Identification and Pathway Analysis of SNP Loci Affecting Abdominal Fat Deposition in Broilers
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. RNA-Seq-Based SNP Detection and Quality Control
2.2.2. Screening SNPs with Combined Genome Resequencing Data
2.2.3. Screening for SNPs Potentially Affecting the Expression of Genes Related to Abdominal Fat Deposition
2.2.4. Prediction of SNP-Regulated Gene Expression Pathways
2.3. Statistical and Bioinformatics Analysis
2.3.1. Principal Component Analysis
2.3.2. Additional Statistical Methods
3. Results
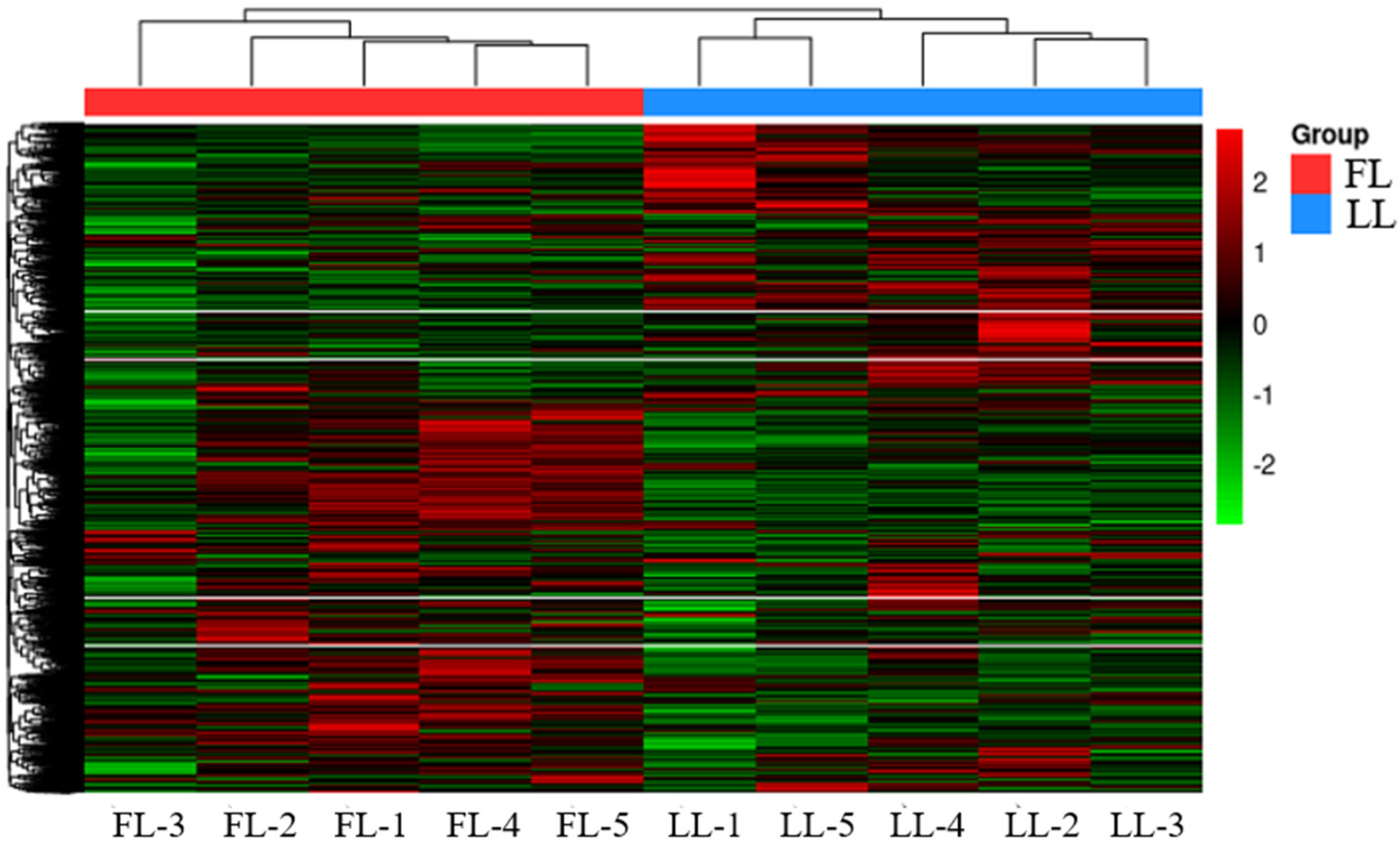
3.1. Screening Differential SNPs Between High- and Low-Fat Lines in Abdominal Adipose Tissue Based on Transcriptomic Data
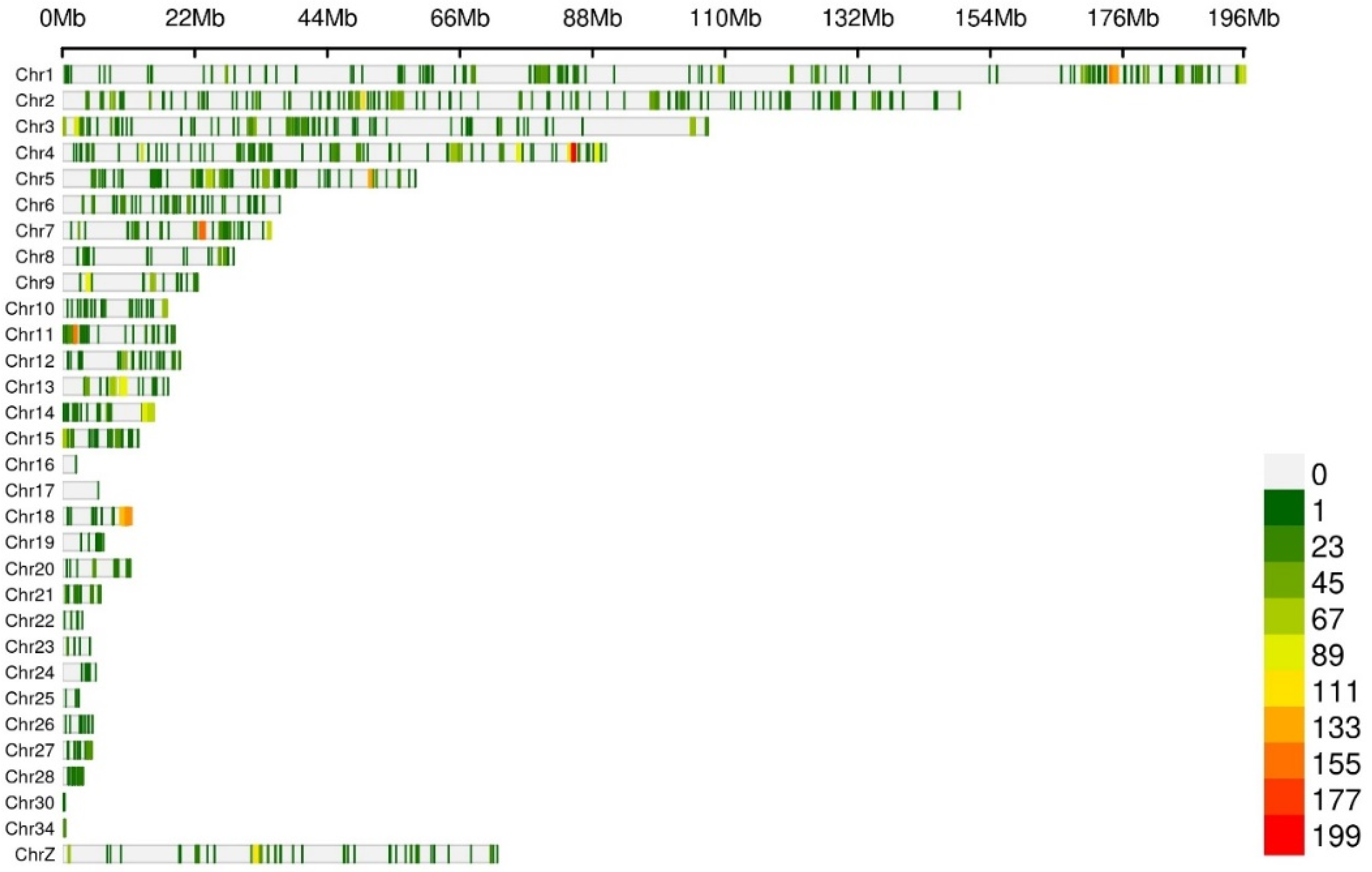
3.2. Identification of SNPs Associated with Abdominal Fat Deposition in Broilers Based on Genome Resequencing Data
3.2.1. Allele Frequency Analysis
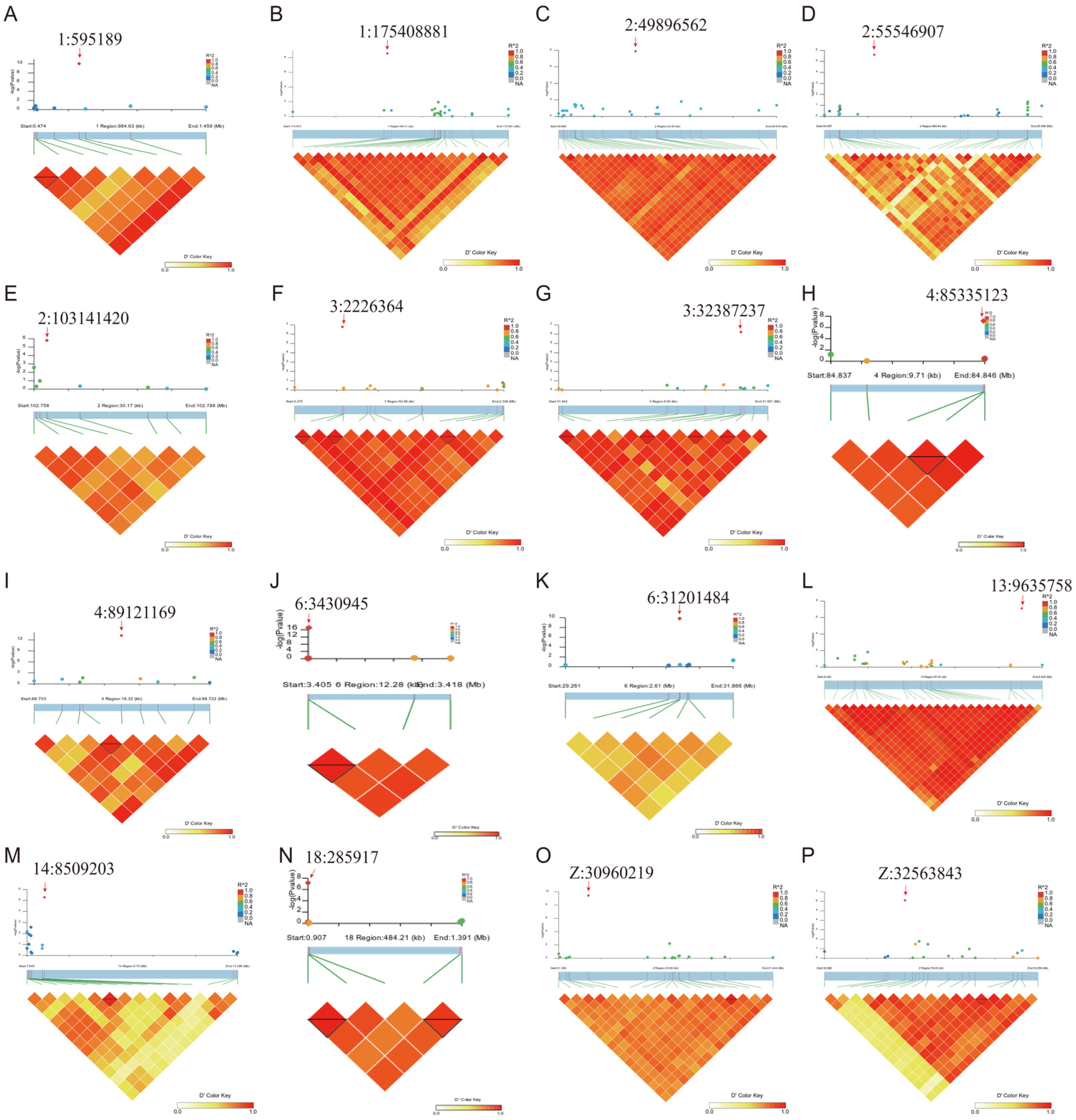
3.2.2. Genome-Wide Association Analysis (GWAS) of Abdominal Fat Weight
3.2.3. Linkage Disequilibrium (LD) Analysis
3.3. Identification of Target Genes Regulated by SNPs Associated with Abdominal Fat Deposition in Broilers
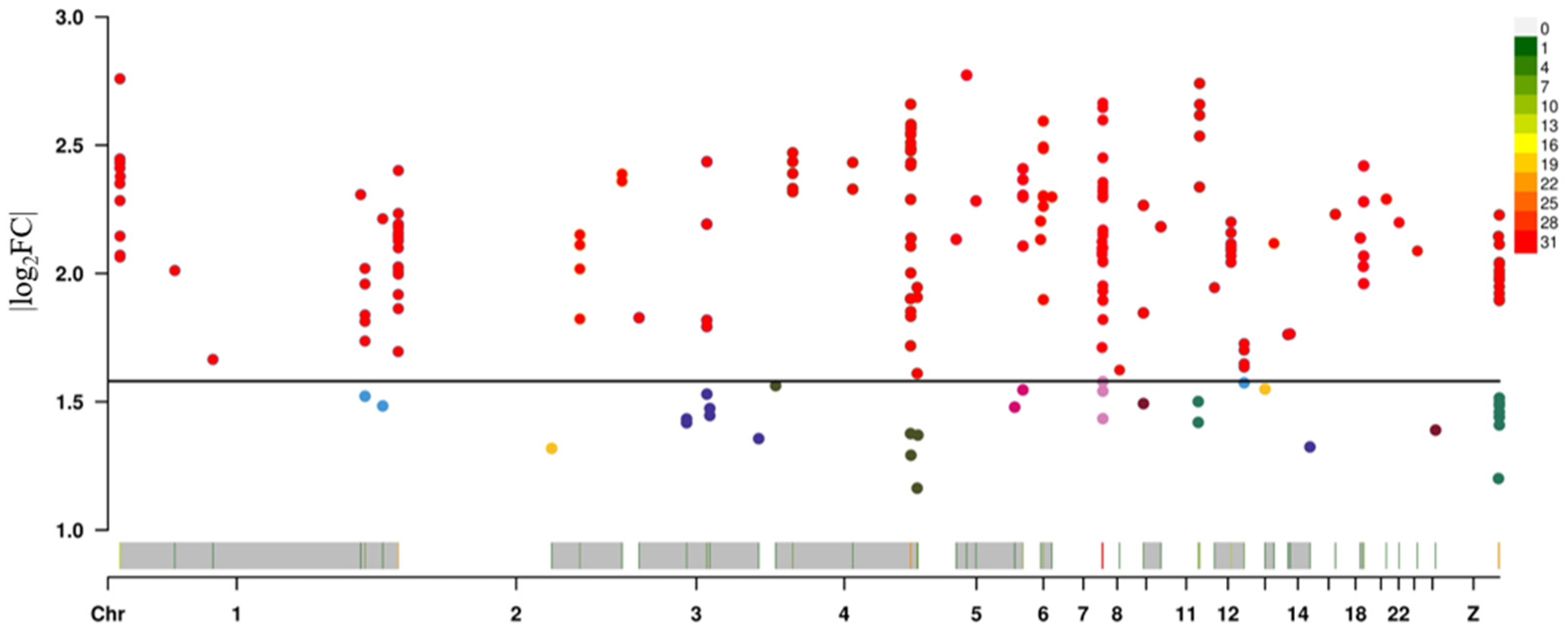
3.3.1. Identification of Differentially Expressed Genes Between Abdominal Adipose Tissues of Broilers from High- and Low-Fat Lines
3.3.2. SNPs Affect Broiler Abdominal Fat Deposition by Regulating Their Own Gene Expression
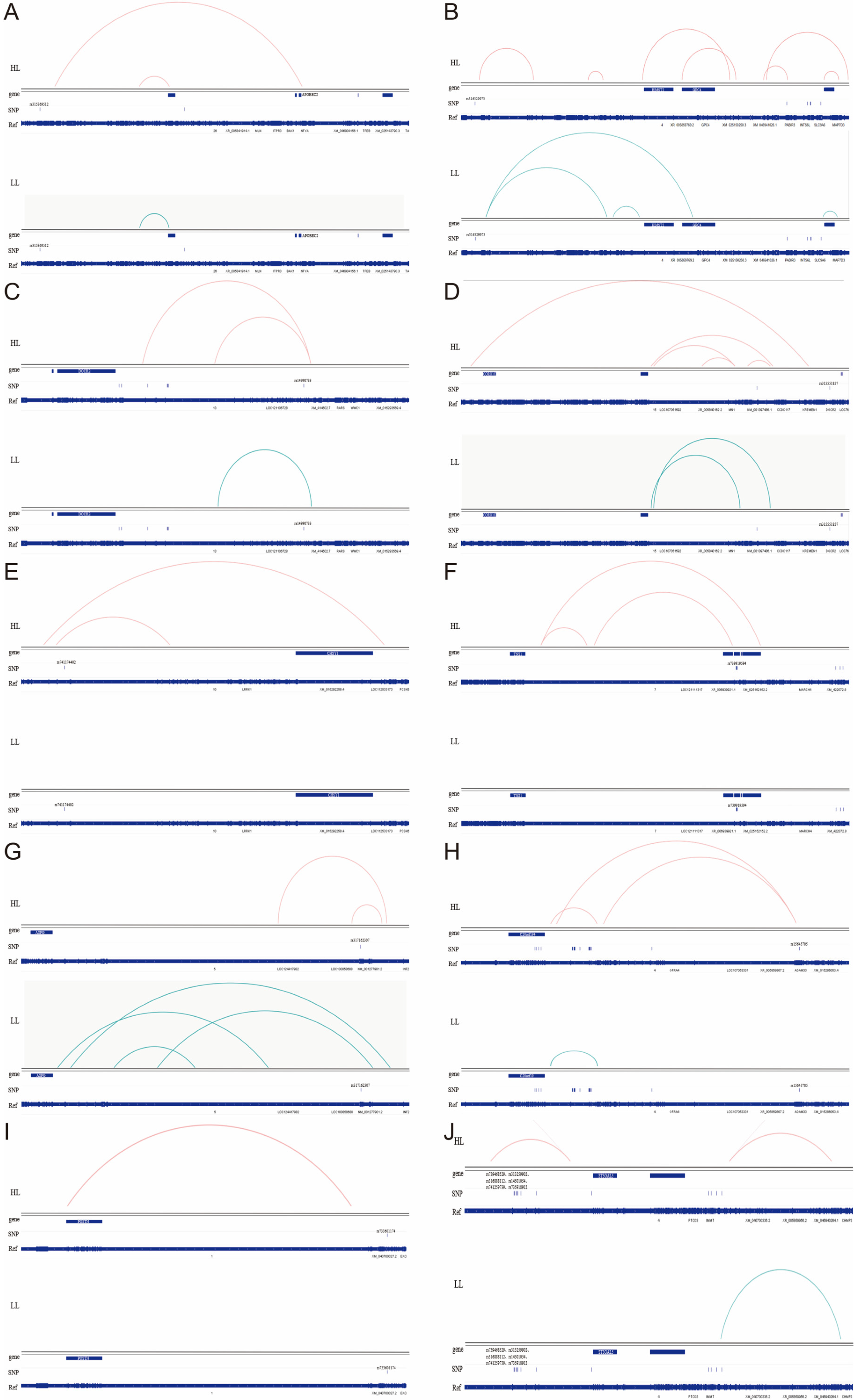
3.3.3. SNPs Affect Broiler Abdominal Fat Deposition Through Long-Distance Regulation of Target Gene Expression
3.4. Bioinformatics Analysis of the Pathways by Which SNPs Regulate Target Gene Expression
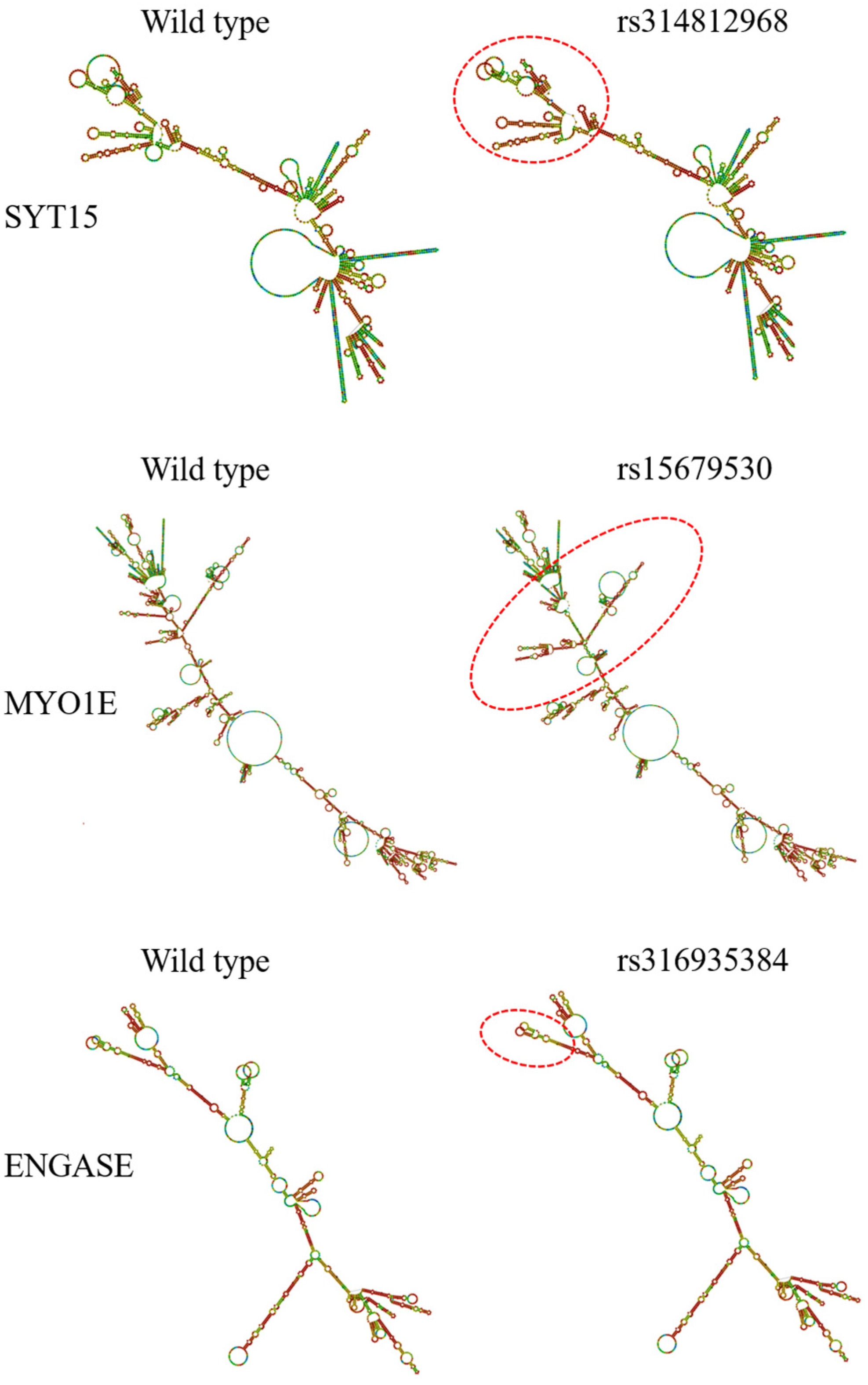
3.4.1. SNPs Regulate Their Own Gene Expression by Altering Protein Expression
Codon Translation Rate Analysis
Analysis of mRNA Secondary Structure
3.4.2. SNPs Regulate Their Own Gene Expression by Modulating Transcription Factor Binding Site Activity
3.4.3. SNPs Regulate Distal Gene Expression by Modulating Transcription Factor Binding Site Activity
3.5. Validation of the Effects of SNPs on Phenotype
4. Discussion
4.1. Identification of SNP Loci Associated with Abdominal Fat Deposition in Broiler Chickens by Integrating Transcriptomic and Genomic Data
4.2. Identification of Target Genes for SNP Regulation Related to Abdominal Fat Deposition in Broiler Chickens
4.3. Pathways of SNPs Regulating Target Gene Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, S.; Wang, Y.; Wang, L.; Shi, Z.; Ou, X.; Wu, D.; Zhang, X.; Hu, H.; Yuan, J.; Wang, W.; et al. RNA-Seq Reveals Differentially Expressed Genes Affecting Polyunsaturated Fatty Acids Percentage in the Huangshan Black Chicken Population. PLoS ONE 2018, 13, e0195132. [Google Scholar] [CrossRef]
- Leclercq, B.; Saadoun, A.J. Selecting Broilers for Low or High Abdominal Fat: Comparison of Energy Metabolism of the Lean and Fat Lines. Poult. Sci. 1982, 61, 1799–1803. [Google Scholar] [CrossRef]
- Lilburn, M.S.; Leach, R.M.; Buss, E.G.; Martin, R.J. The Developmental Characteristics of Two Strains of Chickens Selected for Differences in Mature Abdominal Fat Pad Size. Growth 1982, 46, 171–181. [Google Scholar]
- Griffin, H.D.; Whitehead, C.C. Plasma Lipoprotein Concentration as an Indicator of Fatness in Broilers: Development and Use of a Simple Assay for Plasma Very Low Density Lipoproteins. Br. Poult. Sci. 1982, 23, 307–313. [Google Scholar] [CrossRef]
