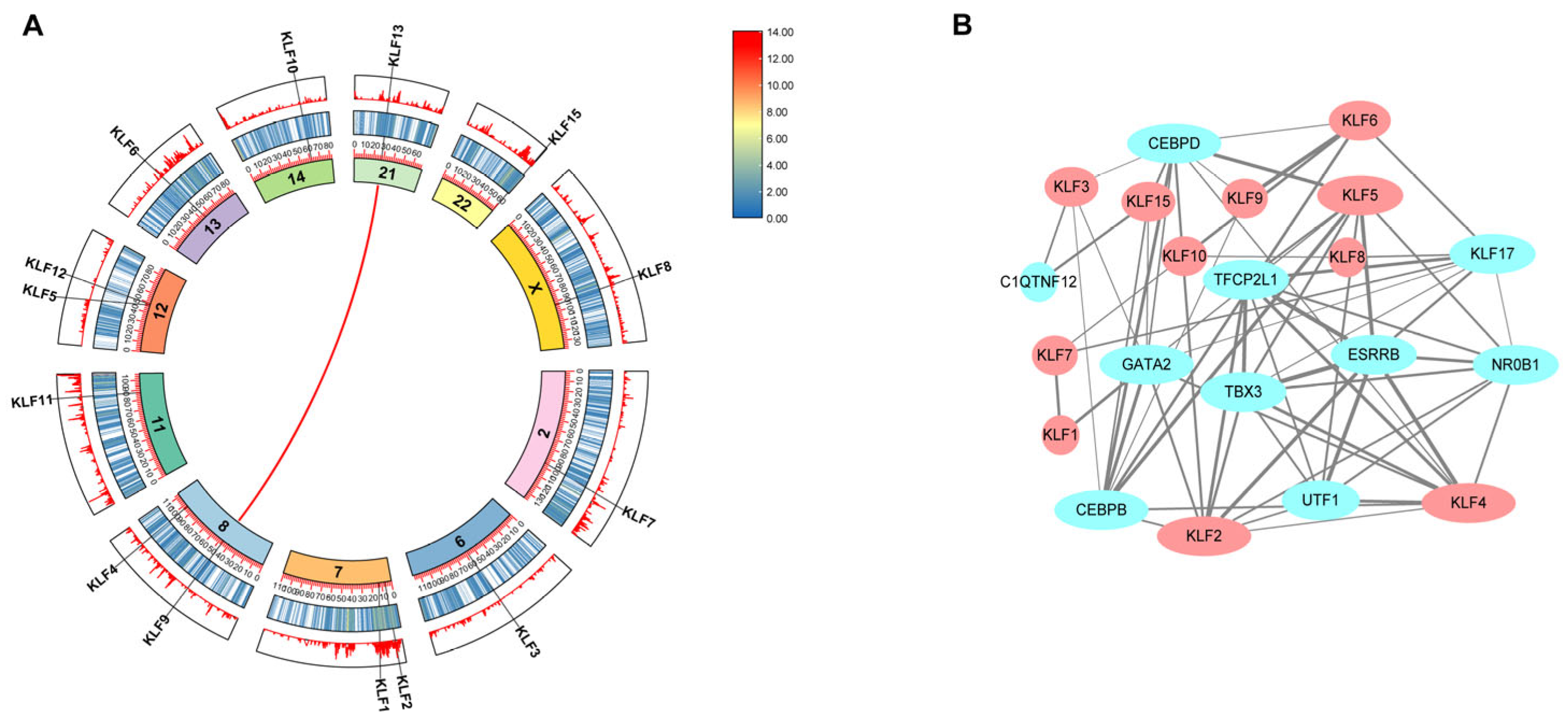
2.2. Structural and Evolutionary Analysis of KLF Genes: Protein Alignment, Phylogeny, and Chromosomal Location
2.3. Isolation of Nuclei from Bovine LD Tissue
2.4. Preparation of Single-Nuclei Libraries and Sequencing
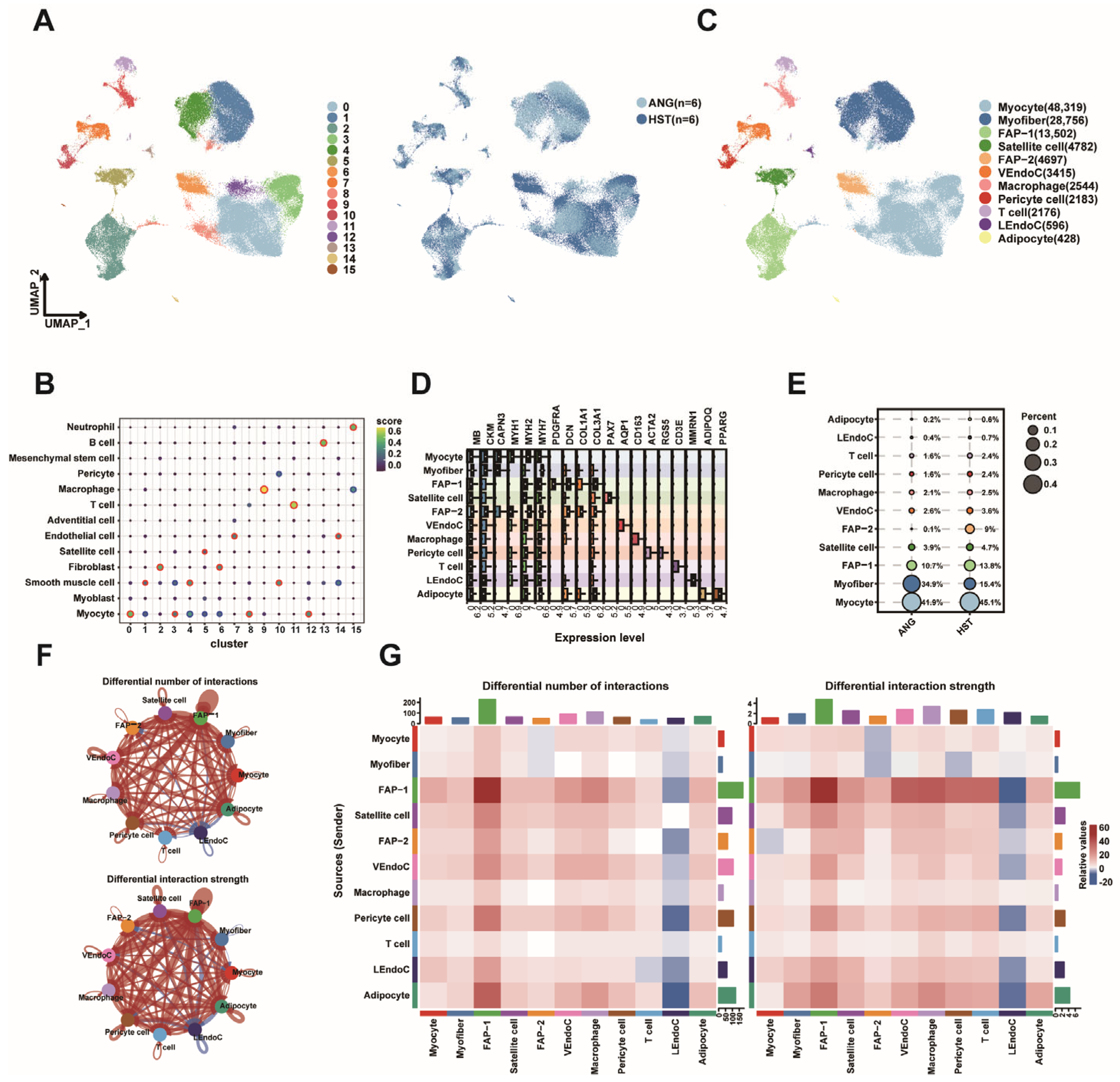
2.5. Single-Nucleus Data Analysis
2.6. Cell Type Annotation
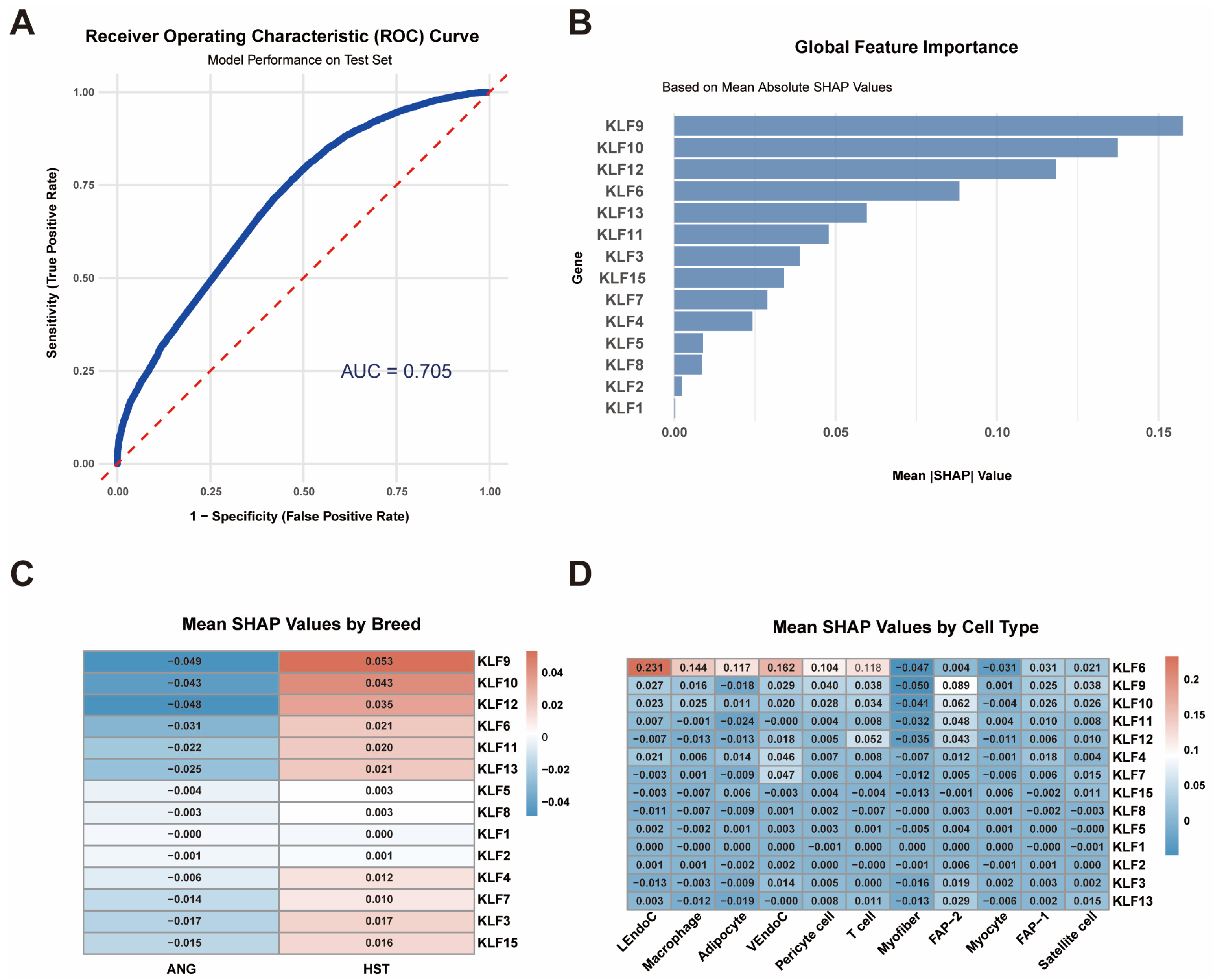
2.7. Explainable Machine Learning
3. Results
3.1. Characterization and Evolutionary Analysis of the KLF Gene Family
3.2. Genomic Distribution of Bovine KLF Genes
3.3. Analysis of Conserved Motifs and Protein Domains
3.4. Analysis of Evolutionary Conservation and Functional Interaction Maps
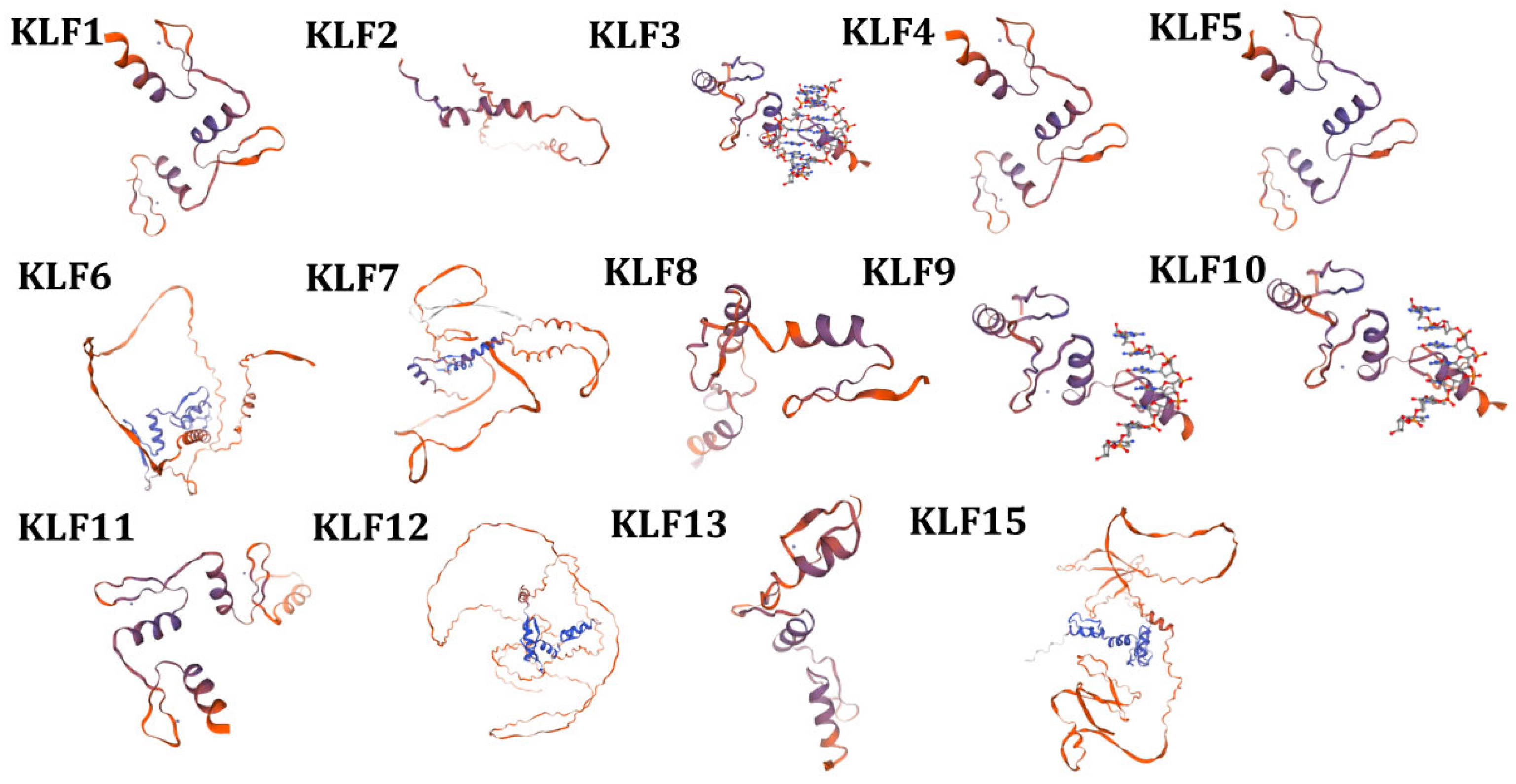
3.5. Prediction of the Tertiary Structure and Transmembrane Region of KLF Family Proteins
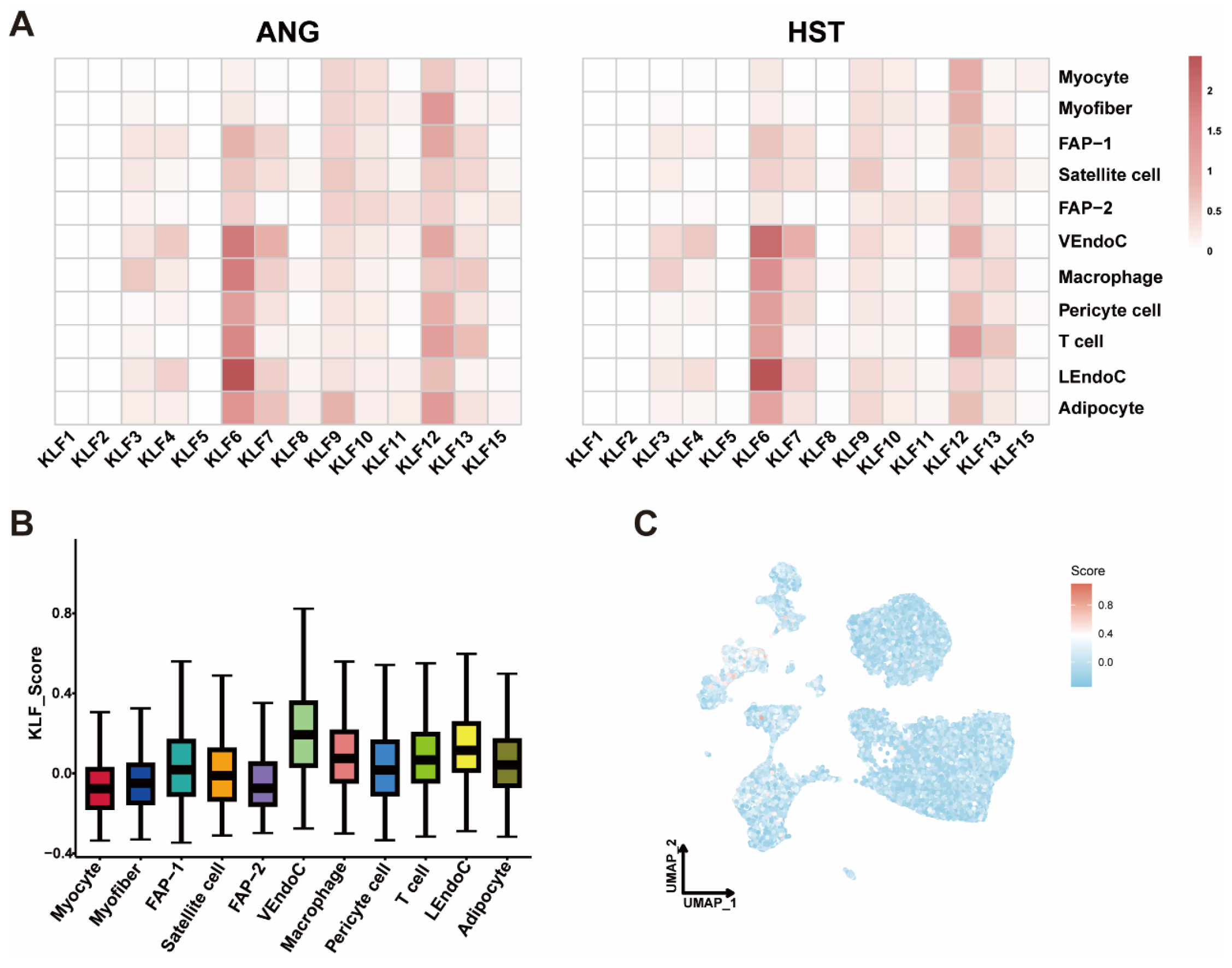
3.6. Overview of KLF in the snRNA Seq Dataset of Bovine LD
3.7. SHAP Explains the Contribution of the KLF Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANG | Angus |
| HST | Holstein |
| LEndoC | Lymphatic endothelial cells |
| VEndoC | Vascular endothelial cell |
| FAP | Fibro/Adipogenic Progenitor |
| snRNA-seq | Single-nucleus RNA sequencing |
| LD | Longissimus dorsi |
