Tissue-Specific Mitochondrial Functionality and Mitochondrial-Related Gene Profiles in Response to Maternal Nutrition and One-Carbon Metabolite Supplementation During Early Pregnancy in Heifers
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Animals and Ethics
2.2. Mitochondria Isolation
2.3. Oxygen Consumption Rate (OCR) Determination
2.4. Relative Mitochondrial DNA Copy Number Determination
2.5. RNA Extraction and Sequencing
2.6. Statistical Analysis
3. Results
3.1. Mitochondrial Respiration Profiles in Fetal Liver and Muscle Tissues
3.2. mtDNA Copy Number Variations in Fetal Liver and Muscle
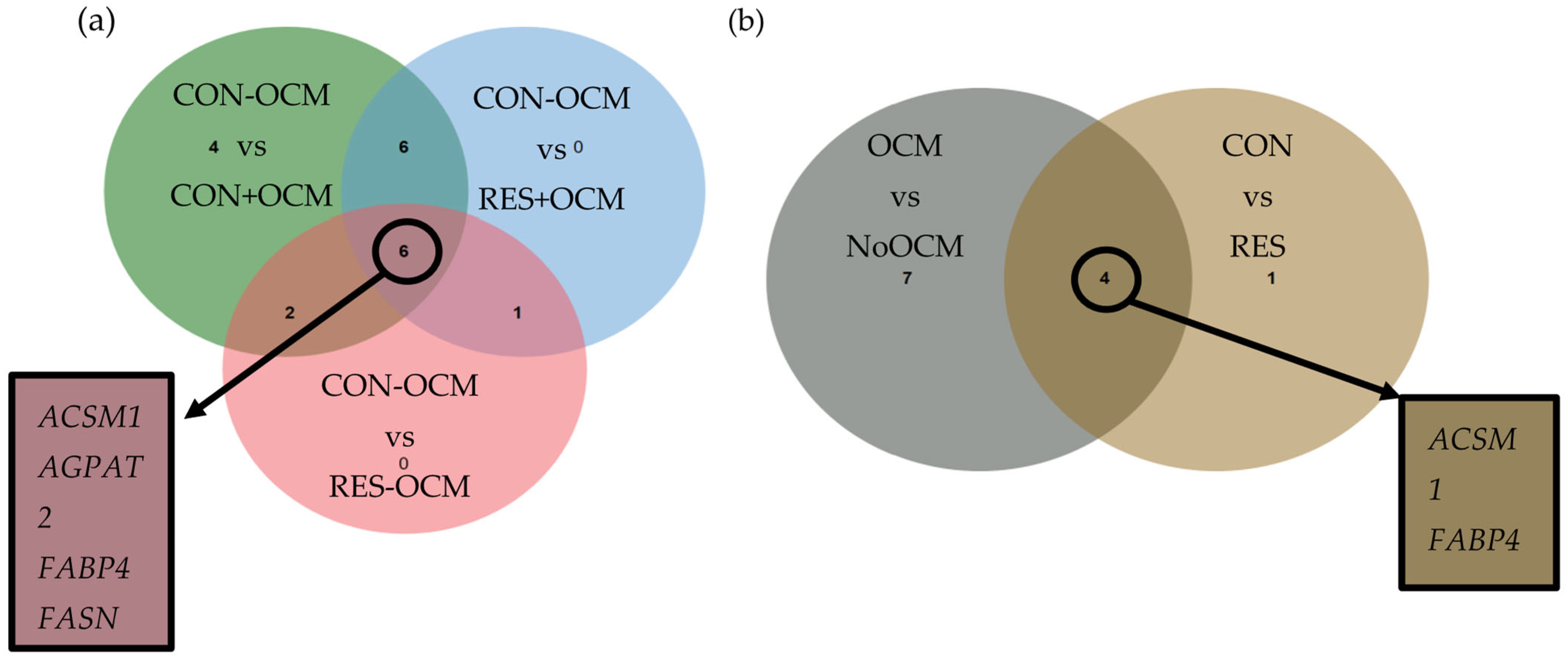
3.3. Mitochondria-Related Differentially Expressed Genes (mtDEGs) in Fetal Liver
3.4. Mitochondria-Related Differentially Expressed Genes (mtDEGs) in Fetal Muscle
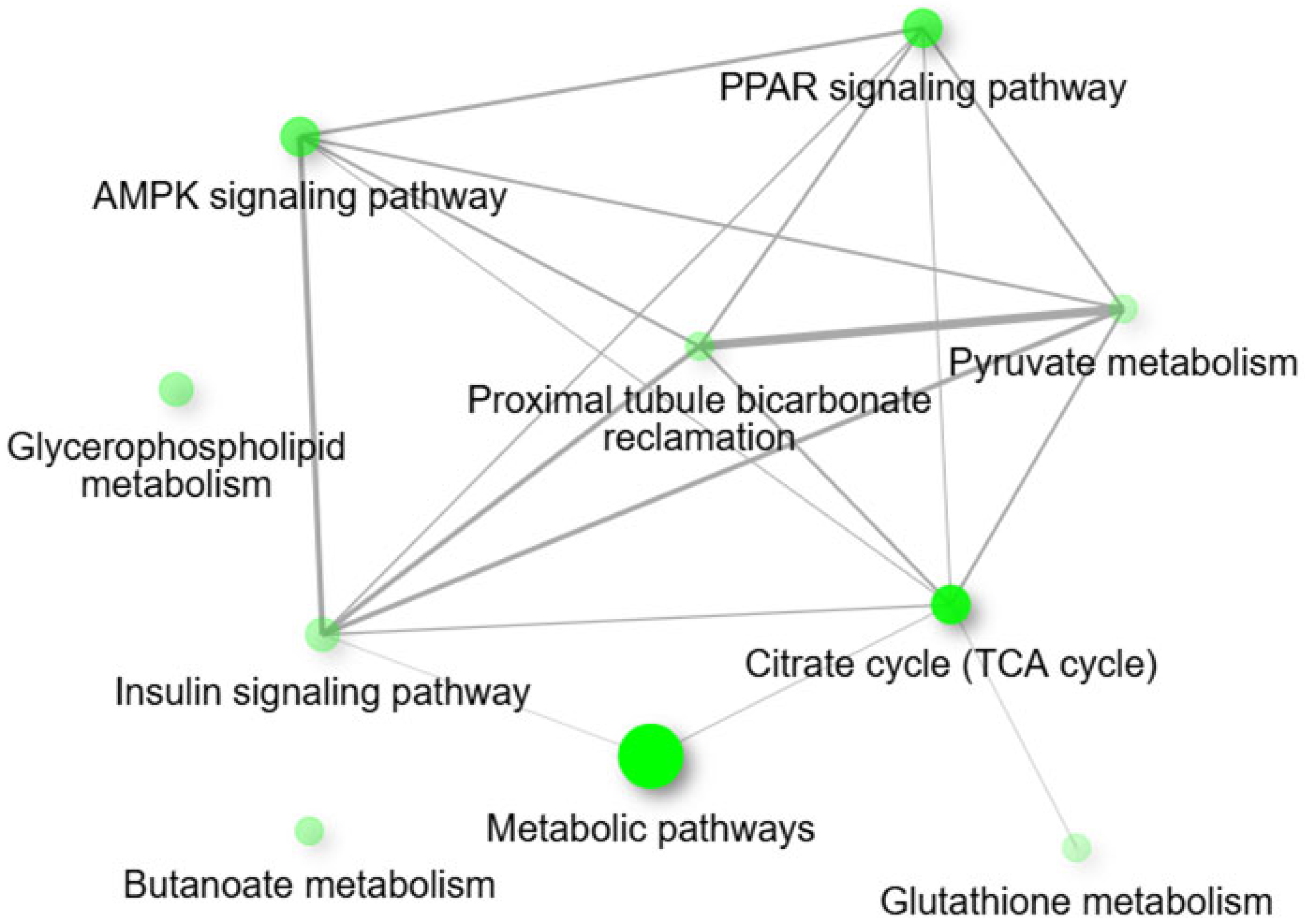
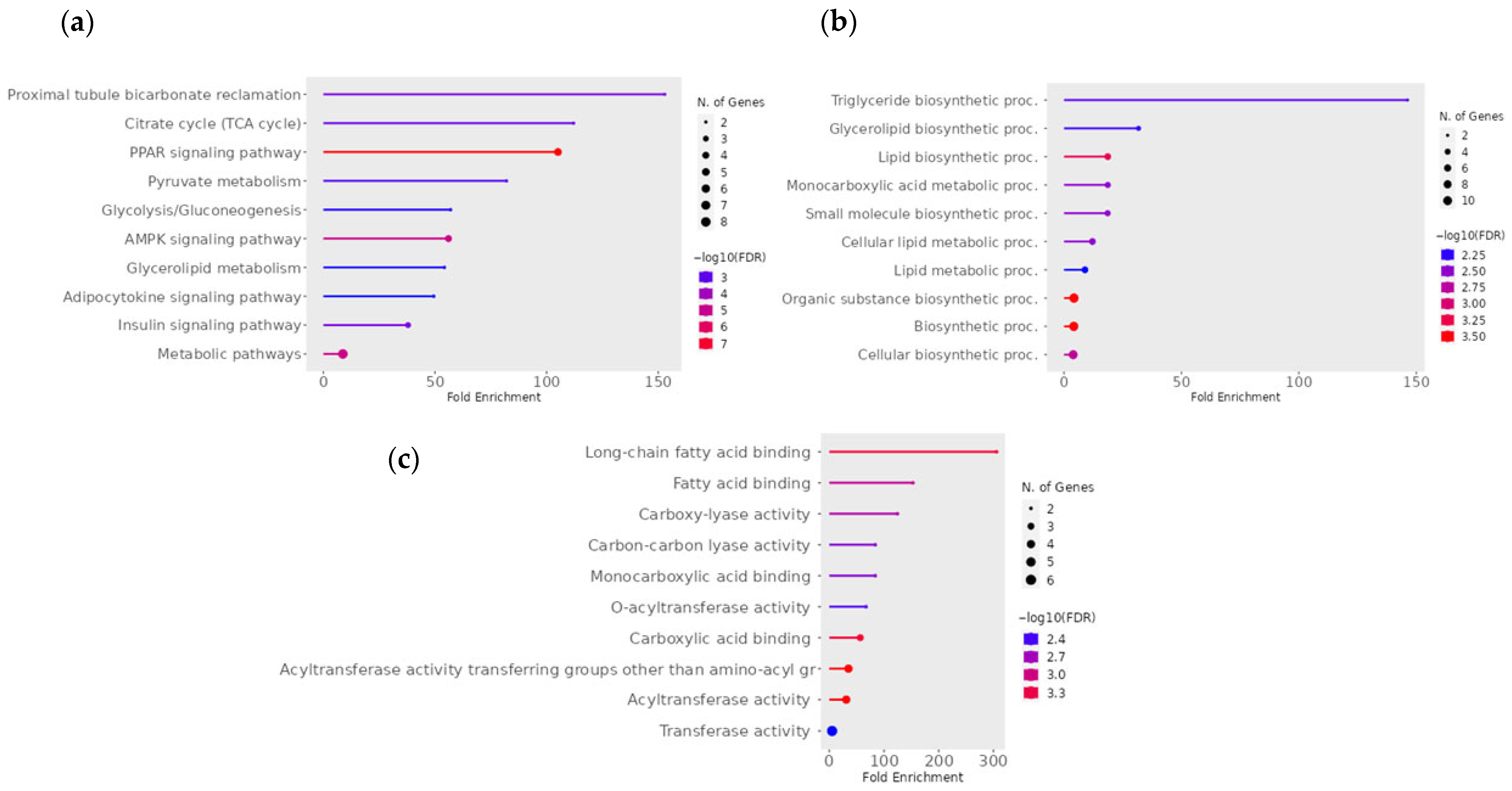
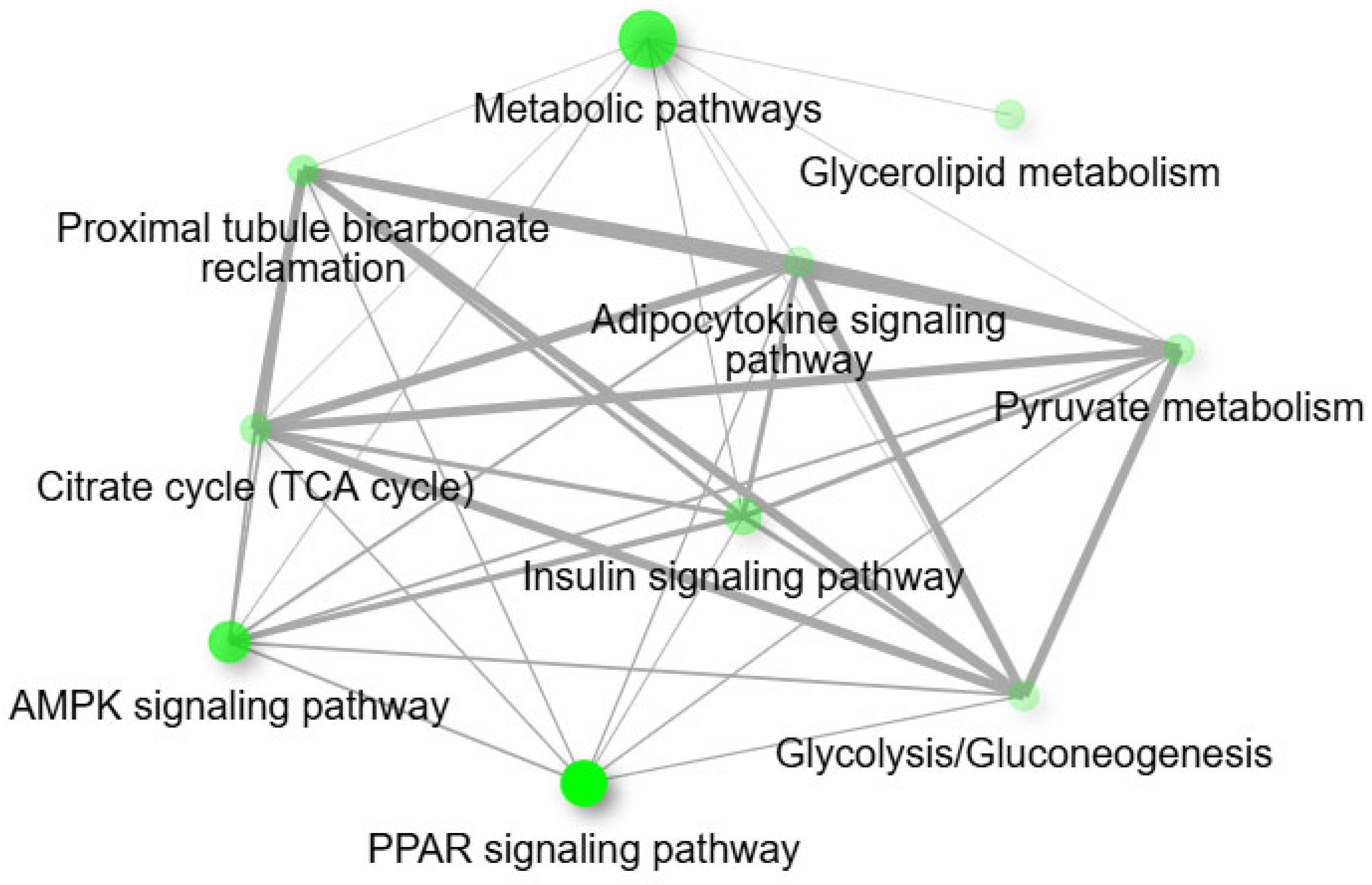
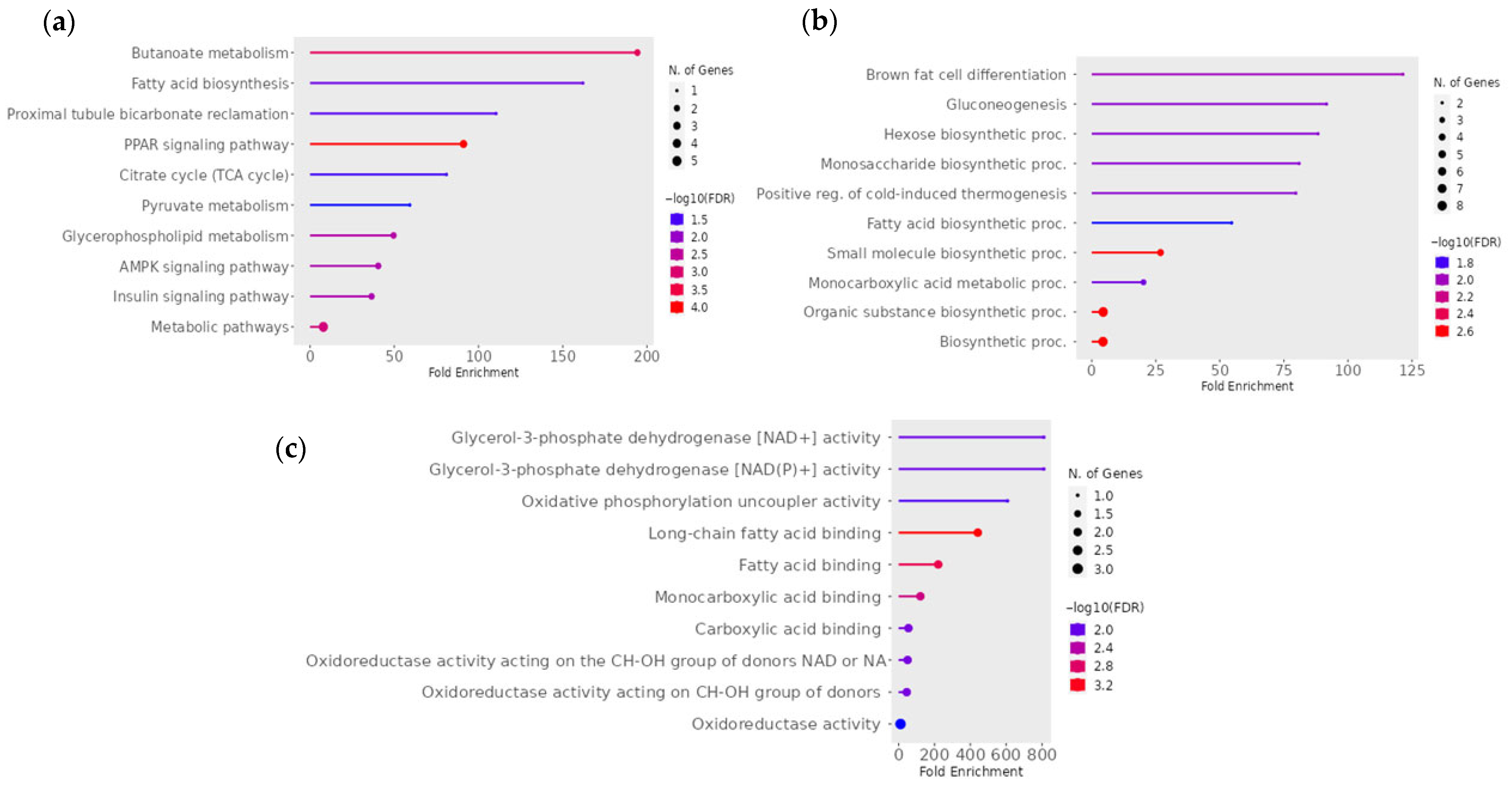