- Whitehead, C.C.; Griffin, H.D. Development of Divergent Lines of Lean and Fat Broilers Using Plasma Very Low Density Lipoprotein Concentration as Selection Criterion: The First Three Generations. Br. Poult. Sci. 1984, 25, 573–582. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, R.; Lu, X.; Hu, Y.; Zhao, G.; Zheng, M.; Chen, J.; Wang, H.; Wen, J. Associations of Polymorphisms in Four Candidate Genes with Carcass and/or Meat-Quality Traits in Two Meat-Type Chicken Lines. Anim. Biotechnol. 2013, 24, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Luo, N.; Shu, J.; Yuan, X.; Jin, Y.; Cui, H.; Zhao, G.; Wen, J. Differential Regulation of Intramuscular Fat and Abdominal Fat Deposition in Chickens. BMC Genomics 2022, 23, 308. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhong, H.; Lin, S.; Liang, L.; Ye, S.; Xu, Z.; Ji, C.; Zhang, Z.; Zhang, D.; Zhang, X. Polymorphisms of AMY1A Gene and Their Association with Growth, Carcass Traits and Feed Intake Efficiency in Chickens. Genomics 2021, 113, 583–594. [Google Scholar] [CrossRef]
- Cheng, B.; Zhang, H.; Liu, C.; Chen, X.; Chen, Y.; Sun, Y.; Leng, L.; Li, Y.; Luan, P.; Li, H. Functional Intronic Variant in the Retinoblastoma 1 Gene Underlies Broiler Chicken Adiposity by Altering Nuclear Factor-kB and SRY-Related HMG Box Protein 2 Binding Sites. J. Agric. Food Chem. 2019, 67, 9727–9737. [Google Scholar] [CrossRef] [PubMed]
- Trevisoli, P.A.; Moreira, G.C.M.; Boschiero, C.; Cesar, A.S.M.; Petrini, J.; Margarido, G.R.A.; Ledur, M.C.; Mourão, G.B.; Garrick, D.; Coutinho, L.L. A Missense Mutation in the MYBPH Gene Is Associated with Abdominal Fat Traits in Meat-Type Chickens. Front. Genet. 2021, 12, 698163. [Google Scholar] [CrossRef] [PubMed]
- Goswami, A.M. Structural Modeling and in Silico Analysis of Non-Synonymous Single Nucleotide Polymorphisms of Human 3β-Hydroxysteroid Dehydrogenase Type 2. Meta Gene 2015, 5, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Dávila, S.G.; Gil, M.G.; Resino-Talaván, P.; Campo, J.L. Association between Polymorphism in the Melanocortin 1 Receptor Gene and E Locus Plumage Color Phenotype. Poult. Sci. 2014, 93, 1089–1096. [Google Scholar] [CrossRef] [PubMed]
- Faheem, M.; Zhang, C.J.; Morris, M.N.; Pleiss, J.; Oelschlaeger, P. Role of Synonymous Mutations in the Evolution of TEM β-Lactamase Genes. Antimicrob. Agents Chemother. 2021, 65, e00018–e00021. [Google Scholar] [CrossRef]
- Zhang, D.; Xia, J. Somatic Synonymous Mutations in Regulatory Elements Contribute to the Genetic Aetiology of Melanoma. BMC Med. Genomics 2020, 13, 43. [Google Scholar] [CrossRef]
- Li, Q.; Li, J.; Yu, C.-P.; Chang, S.; Xie, L.-L.; Wang, S. Synonymous Mutations That Regulate Translation Speed Might Play a Non-Negligible Role in Liver Cancer Development. BMC Cancer 2021, 21, 388. [Google Scholar] [CrossRef]
- Goddard, K.A.; Hopkins, P.J.; Hall, J.M.; Witte, J.S. Linkage Disequilibrium and Allele-Frequency Distributions for 114 Single-Nucleotide Polymorphisms in Five Populations. Am. J. Hum. Genet. 2000, 66, 216–234. [Google Scholar] [CrossRef]
- Orozco, G.; Schoenfelder, S.; Walker, N.; Eyre, S.; Fraser, P. 3D Genome Organization Links Non-Coding Disease-Associated Variants to Genes. Front. Cell Dev. Biol. 2022, 10, 995388. [Google Scholar] [CrossRef]
- Guo, L.; Sun, B.; Shang, Z.; Leng, L.; Wang, Y.; Wang, N.; Li, H. Comparison of Adipose Tissue Cellularity in Chicken Lines Divergently Selected for Fatness. Poult. Sci. 2011, 90, 2024–2034. [Google Scholar] [CrossRef]
- National Research Council. Nutrient Requirements of Poultry; 9th Revised Ed. The National Academies Press: Washington, DC, USA, 1994. [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Sirén, J.; Välimäki, N.; Mäkinen, V. Indexing Graphs for Path Queries with Applications in Genome Research. IEEE/ACM Trans. Comput. Biol. Bioinformatics 2014, 11, 375–388. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A Package for the next Level of Genome-Wide Association Studies with Both Individuals and Markers in the Millions. GigaScience 2019, 8, giy154. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-Level Expression Analysis of RNA-Seq Experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Blom, N.; Gammeltoft, S.; Brunak, S. Sequence and Structure-Based Prediction of Eukaryotic Protein Phosphorylation Sites. J. Mol. Biol. 1999, 294, 1351–1362. [Google Scholar] [CrossRef] [PubMed]