| SHAP | Shapley Additive exPlanations |
| KLF | Krüppel-like factor |
| PPI | Protein–protein interaction |
References
- Ponnampalam, E.N.; Priyashantha, H.; Vidanarachchi, J.K.; Kiani, A.; Holman, B.W.B. Effects of Nutritional Factors on Fat Content, Fatty Acid Composition, and Sensorial Properties of Meat and Milk from Domesticated Ruminants: An Overview. Animals 2024, 14, 840. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Jiang, H. Molecular and Cellular Mechanisms of Intramuscular Fat Development and Growth in Cattle. Int. J. Mol. Sci. 2024, 25, 2520. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, E.; Li, Q.; Peng, Y.; Jin, H.; Naseem, S.; Sun, B.; Park, S.; Choi, S.; Li, X. GSK3 regulation Wnt/β-catenin signaling affects adipogenesis in bovine skeletal muscle fibro/adipogenic progenitors. Int. J. Biol. Macromol. 2024, 275, 133639. [Google Scholar] [CrossRef]
- Ferreira, K.K.S.; de Morais Gomes, E.R.; de Lima Filho, J.L.; Castelletti, C.H.M.; Martins, D.B.G. Bioinformatics analysis of non-synonymous variants in the KLF genes related to cardiac diseases. Gene 2018, 650, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Yuce, K.; Ozkan, A.I. The kruppel-like factor (KLF) family, diseases, and physiological events. Gene 2024, 895, 148027. [Google Scholar] [CrossRef]