3.5. Functional and Pathway Analysis of mtDEGs in Fetal Muscle
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Colleoni, F.; Lattuada, D.; Garretto, A.; Massari, M.; Mandò, C.; Somigliana, E.; Cetin, I. Maternal blood mitochondrial DNA content during normal and intrauterine growth restricted (IUGR) pregnancy. Am. J. Obstet. Gynecol. 2010, 203, 365.e1–365.e6. [Google Scholar] [CrossRef]
- Harvey, A.J. Mitochondria in early development: Linking the microenvironment, metabolism and the epigenome. Reproduction 2019, 157, R159–R179. [Google Scholar] [CrossRef]
- Priliani, L.; Prado, E.L.; Restuadi, R.; Waturangi, D.E.; Shankar, A.H.; Malik, S.G. Maternal multiple micronutrient supplementation stabilizes mitochondrial DNA copy number in pregnant women in Lombok, Indonesia. J. Nutr. 2019, 149, 1309–1316. [Google Scholar] [CrossRef] [PubMed]
- Kunz, W.S. Different Metabolic Properties of Mitochondrial Oxidative Phosphorylation in Different Cell Types-Important Implications for Mitochondrial Cytopathies. Exp. Physiol. 2003, 88, 149–154. [Google Scholar] [CrossRef]