- Blom, N.; Sicheritz-Pontén, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of Post-Translational Glycosylation and Phosphorylation of Proteins from the Amino Acid Sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar] [CrossRef]
- Gupta, R.; Brunak, S. Prediction of Glycosylation across the Human Proteome and the Correlation to Protein Function. Pac. Symp. Biocomput. 2002, 7, 310–322. [Google Scholar] [CrossRef]
- Tsunoda, T.; Takagi, T. Estimating Transcription Factor Bindability on DNA. Bioinformatics 1999, 15, 622–630. [Google Scholar] [CrossRef]
- Jehl, F.; Degalez, F.; Bernard, M.; Lecerf, F.; Lagoutte, L.; Désert, C.; Coulée, M.; Bouchez, O.; Leroux, S.; Abasht, B.; et al. RNA-Seq Data for Reliable SNP Detection and Genotype Calling: Interest for Coding Variant Characterization and Cis-Regulation Analysis by Allele-Specific Expression in Livestock Species. Front. Genet. 2021, 12, 655707. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, R.; Liu, X.; Shi, G.; Liu, H.; Wang, L.; Hou, X.; Shi, L.; Wang, L.; Zhang, L. Integrating Genome-Wide Association Study with RNA-Seq Revealed DBI as a Good Candidate Gene for Intramuscular Fat Content in Beijing Black Pigs. Anim. Genet. 2023, 54, 24–34. [Google Scholar] [CrossRef]
- Jiang, Y.; Xie, M.; Chen, W.; Talbot, R.; Maddox, J.F.; Faraut, T.; Wu, C.; Muzny, D.M.; Li, Y.; Zhang, W.; et al. The Sheep Genome Illuminates Biology of the Rumen and Lipid Metabolism. Science 2014, 344, 1168–1173. [Google Scholar] [CrossRef]
- Ullah, M.; Sittinger, M.; Ringe, J. Extracellular Matrix of Adipogenically Differentiated Mesenchymal Stem Cells Reveals a Network of Collagen Filaments, Mostly Interwoven by Hexagonal Structural Units. Matrix Biol. J. Int. Soc. Matrix Biol. 2013, 32, 452–465. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, C.; Sun, Y.; Li, Y.; Kang, L.; Jiang, Y. Dynamic Transcriptome and DNA Methylome Analyses on Longissimus Dorsi to Identify Genes Underlying Intramuscular Fat Content in Pigs. BMC Genomics 2017, 18, 780. [Google Scholar] [CrossRef]
- Cai, C.; Li, M.; Zhang, Y.; Meng, S.; Yang, Y.; Gao, P.; Guo, X.; Cao, G.; Li, B. Comparative Transcriptome Analyses of Longissimus Thoracis between Pig Breeds Differing in Muscle Characteristics. Front. Genet. 2020, 11, 526309. [Google Scholar] [CrossRef]
- Chao, C.C.-K.; Hung, F.-C.; Chao, J.J. Gas7 Is Required for Mesenchymal Stem Cell-Derived Bone Development. Stem Cells Int. 2013, 2013, 137010. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Wang, R.; Wang, R.; Tian, Y.; Shao, C.; Jia, X.; Chen, S. The Integrated Analysis of RNA-Seq and microRNA-Seq Depicts miRNA-mRNA Networks Involved in Japanese Flounder (Paralichthys olivaceus) Albinism. PLoS ONE 2017, 12, e0181761. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Guo, W.; Liang, S.; Lu, H.; Sun, W.; Ren, X.; Sun, Q.; Yang, X. Systematic Analysis of the Regulatory Roles of microRNAs in Postnatal Maturation and Metergasis of Liver of Breeder Cocks. Sci. Rep. 2018, 8, 61. [Google Scholar] [CrossRef]
- Xu, C.; Liu, Q.; Zhou, J.; Xie, M.; Feng, J.; Jiang, T. Quantifying Functional Impact of Non-Coding Variants with Multi-Task Bayesian Neural Network. Bioinformatics 2020, 36, 1397–1404. [Google Scholar] [CrossRef]
- Dominguez-Alonso, S.; Carracedo, A.; Rodriguez-Fontenla, C. The Non-Coding Genome in Autism Spectrum Disorders. Eur. J. Med. Genet. 2023, 66, 104752. [Google Scholar] [CrossRef] [PubMed]
- Kikutake, C.; Yoshihara, M.; Suyama, M. Pan-Cancer Analysis of Non-Coding Recurrent Mutations and Their Possible Involvement in Cancer Pathogenesis. NAR Cancer 2021, 3, zcab008. [Google Scholar] [CrossRef]