- Choi, J.; Baldwin, T.M.; Wong, M.; Bolden, J.E.; Fairfax, K.A.; Lucas, E.C.; Cole, R.; Biben, C.; Morgan, C.; Ramsay, K.A.; et al. Haemopedia RNA-seq: A database of gene expression during haematopoiesis in mice and humans. Nucleic Acids Res. 2019, 47, D780–D785. [Google Scholar] [CrossRef]
- Dasgupta, A.; Gibbard, D.F.; Schmitt, R.E.; Arneson-Wissink, P.C.; Ducharme, A.M.; Bruinsma, E.S.; Hawse, J.R.; Jatoi, A.; Doles, J.D. A TGF-β/KLF10 signaling axis regulates atrophy-associated genes to induce muscle wasting in pancreatic cancer. Proc. Natl. Acad. Sci. USA 2023, 120, e2215095120. [Google Scholar] [CrossRef]
- Himeda, C.L.; Ranish, J.A.; Pearson, R.C.M.; Crossley, M.; Hauschka, S.D. KLF3 Regulates Muscle-Specific Gene Expression and Synergizes with Serum Response Factor on KLF Binding Sites. Mol. Cell Biol. 2010, 30, 3430–3443. [Google Scholar] [CrossRef]
- Chen, Z.; Lei, T.; Chen, X.; Zhang, J.; Yu, A.; Long, Q.; Long, H.; Jin, D.; Gan, L.; Yang, Z. Porcine KLF gene family: Structure, mapping, and phylogenetic analysis. Genomics 2010, 95, 111–119. [Google Scholar] [CrossRef]