- Jovandaric, M.Z.; Babic, S.; Raus, M.; Medjo, B. The importance of metabolic and environmental factors in the occurrence of oxidative stress during pregnancy. Int. J. Mol. Sci. 2023, 24, 11964. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Sferruzzi-Perri, A.N. Placental mitochondrial function in response to gestational exposures. Placenta 2021, 104, 124–137. [Google Scholar] [CrossRef]
- Bazer, F.W.; Spencer, T.E.; Wu, G.; Cudd, T.A.; Meininger, C.J. Maternal nutrition and fetal development. J. Nutr. 2004, 134, 2169–2172. [Google Scholar] [CrossRef] [PubMed]
- Lyon, P. B Vitamins and one-carbon metabolism: Implications in human health and disease. Nutrients 2020, 12, 2867. [Google Scholar] [CrossRef]
- Xiu, Y.; Field, M.S. The roles of mitochondrial folate metabolism in supporting mitochondrial DNA synthesis, oxidative phosphorylation, and cellular function. Curr. Dev. Nutr. 2020, 4, nzaa153. [Google Scholar] [CrossRef]
- Jiang, Q.; Sherlock, D.N.; Zhang, H.; Guyader, J.; Pan, Y.-X.; Loor, J.J. One-carbon metabolism and related pathways in ruminal and small intestinal epithelium of lactating dairy cows. J. Anim. Sci. 2023, 101, skad062. [Google Scholar] [CrossRef]
- Cai, S.; Quan, S.; Yang, G.; Ye, Q.; Chen, M.; Yu, H.; Wang, G.; Wang, Y.; Zeng, X.; Qiao, S. One carbon metabolism and mammalian pregnancy outcomes. Mol. Nutr. Food Res. 2021, 65, 2000734. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Cano, A.M.; Calzada-Mendoza, C.C.; Estrada-Gutierrez, G.; Mendoza-Ortega, J.A.; Perichart-Perera, O. Nutrients, mitochondrial function, and perinatal health. Nutrients 2020, 12, 2166. [Google Scholar] [CrossRef] [PubMed]
- Owens, F.N.; Basalan, M. Ruminal Fermentation. In Rumenology; Springer Nature: Berlin/Heidelberg, Germany, 2016; pp. 63–102. [Google Scholar]