- Li, B.; Yang, J.; Gong, Y.; Xiao, Y.; Chen, W.; Zeng, Q.; Xu, K.; Duan, Y.; Ma, H. Effects of Age on Subcutaneous Adipose Tissue Proteins in Chinese Indigenous Ningxiang Pig by TMT-Labeled Quantitative Proteomics. J. Proteomics 2022, 265, 104650. [Google Scholar] [CrossRef]
- Ma, W.-W.; Ding, B.-J.; Yuan, L.-H.; Zhao, L.; Yu, H.-L.; Xi, Y.; Xiao, R. Neurocalcin-Delta: A Potential Memory-Related Factor in Hippocampus of Obese Rats Induced by High-Fat Diet. Afr. Health Sci. 2017, 17, 1211–1221. [Google Scholar] [CrossRef]
- Lu, H.; Ye, Z.; Zhai, Y.; Wang, L.; Liu, Y.; Wang, J.; Zhang, W.; Luo, W.; Lu, Z.; Chen, J. QKI Regulates Adipose Tissue Metabolism by Acting as a Brake on Thermogenesis and Promoting Obesity. EMBO Rep. 2020, 21, e47929. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; Yang, M.; Quan, J.; Li, S.; Zhuang, Z.; Zhou, S.; Zheng, E.; Hong, L.; Li, Z.; Cai, G.; et al. Single-Locus and Multi-Locus Genome-Wide Association Studies for Intramuscular Fat in Duroc Pigs. Front. Genet. 2019, 10, 619. [Google Scholar] [CrossRef]
- Heidaritabar, M.; Bink, M.C.A.M.; Dervishi, E.; Charagu, P.; Huisman, A.; Plastow, G.S. Genome-Wide Association Studies for Additive and Dominance Effects for Body Composition Traits in Commercial Crossbred Piétrain Pigs. J. Anim. Breed. Genet. 2023, 140, 413–430. [Google Scholar] [CrossRef] [PubMed]
- Minchenko, O.H.; Bashta, Y.M.; Minchenko, D.O.; Ratushna, O.O. Glucose Tolerance in Obese Men Is Associated with Dysregulation of Some Angiogenesis-Related Gene Expressions in Subcutaneous Adipose Tissue. Fiziol. Zh. 2016, 62, 12–23. [Google Scholar] [CrossRef]
- De la Cruz-Color, L.; Hernández-Nazará, Z.H.; Maldonado-González, M.; Navarro-Muñíz, E.; Domínguez-Rosales, J.A.; Torres-Baranda, J.R.; Ruelas-Cinco, E.D.C.; Ramírez-Meza, S.M.; Ruíz-Madrigal, B. Association of the PNPLA2, SCD1 and Leptin Expression with Fat Distribution in Liver and Adipose Tissue from Obese Subjects. Exp. Clin. Endocrinol. Diabetes. 2020, 128, 715–722. [Google Scholar] [CrossRef]
- Xiao, Z.; Chu, Y.; Qin, W. IGFBP5 Modulates Lipid Metabolism and Insulin Sensitivity through Activating AMPK Pathway in Non-Alcoholic Fatty Liver Disease. Life Sci. 2020, 256, 117997. [Google Scholar] [CrossRef]
- Vander Velden, J.W.; Osborne, D.M. Prolonged Diet-Induced Obesity Modifies DNA Methylation and Gene Expression in the Hippocampus. Neurosci. Lett. 2022, 780, 136656. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, A.; Kitamoto, T.; Nakamura, T.; Matsuo, T.; Nakata, Y.; Hyogo, H.; Ochi, H.; Kamohara, S.; Miyatake, N.; Kotani, K.; et al. CDH13 Polymorphisms Are Associated with Adiponectin Levels and Metabolic Syndrome Traits Independently of Visceral Fat Mass. J. Atheroscler. Thromb. 2016, 23, 309–319. [Google Scholar] [CrossRef]
- Zhang, Q.-H.; Cui, X.-Y.; Wang, D.; Jin, Y.; Guan, Y.-X. Anti-Obesity Effect of Escin: A Study on High-Fat Diet-Induced Obese Mice. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 7797–7812. [Google Scholar] [CrossRef]
- Ji, S.; Sun, J.; Bian, C.; Huang, X.; Chang, Z.; Yang, M.; Lu, R.-H.; Ji, H. cAMP-Dependent Protein Kinase a in Grass Carp Ctenopharyngodon Idella: Molecular Characterization, Gene Structure, Tissue Distribution and mRNA Expression in Endoplasmic Reticulum Stress-Induced Adipocyte Lipolysis. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2020, 250, 110479. [Google Scholar] [CrossRef]
- Ten Klooster, J.P.; Sotiriou, A.; Boeren, S.; Vaessen, S.; Vervoort, J.; Pieters, R. Type 2 Diabetes-Related Proteins Derived from an in Vitro Model of Inflamed Fat Tissue. Arch. Biochem. Biophys. 2018, 644, 81–92. [Google Scholar] [CrossRef]