- Li, Z.; Zheng, M.; Mo, J.; Li, K.; Yang, X.; Guo, L.; Zhang, X.; Abdalla, B.A.; Nie, Q. Single-cell RNA sequencing of preadipocytes reveals the cell fate heterogeneity induced by melatonin. J. Pineal Res. 2021, 70, e12725. [Google Scholar] [CrossRef]
- Liu, H.; Wei, W.; Lin, W.; Yu, W.; Luo, W.; Niu, Y.; Zhang, L.; Chen, J. miR-32-5p Regulates Lipid Accumulation in Intramuscular Fat of Erhualian Pigs by Suppressing KLF3. Lipids 2021, 56, 279–287. [Google Scholar] [CrossRef]
- Guo, H.; Khan, R.; Raza, S.H.A.; Ning, Y.; Wei, D.; Wu, S.; Hosseini, S.M.; Ullah, I.; Garcia, M.D.; Zan, L. KLF15 promotes transcription of KLF3 gene in bovine adipocytes. Gene 2018, 659, 77–83. [Google Scholar] [CrossRef]
- Song, C.; Fang, X.; Yang, Z.; Wang, Q.; Meng, F.; Chen, Y.; Chen, J.; Zhao, B.; Wang, Y.; Fang, X.; et al. miR-152 Regulates Bovine Myoblast Proliferation by Targeting KLF6. Animals 2021, 11, 3001. [Google Scholar] [CrossRef]
- Raza, S.H.A.; Khan, R.; Schreurs, N.M.; Guo, H.; Gui, L.; Mei, C.; Zan, L. Expression of the bovine KLF6 gene polymorphisms and their association with carcass and body measures in Qinchuan cattle (Bos taurus). Genomics 2020, 112, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hao, J.; Feng, Y.; Qiu, T.; Wu, J.; Zhou, X.; Fan, H.; Chang, Y. Klf9 Loss of Function Protects Against Glucocorticoids Induced Skeletal Muscle Wasting. J. Cachexia Sarcopenia Muscle 2025, 16, e70020. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Koike, H.; Ono, T.; Hayashi, S.; Kudo, F.; Kaneda, A.; Kagechika, H.; Manabe, I.; Nakashima, T.; Oishi, Y. Identification of a KLF5-dependent program and drug development for skeletal muscle atrophy. Proc. Natl. Acad. Sci. USA 2021, 118, e2102895118. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Sweet, D.R.; Fan, E.K.; Prosdocimo, D.A.; Madera, A.; Jiang, Z.; Padmanabhan, R.; Haldar, S.M.; Vinayachandran, V.; Jain, M.K. Transcription factors KLF15 and PPARδ cooperatively orchestrate genome-wide regulation of lipid metabolism in skeletal muscle. J. Biol. Chem. 2022, 298, 101926. [Google Scholar] [CrossRef]
- Lu, S.; Jolly, A.J.; Strand, K.A.; Dubner, A.M.; Mutryn, M.F.; Moulton, K.S.; Nemenoff, R.A.; Majesky, M.W.; Weiser-Evans, M.C.M. Smooth muscle–derived progenitor cell myofibroblast differentiation through KLF4 downregulation promotes arterial remodeling and fibrosis. JCI Insight 2020, 5, e139445. [Google Scholar] [CrossRef]
- Jovic, D.; Liang, X.; Zeng, H.; Lin, L.; Xu, F.; Luo, Y. Single-cell RNA sequencing technologies and applications: A brief overview. Clin. Transl. Med. 2022, 12, e694. [Google Scholar] [CrossRef]