- Aon, M.A.; Cortassa, S.; Juhaszova, M.; Sollott, S.J. Mitochondrial health, the epigenome and healthspan. Clin. Sci. 2016, 130, 1285–1305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N. Role of methionine on epigenetic modification of DNA methylation and gene expression in animals. Anim. Nutr. 2018, 4, 11–16. [Google Scholar] [CrossRef]
- Zeisel, S.H. The supply of choline is important for fetal progenitor cells. Semin. Cell Dev. Biol. 2011, 22, 624–628. [Google Scholar] [CrossRef]
- Crouse, M.S.; Cushman, R.A.; Redifer, C.A.; Neville, B.W.; Dahlen, C.R.; Caton, J.S.; Diniz, W.J.; Ward, A.K. International Symposium on Ruminant Physiology: One-carbon metabolism in beef cattle throughout the production cycle. J. Dairy Sci. 2025, 108, 7615–7630. [Google Scholar] [CrossRef]
- Kalhan, S.C.; Marczewski, S.E. Methionine, homocysteine, one carbon metabolism and fetal growth. Rev. Endocr. Metab. Disord. 2012, 13, 109–119. [Google Scholar] [CrossRef]
- Solé-Navais, P.; Cavallé-Busquets, P.; Fernandez-Ballart, J.D.; Murphy, M.M. Early pregnancy B vitamin status, one carbon metabolism, pregnancy outcome and child development. Biochimie 2016, 126, 91–96. [Google Scholar] [CrossRef]