- Akter, F.; Tsuyama, T.; Yoshizawa, T.; Sobuz, S.U.; Yamagata, K. SIRT7 Regulates Lipogenesis in Adipocytes through Deacetylation of PPARγ2. J. Diabetes Investig. 2021, 12, 1765–1774. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, S.; Guo, Y.; Gao, L.; Zhang, H.; Chen, W.; Huang, Y. Chicken CDS2 Isoforms Presented Distinct Spatio-Temporal Expression Pattern and Regulated by Insulin in a Breed-Specific Manner. Poult. Sci. 2022, 101, 101893. [Google Scholar] [CrossRef]
- Choi, J.H.; Lee, H. Histone Demethylase KDM4D Cooperates with NFIB and MLL1 Complex to Regulate Adipogenic Differentiation of C3H10T1/2 Mesenchymal Stem Cells. Sci. Rep. 2020, 10, 3050. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Spiegelman, B.M. What We Talk about When We Talk about Fat. Cell 2014, 156, 20–44. [Google Scholar] [CrossRef] [PubMed]
- Tontonoz, P.; Spiegelman, B.M. Fat and beyond: The Diverse Biology of PPARgamma. Annu. Rev. Biochem. 2008, 77, 289–312. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, Y.; Zhou, X.; Chen, D.; Ouyang, G.; Liu, Y.; Cui, D. Periostin Deficiency Attenuates Lipopolysaccharide- and Obesity-Induced Adipose Tissue Fibrosis. FEBS Lett. 2021, 595, 2099–2112. [Google Scholar] [CrossRef]
- Liu, L.; Gu, H.; Zhao, Y.; An, L.; Yang, J. Glypican 4 May Be Involved in the Adipose Tissue Redistribution in High-Fat Feeding C57BL/6J Mice with Peroxisome Proliferators-Activated Receptor γ Agonist Rosiglitazone Treatment. Exp. Ther. Med. 2014, 8, 1813–1818. [Google Scholar] [CrossRef]
- Wentworth, J.M.; Naselli, G.; Ngui, K.; Smyth, G.K.; Liu, R.; O’Brien, P.E.; Bruce, C.; Weir, J.; Cinel, M.; Meikle, P.J.; et al. GM3 Ganglioside and Phosphatidylethanolamine-Containing Lipids Are Adipose Tissue Markers of Insulin Resistance in Obese Women. Int. J. Obes. 2016, 40, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Li, F.; Xu, Z.; Yin, A.; Yin, H.; Li, C.; Chen, S.-Y. DOCK2 Deficiency Mitigates HFD-Induced Obesity by Reducing Adipose Tissue Inflammation and Increasing Energy Expenditure. J. Lipid Res. 2017, 58, 1777–1784. [Google Scholar] [CrossRef] [PubMed]
- Joshi, H.; Vastrad, B.; Joshi, N.; Vastrad, C.; Tengli, A.; Kotturshetti, I. Identification of Key Pathways and Genes in Obesity Using Bioinformatics Analysis and Molecular Docking Studies. Front. Endocrinol. 2021, 12, 628907. [Google Scholar] [CrossRef]
- Hachero-Cruzado, I.; Rodríguez-Rua, A.; Román-Padilla, J.; Ponce, M.; Fernández-Díaz, C.; Manchado, M. Characterization of the Genomic Responses in Early Senegalese Sole Larvae Fed Diets with Different Dietary Triacylglycerol and Total Lipids Levels. Comp. Biochem. Physiol. D Genom. Proteom. 2014, 12, 61–73. [Google Scholar] [CrossRef]
- Chakraborty, M.; Jandhyam, H.; Basak, S.K.; Das, S.; Alone, D.P. Intergenic Variants, Rs1200114 and Rs1200108 Are Genetically Associated along with a Decreased ATP1B1 Expression in Fuchs Endothelial Corneal Dystrophy. Exp. Eye Res. 2023, 228, 109403. [Google Scholar] [CrossRef]
- Chen, H.; Yang, P.; Yang, D.; Wang, D.; Lu, M.; Li, Y.; Zhong, Z.; Zhang, J.; Zeng, Z.; Liu, Z.; et al. The PER3rs772027021 SNP Induces Pigmentation Phenotypes of Dyschromatosis Universalis Hereditaria. J. Mol. Med. 2023, 101, 279–294. [Google Scholar] [CrossRef]
- Wang, N.-N.; Zhang, Y.; Jiang, F.; Zhu, D.-L.; Di, C.-X.; Hu, S.-Y.; Chen, X.-F.; Zhi, L.-Q.; Rong, Y.; Ke, X.; et al. Enhancer Variants on Chromosome 2p14 Regulating SPRED2 and ACTR2 Act as a Signal Amplifier to Protect against Rheumatoid Arthritis. Am. J. Hum. Genet. 2023, 110, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Stülke, J. More than Just Activity Control: Phosphorylation May Control All Aspects of a Protein’s Properties. Mol. Microbiol. 2010, 77, 273–275. [Google Scholar] [CrossRef]
- Crowley, S.T.; Brownlee, M.; Edelstein, D.; Satriano, J.A.; Mori, T.; Singhal, P.C.; Schlondorff, D.O. Effects of Nonenzymatic Glycosylation of Mesangial Matrix on Proliferation of Mesangial Cells. Diabetes 1991, 40, 540–547. [Google Scholar] [CrossRef]