- Potter, S.S. Single-cell RNA sequencing for the study of development, physiology and disease. Nat. Rev. Nephrol. 2018, 14, 479–492. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun. 2022, 13, 6968. [Google Scholar] [CrossRef]
- Haber, A.L.; Biton, M.; Rogel, N.; Herbst, R.H.; Shekhar, K.; Smillie, C.; Burgin, G.; Delorey, T.M.; Howitt, M.R.; Katz, Y.; et al. A single-cell survey of the small intestinal epithelium. Nature 2017, 551, 333–339. [Google Scholar] [CrossRef]
- Tian, Y.; Liu, S.; Shi, H.; Li, J.; Wan, X.; Sun, Y.; Li, H.; Cao, N.; Feng, Z.; Zhang, T.; et al. Revealing the Transcriptional and Metabolic Characteristics of Sebocytes Based on the Donkey Cell Transcriptome Atlas. Adv. Sci. 2025, 12, 2413819. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Wei, X.; Liu, C.; Volpe, G.; Zhuang, Z.; Zou, X.; Wang, Z.; Pan, T.; Yuan, Y.; Zhang, X.; et al. Cell transcriptomic atlas of the non-human primate Macaca fascicularis. Nature 2022, 604, 723–731. [Google Scholar] [CrossRef] [PubMed]
- A Portable and Cost-Effective Microfluidic System for Massively Parallel Single-Cell Transcriptome Profiling|bioRxiv. Available online: https://www.biorxiv.org/content/10.1101/818450v1.full (accessed on 22 September 2025).