- Pejznochova, M.; Tesarova, M.; Hansikova, H.; Magner, M.; Honzik, T.; Vinsova, K.; Hajkova, Z.; Havlickova, V.; Zeman, J. Mitochondrial DNA content and expression of genes involved in mtDNA transcription, regulation and maintenance during human fetal development. Mitochondrion 2010, 10, 321–329. [Google Scholar] [CrossRef]
- Papacleovoulou, G.; Nikolova, V.; Oduwole, O.; Chambers, J.; Vazquez-Lopez, M.; Jansen, E.; Nicolaides, K.; Parker, M.; Williamson, C. Gestational disruptions in metabolic rhythmicity of the liver, muscle, and placenta affect fetal size. FASEB J. 2017, 31, 1698. [Google Scholar] [CrossRef]
- Zaret, K.S. Regulatory phases of early liver development: Paradigms of organogenesis. Nat. Rev. Genet. 2002, 3, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Diniz, W.J.; Crouse, M.S.; Cushman, R.A.; McLean, K.J.; Caton, J.S.; Dahlen, C.R.; Reynolds, L.P.; Ward, A.K. Cerebrum, liver, and muscle regulatory networks uncover maternal nutrition effects in developmental programming of beef cattle during early pregnancy. Sci. Rep. 2021, 11, 2771. [Google Scholar] [CrossRef]
- Safain, K.S.; Crouse, M.S.; Syring, J.G.; Entzie, Y.L.; King, L.E.; Hirchert, M.R.; Ward, A.K.; Reynolds, L.P.; Borowicz, P.P.; Dahlen, C.R. One-carbon metabolites supplementation and nutrient restriction alter the fetal liver metabolomic profile during early gestation in beef heifers. J. Anim. Sci. 2024, 102, skae258. [Google Scholar] [CrossRef]
- King, L.E.; Syring, J.; Entzie, Y.; Hirchert, M.; Crouse, M.S.; Caton, J.S.; Dahlen, C.R.; Ward, A.K. Supplementation of One-Carbon Metabolites to Beef Heifers During Early Gestation Effects on Fetal Liver and Muscle Mitochondrial Oxygen Consumption Rate Ratios at Day 63 of Gestation. 2022 North Dakota Livestock Research Report. 2022. Available online: https://www.ndsu.edu/agriculture/extension/publications/2022-north-dakota-livestock-research-report (accessed on 2 January 2025).