- Tang, S.; Li, J.; Huang, G.; Yan, L. Predicting Protein Surface Property with Its Surface Hydrophobicity. Protein Pept. Lett. 2021, 28, 938–944. [Google Scholar] [CrossRef]
- Ji, Y.-Y.; Li, Y.-Q. The Role of Secondary Structure in Protein Structure Selection. Eur. Phys. J.E. Soft Matter 2010, 32, 103–107. [Google Scholar] [CrossRef]
- O’Brien, E.P.; Ciryam, P.; Vendruscolo, M.; Dobson, C.M. Understanding the Influence of Codon Translation Rates on Cotranslational Protein Folding. Acc. Chem. Res. 2014, 47, 1536–1544. [Google Scholar] [CrossRef]
- Faure, G.; Ogurtsov, A.Y.; Shabalina, S.A.; Koonin, E.V. Adaptation of mRNA Structure to Control Protein Folding. RNA Biol. 2017, 14, 1649–1654. [Google Scholar] [CrossRef]
- Šedová, L.; Pravenec, M.; Křenová, D.; Kazdová, L.; Zídek, V.; Krupková, M.; Liška, F.; Křen, V.; Šeda, O. Isolation of a Genomic Region Affecting Most Components of Metabolic Syndrome in a Chromosome-16 Congenic Rat Model. PLoS ONE 2016, 11, e0152708. [Google Scholar] [CrossRef]
- Sheth, N.; Roca, X.; Hastings, M.L.; Roeder, T.; Krainer, A.R.; Sachidanandam, R. Comprehensive Splice-Site Analysis Using Comparative Genomics. Nucleic Acids Res. 2006, 34, 3955–3967. [Google Scholar] [CrossRef]
- Wong, E.S.; Thybert, D.; Schmitt, B.M.; Stefflova, K.; Odom, D.T.; Flicek, P. Decoupling of Evolutionary Changes in Transcription Factor Binding and Gene Expression in Mammals. Genome Res. 2015, 25, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Algarra, D.; Souaid, C.; Singh, H.; Dao, L.T.M.; Hussain, S.; Medina-Rivera, A.; Ramirez-Navarro, L.; Castro-Mondragon, J.A.; Sadouni, N.; Charbonnier, G.; et al. Epromoters Function as a Hub to Recruit Key Transcription Factors Required for the Inflammatory Response. Nat. Commun. 2021, 12, 6660. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Sun, W.; Han, H.; Chu, W.; Zhang, L.; Chen, J. miR-130a Regulates Differential Lipid Accumulation between Intramuscular and Subcutaneous Adipose Tissues of Pigs via Suppressing PPARG Expression. Gene 2017, 636, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.P.; Kim, D.-Y.; Lee, Y.-G.; Lee, Y.-S.; Truong, X.T.; Lee, J.-H.; Song, D.-K.; Kwon, T.K.; Park, S.-H.; Jung, C.H.; et al. SREBP-1c Impairs ULK1 Sulfhydration-Mediated Autophagic Flux to Promote Hepatic Steatosis in High-Fat-Diet-Fed Mice. Mol. Cell 2021, 81, 3820–3832.e7. [Google Scholar] [CrossRef]
- Gui, L.; Raza, S.H.A.; Ma, B.; Easa, A.A.; Althobaiti, F.; Shukry, M.; Alotaibi, M.A.; Al Hazani, T.M.I.; Dawood, M.A.O.; Khan, R.; et al. CEBPβ Binding Directly to the Promoter Region Drives CEBPɑ Transcription and Improves FABP4 Transcriptional Activity in Adipose Tissue of Yak (Bos grunniens). Res. Vet. Sci. 2021, 141, 174–179. [Google Scholar] [CrossRef]
- Lavrador, M.S.F.; Afonso, M.S.; Cintra, D.E.; Koike, M.; Nunes, V.S.; Demasi, M.; Lin, C.J.; Beda, L.M.M.; Gioielli, L.A.; Bombo, R. de P.A.; et al. Interesterified Fats Induce Deleterious Effects on Adipose Tissue and Liver in LDLr-KO Mice. Nutrients 2019, 11, 466. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Wang, Y.; Zhang, Y.; Zhu, J.; Lin, Y. RXRα Cooperates with KLF8 to Promote the Differentiation of Intramuscular Preadipocytes in Goat. Anim. Biotechnol. 2021, 32, 580–590. [Google Scholar] [CrossRef]
- Hwang, J.S.; Lee, S.B.; Choi, M.-J.; Kim, J.T.; Seo, H.G. Anti-Adipogenic Effect of a Turmeric Extract-Loaded Nanoemulsion in 3T3-L1 Preadipocytes and High Fat Diet-Fed Mice. Food Sci. Technol. 2019, 39, 439–447. [Google Scholar] [CrossRef]
- Yoon, Y.-S.; Liu, W.; Van de Velde, S.; Matsumura, S.; Wiater, E.; Huang, L.; Montminy, M. Activation of the Adipocyte CREB/CRTC Pathway in Obesity. Commun. Biol. 2021, 4, 1214. [Google Scholar] [CrossRef]