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.E29. [Google Scholar] [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef]
- Yang, L.; Ng, Y.E.; Sun, H.; Li, Y.; Chini, L.C.S.; LeBrasseur, N.K.; Chen, J.; Zhang, X. Single-cell Mayo Map (scMayoMap): An easy-to-use tool for cell type annotation in single-cell RNA-sequencing data analysis. BMC Biol. 2023, 21, 223. [Google Scholar] [CrossRef]
- Cai, C.; Yue, Y.; Yue, B. Single-cell RNA sequencing in skeletal muscle developmental biology. Biomed. Pharmacother. 2023, 162, 114631. [Google Scholar] [CrossRef]
- Syafruddin, S.E.; Mohtar, M.A.; Wan Mohamad Nazarie, W.F.; Low, T.Y. Two Sides of the Same Coin: The Roles of KLF6 in Physiology and Pathophysiology. Biomolecules 2020, 10, 1378. [Google Scholar] [CrossRef]
- Shao, M.; Ge, G.-Z.; Liu, W.-J.; Xiao, J.; Xia, H.-J.; Fan, Y.; Zhao, F.; He, B.-L.; Chen, C. Characterization and phylogenetic analysis of Krüppel-like transcription factor (KLF) gene family in tree shrews (Tupaia belangeri chinensis). Oncotarget 2016, 8, 16325–16339. [Google Scholar] [CrossRef]
- Abe, M.; Saeki, N.; Ikeda, Y.; Ohba, S. Kruppel-like Factors in Skeletal Physiology and Pathologies. Int. J. Mol. Sci. 2022, 23, 15174. [Google Scholar] [CrossRef]
- Li, Z.-Y.; Zhu, Y.-X.; Chen, J.-R.; Chang, X.; Xie, Z.-Z. The role of KLF transcription factor in the regulation of cancer progression. Biomed. Pharmacother. 2023, 162, 114661. [Google Scholar] [CrossRef] [PubMed]
- Salmon, J.M.; Adams, H.; Magor, G.W.; Perkins, A.C. KLF feedback loops in innate immunity. Front. Immunol. 2025, 16, 1606277. [Google Scholar] [CrossRef] [PubMed]
- Pearson, R.; Fleetwood, J.; Eaton, S.; Crossley, M.; Bao, S. Krüppel-like transcription factors: A functional family. Int. J. Biochem. Cell Biol. 2008, 40, 1996–2001. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; MacDougald, O.A. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 2006, 7, 885–896. [Google Scholar] [CrossRef]
- Botella, L.M.; Sánchez-Elsner, T.; Sanz-Rodriguez, F.; Kojima, S.; Shimada, J.; Guerrero-Esteo, M.; Cooreman, M.P.; Ratziu, V.; Langa, C.; Vary, C.P.H.; et al. Transcriptional activation of endoglin and transforming growth factor-β signaling components by cooperative interaction between Sp1 and KLF6: Their potential role in the response to vascular injury. Blood 2002, 100, 4001–4010. [Google Scholar] [CrossRef]
- Zhang, Y.; Qin, L.; Liu, J. Bioinformatics and machine learning approaches to explore key biomarkers in muscle aging linked to adipogenesis. BMC Musculoskelet. Disord. 2025, 26, 285. [Google Scholar] [CrossRef]
- Lee, E.-J.; Kim, M.; Kim, Y.D.; Chung, M.-J.; Elfadl, A.; Ulah, H.M.A.; Park, D.; Lee, S.; Park, H.-S.; Kim, T.-H.; et al. Establishment of stably expandable induced myogenic stem cells by four transcription factors. Cell Death Dis. 2018, 9, 1092. [Google Scholar] [CrossRef]


| Gene Name | Gene ID | Protein ID | CDS ID | Chr | Genomic Location | Exon | AA | Mass (Da) |
|---|---|---|---|---|---|---|---|---|