- Jin, J.-H.; Wen, D.-T.; Chen, Y.-L.; Hou, W.-Q. Muscle FOXO-Specific Overexpression and Endurance Exercise Protect Skeletal Muscle and Heart from Defects Caused by a High-Fat Diet in Young Drosophila. Front. Biosci. Landmark 2023, 28, 16. [Google Scholar] [CrossRef]
- Zhu, D.-L.; Chen, X.-F.; Zhou, X.-R.; Hu, S.-Y.; Tuo, X.-M.; Hao, R.-H.; Dong, S.-S.; Jiang, F.; Rong, Y.; Yang, T.-L.; et al. An Osteoporosis Susceptibility Allele at 11p15 Regulates SOX6 Expression by Modulating TCF4 Chromatin Binding. J. Bone Miner. Res. 2022, 37, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-Y.; Han, J.-X.; Zhang, J.; Jiang, P.; Shen, C.; Guo, F.; Tang, J.; Yan, T.; Tian, X.; Zhu, X.; et al. A 16q22.1 Variant Confers Susceptibility to Colorectal Cancer as a Distal Regulator of ZFP90. Oncogene 2020, 39, 1347–1360. [Google Scholar] [CrossRef] [PubMed]





| Chr | rs | WT | MT | FL_1 | FL_2 | FL_3 | FL_4 | FL_5 | LL_1 | LL_2 | LL_3 | LL_4 | LL_5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 337683 | A | G | G/G | G/A | G/G | G/G | G/G | A/A | A/A | A/A | A/A | A/A |
| 2 | 3908887 | T | C | T/T | T/T | T/T | T/T | T/T | C/C | C/C | C/C | C/C | C/C |
| 3 | 2127089 | C | A | A/A | A/A | A/A | A/A | A/A | C/C | C/C | C/C | C/C | C/C |
| 4 | 31488349 | C | T | T/T | T/T | T/T | T/T | T/T | C/C | C/C | C/T | C/C | C/C |
| 5 | 9960422 | G | A | G/G | G/G | G/G | G/G | G/G | A/A | A/A | A/A | A/A | A/A |
| 6 | 12977377 | G | T | T/T | T/T | T/T | T/T | T/T | G/G | G/G | G/G | G/G | G/T |
| 7 | 2716815 | A | G | G/G | G/G | G/G | A/G | G/G | A/A | A/A | A/A | A/A | A/A |
| 8 | 27654376 | G | T | G/G | G/G | G/G | G/G | G/G | T/T | T/T | T/T | T/T | G/T |
| 9 | 3548057 | G | A | G/A | G/G | G/G | G/G | G/G | A/A | A/A | A/A | A/A | A/A |
| Z | 70803755 | A | C | A/A | A/A | A/A | A/A | A/A | C/C | C/C | C/C | C/C | C/C |
| Chr | rs | rsID | WT | MT | Gene |
|---|---|---|---|---|---|
| 1 | 173675555 | rs733601174 | C | T | POSTN |
| 4 | 3022801 | rs316329973 | G | A | GPC4, HS6ST2 |
| 4 | 85315858 | rs739468529 | T | C | ST3GAL5 |
| 4 | 85317113 | rs313259902 | A | T | ST3GAL5 |
| 4 | 85317787 | rs316888112 | G | A | ST3GAL5 |
| 4 | 85318002 | rs14501054 | C | T | ST3GAL5 |
| 4 | 85318751 | rs741259739 | C | T | ST3GAL5 |
| 4 | 85321374 | rs735918912 | G | A | ST3GAL5 |
| 4 | 89576514 | rs15645705 | G | A | C20orf194 |
| 5 | 51579773 | rs317162307 | A | G | ASPG |
| 7 | 23241529 | rs739919594 | T | C | TNS1 |
| 10 | 17802386 | rs741174402 | A | T | CHSY1 |
| 13 | 5392030 | rs14990733 | T | C | DOCK2 |
| 15 | 8176792 | rs315551857 | A | G | CORO1C |
| 26 | 3856885 | rs315369512 | G | C | APOBEC2 |
| rsID | WT | MT | CAI Value Before Mutation | CAI Value After Mutation | Gene |
|---|---|---|---|---|---|
| rs315605586 | C | T | 0.75 | 0.76 | PAK3 |
| rs15674853 | T | C | 0.78 | 0.77 | STBD1 |
| rs314812968 | C | T | 0.76 | 0.75 | SYT15 |
| rs | rsID | WT | MT | Specific Transcription Factor Binding Sites Before Mutation | Specific Transcription Factor Binding Sites After Mutation |
|---|---|---|---|---|---|
| 1:173675555 | rs733601174 | C | T | PLAGL2, Plagl1, PLAG1, ZNF692, Zbtb41, Zfp711, ZNF134, ZNF707, Hnf4g, ZKSCAN3, ZBTB6, ZNF454 | Prdm5, Zfp37, Hes7, ZNF135, Zfp691, TFAP2E |
| 4:3022801 | rs316329973 | G | A | Zfp3, Zbtb20, Zfp184, Znf431 | AR, Dmrtb1, ZNF189, Mafb |
| 4:85315858 | rs739468529 | T | C | Zfp689, Rfx7, Hic1, Rfx4, ZNF527, ZNF582, RFX5, RFX2, Rfx3 | |
| 4:85317113 | rs313259902 | A | T | FOXC1, Pax5 | REST, Sall2 |
| 4:85317787 | rs316888112 | G | A | ZNF677 | POU2F2, POU5F1, POU1F1, POU3F4, ZNF84, MAFF, POU6F1, PATZ1, Pou2f1, POU2F3, POU3F2, ZNF394, FEZF1, Foxm1, POU3F1, ZNF260 |
| 4:85318002 | rs14501054 | C | T | SREBF2, Rara, Nkx2-2, NKX2-1, RARG | YY1, MXI1, Prdm5, RFX5, BCL11A, Rfx2, Rfx1 |
| 4:85318751 | rs741259739 | C | T | PAX5, Nr1h4, ZNF791, ZNF383, FERD3L, ZNF681, ATF1 | ZNF136, ZNF324, Npas4 |
| 4:85321374 | rs735918912 | G | A | JUND, NFE2, Npas4, JUNB | Sox4, ZNF768, ZNF212, Sox11, SOX3, ZNF652, SOX10, AR, ZNF223 |
| 4:89576514 | rs15645705 | G | A | HIC2, Mtf1, HIC1, Nr1h4, ATF6, ZNF141, ZNF93, ATF6B, Zbtb41 | RFX3, ZNF429, THAP1, Rfx4, ZNF320, Npas4 |
| 5:51579773 | rs317162307 | A | G | ZNF35, ZNF280A, Zfp583 | ZNF212, SOX18, ZNF180, Rela |
| 7:23241529 | rs739919594 | T | C | ZNF3, ZNF768 | MYBL1, ESR2, Myb, Mybl2, ZNF324 |
| 10:17802386 | rs741174402 | A | T | NR1D2 | IRF4, Zfp41, Zfp287, Zfp768 |
| 13:5392030 | rs14990733 | T | C | ZIM3, Stat6, ZNF274 | RELA, NFKB1, REL |
| 15:8176792 | rs315551857 | A | G | PITX2, Myb | GSX2, GSX1, PBX4, Lhx2, POU6F2, Uncx, NKX6-2, LBX1, PRRX2, SHOX, ISX, HESX1, LHX9, ARX, VENTX, Pax6, PROP1, MEIS2, RORA, PRRX1, PAX4, MIXL1, NKX6-1, Nobox, HOXC4, NOTO, HOXD13, Nkx6-3, ZNF396, Lhx8, ESX1, LHX6, VSX1, Hlx, ZFHX2, En2, Phox2b, Phox2a, DLX3, Lhx1, RORC, Shox2, RAX2, Rhox6, HOXC13, MEOX1, EMX1, RAX, Otp, Gbx1 |
| 26:3856885 | rs315369512 | G | C | PAX6, ZNF621, ZNF768 | THAP1, FOXO3, ZNF75A, Zfp770 |
| rs | rsID | WT | MT | Specific Transcription Factor Binding Sites Before Mutation | Specific Transcription Factor Binding Sites After Mutation |
|---|---|---|---|---|---|
| 1:173675555 | rs733601174 | C | T | HNF4, COUP, USF_C | ZID |
| 4:3022801 | rs316329973 | G | A | ZID | PAX2, CEBP_C, CEBPB, IRF1, CHOP, R |
| 4:85315858 | rs739468529 | T | C | BRACH, GATA3, EVI1, SRY | STAF, PAX5, RFX1, CMYB |
| 4:85317113 | rs313259902 | A | T | GATA1, CREL, CDP, S8, ISRE, IRF1, IRF2, HFH1, CEBP, PBX1 | OCT1_06, HNF1_C, E4BP4, VBP, HLF, EVI1, NRSF, CMYB |
| 4:85317787 | rs316888112 | G | A | GR, AHRARNT, IRF2, PAX5 | EVI1 |
| 4:85318002 | rs14501054 | C | T | CMYB, MYB, VMYB | COMP1, RFX1, AP1FJ |
| 4:85318751 | rs741259739 | C | T | ER, T3R, P300 | USF, CDPCR3, MZF1, GATA1, GATA2, GATA3 |
| 4:85321374 | rs735918912 | G | A | PAX5, STAF, NRF2, ISRE | MZF1, HNF4, GR, CDXA |
| 4:89576514 | rs15645705 | G | A | CMYB, E2, E2F, IK1 | CDPCR3, CREB, USF_C, ER, GRE_C |
| 5:51579773 | rs317162307 | A | G | NFKB_C, NFKAPPAB, GR, R, MYCMAX, LMO2COM, AML1 | TAXCREB, VMYB |
| 7:23241529 | rs739919594 | T | C | TAL1ALPHAE47, TAL1BETAITF2, GATA1, SRY | GR, GRE_C, COMP1 |
| 10:17802386 | rs741174402 | A | T | GRE_C, EVI1, CREL, USF, SP1 | COMP1, IK3, NFE2, ELK1 |
| 13:5392030 | rs14990733 | T | C | POLY_C, EVI1 | STAT1, CREL, DELTAEF1, STAT, CAP |
| 15:8176792 | rs315551857 | A | G | PBX1, CREBP1 | RORA1, ER |
| 26:3856885 | rs315369512 | G | C | OLF1, CAAT | CEBPA, CEBP, GFI1, MZF1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dou, D.; Chen, H.; Ge, Y.; Zhou, J.; Chang, C.; Zhang, F.; Yang, S.; Cao, Z.; Luan, P.; Li, Y.; et al. Identification and Pathway Analysis of SNP Loci Affecting Abdominal Fat Deposition in Broilers. Animals 2025, 15, 2811. https://doi.org/10.3390/ani15192811
Dou D, Chen H, Ge Y, Zhou J, Chang C, Zhang F, Yang S, Cao Z, Luan P, Li Y, et al. Identification and Pathway Analysis of SNP Loci Affecting Abdominal Fat Deposition in Broilers. Animals. 2025; 15(19):2811. https://doi.org/10.3390/ani15192811
Chicago/Turabian StyleDou, Dachang, Hengcong Chen, Yaowen Ge, Jiamei Zhou, Cheng Chang, Fuyang Zhang, Shengwei Yang, Zhiping Cao, Peng Luan, Yumao Li, and et al. 2025. "Identification and Pathway Analysis of SNP Loci Affecting Abdominal Fat Deposition in Broilers" Animals 15, no. 19: 2811. https://doi.org/10.3390/ani15192811
APA StyleDou, D., Chen, H., Ge, Y., Zhou, J., Chang, C., Zhang, F., Yang, S., Cao, Z., Luan, P., Li, Y., & Zhang, H. (2025). Identification and Pathway Analysis of SNP Loci Affecting Abdominal Fat Deposition in Broilers. Animals, 15(19), 2811. https://doi.org/10.3390/ani15192811