| KLF1 | ENSBTAG00000006083 | ENSBTAP00000043340 | ENSBTAT00000046003 | 7 | 12,671,273–12,675,059 | 3 | 372 | 39,444 |
| KLF2 | ENSBTAG00000052504 | ENSBTAP00000074931 | ENSBTAT00000108559 | 7 | 6,685,278–6,687,052 | 2 | 78 | 8035 |
| KLF3 | ENSBTAG00000017488 | ENSBTAP00000057281 | ENSBTAT00000085769 | 6 | 57,957,697–57,974,562 | 5 | 346 | 38,943 |
| KLF4 | ENSBTAG00000020355 | ENSBTAP00000072107 | ENSBTAT00000077353 | 8 | 97,175,407–97,178,550 | 3 | 145 | 16,518 |
| KLF5 | ENSBTAG00000002129 | ENSBTAP00000002751 | ENSBTAT00000002751 | 12 | 47,799,775–47,817,130 | 4 | 544 | 50,477 |
| KLF6 | ENSBTAG00000015188 | ENSBTAP00000020207 | ENSBTAT00000020207 | 13 | 44,597,409–44,601,156 | 2 | 309 | 33,456 |
| KLF7 | ENSBTAG00000044097 | ENSBTAP00000066321 | ENSBTAT00000078679 | 2 | 95,426,838–95,439,630 | 2 | 300 | 32,220 |
| KLF8 | ENSBTAG00000005852 | ENSBTAP00000067755 | ENSBTAT00000071858 | X | 93,269,040–93,444,986 | 5 | 248 | 26,365 |
| KLF9 | ENSBTAG00000016229 | ENSBTAP00000021593 | ENSBTAT00000021593 | 8 | 46,587,347–46,611,150 | 2 | 244 | 27,219 |
| KLF10 | ENSBTAG00000014396 | ENSBTAP00000067318 | ENSBTAT00000076027 | 14 | 61,770,518–61,775,159 | 4 | 483 | 53,028 |
| KLF11 | ENSBTAG00000046218 | ENSBTAP00000063288 | ENSBTAT00000080078 | 11 | 87,549,525–87,558,666 | 4 | 523 | 55,735 |
| KLF12 | ENSBTAG00000044007 | ENSBTAP00000053615 | ENSBTAT00000061307 | 12 | 48,526,715–48,823,511 | 7 | 408 | 44,613 |
| KLF13 | ENSBTAG00000051090 | ENSBTAP00000057849 | ENSBTAT00000079732 | 21 | 27,672,129–27,709,397 | 2 | 296 | 31,917 |
| KLF15 | ENSBTAG00000008313 | ENSBTAP00000063598 | ENSBTAT00000125115 | 22 | 60,616,031–60,648,782 | 2 | 394 | 41,542 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, F.; Zhou, L.; Guo, L.; Chang, C.; Dan, D.; Bao, Y.; Han, G.; Gu, M.; Zhu, L.; Na, R.; et al. Comprehensive Identification of the Bovine KLF Gene Family and Its Functional Regulation in Muscle Development: Insights from Single-Nuclei Transcriptomics. Animals 2025, 15, 2930. https://doi.org/10.3390/ani15192930
Ma F, Zhou L, Guo L, Chang C, Dan D, Bao Y, Han G, Gu M, Zhu L, Na R, et al. Comprehensive Identification of the Bovine KLF Gene Family and Its Functional Regulation in Muscle Development: Insights from Single-Nuclei Transcriptomics. Animals. 2025; 15(19):2930. https://doi.org/10.3390/ani15192930
Chicago/Turabian StyleMa, Fengying, Le Zhou, Lili Guo, Chencheng Chang, Dan Dan, Yanchun Bao, Guiting Han, Mingjuan Gu, Lin Zhu, Risu Na, and et al. 2025. "Comprehensive Identification of the Bovine KLF Gene Family and Its Functional Regulation in Muscle Development: Insights from Single-Nuclei Transcriptomics" Animals 15, no. 19: 2930. https://doi.org/10.3390/ani15192930
APA StyleMa, F., Zhou, L., Guo, L., Chang, C., Dan, D., Bao, Y., Han, G., Gu, M., Zhu, L., Na, R., Shi, C., Zhang, J., & Zhang, W. (2025). Comprehensive Identification of the Bovine KLF Gene Family and Its Functional Regulation in Muscle Development: Insights from Single-Nuclei Transcriptomics. Animals, 15(19), 2930. https://doi.org/10.3390/ani15192930