- Bartho, L.A.; Fisher, J.J.; Walton, S.L.; Perkins, A.V.; Cuffe, J.S. The effect of gestational age on mitochondrial properties of the mouse placenta. Reprod. Fertil. 2022, 3, 19–29. [Google Scholar] [CrossRef]
- Daneshi, M.; Borowicz, P.P.; Entzie, Y.L.; Syring, J.G.; King, L.E.; Safain, K.S.; Anas, M.; Reynolds, L.P.; Ward, A.K.; Dahlen, C.R. Influence of Maternal Nutrition and One-Carbon Metabolites Supplementation during Early Pregnancy on Bovine Fetal Small Intestine Vascularity and Cell Proliferation. Vet. Sci. 2024, 11, 146. [Google Scholar] [CrossRef]
- Syring, J.G.; Crouse, M.S.; Entzie, Y.L.; King, L.E.; Hirchert, M.R.; Ward, A.K.; Reynolds, L.P.; Borowicz, P.P.; Dahlen, C.R.; Caton, J.S. One-carbon metabolite supplementation increases vitamin B12, folate, and methionine cycle metabolites in beef heifers and fetuses in an energy dependent manner at day 63 of gestation. J. Anim. Sci. 2024, 102, skae202. [Google Scholar] [CrossRef]
- Claycombe, K.J.; Roemmich, J.N.; Johnson, L.; Vomhof-DeKrey, E.E.; Johnson, W.T. Skeletal muscle Sirt3 expression and mitochondrial respiration are regulated by a prenatal low-protein diet. J. Nutr. Biochem. 2015, 26, 184–189. [Google Scholar] [CrossRef]
- Rogers, G.W.; Brand, M.D.; Petrosyan, S.; Ashok, D.; Elorza, A.A.; Ferrick, D.A.; Murphy, A.N. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS ONE 2011, 6, e21746. [Google Scholar] [CrossRef] [PubMed]
- Reverter, A.; Okimoto, R.; Sapp, R.; Bottje, W.G.; Hawken, R.; Hudson, N.J. Chicken muscle mitochondrial content appears co-ordinately regulated and is associated with performance phenotypes. Biol. Open 2017, 6, 50–58. [Google Scholar] [PubMed]
- Bai, Y.; Carrillo, J.A.; Li, Y.; He, Y.; Song, J. Diet induced the change of mtDNA copy number and metabolism in Angus cattle. J. Anim. Sci. Biotechnol. 2020, 11, 84. [Google Scholar]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Rubini, E.; Baijens, I.M.; Horánszky, A.; Schoenmakers, S.; Sinclair, K.D.; Zana, M.; Dinnyés, A.; Steegers-Theunissen, R.P.; Rousian, M. Maternal one-carbon metabolism during the periconceptional period and human foetal brain growth: A systematic review. Genes 2021, 12, 1634. [Google Scholar] [CrossRef] [PubMed]
- Wathes, D.; Cheng, Z.; Bourne, N.; Taylor, V.; Coffey, M.; Brotherstone, S. Differences between primiparous and multiparous dairy cows in the inter-relationships between metabolic traits, milk yield and body condition score in the periparturient period. Domest. Anim. Endocrinol. 2007, 33, 203–225. [Google Scholar] [CrossRef] [PubMed]
- Hammer, C.J.; Caton, J.S.; Dahlen, C.; Ward, A.K.; Borowicz, P.P.; Reynolds, L. DOHaD: A menagerie of adaptations and perspectives: Large animal models of developmental programming: Sustenance, stress, and sex matter. Reproduction 2023, 165, F1–F13. [Google Scholar] [CrossRef]
- Bromley, P.N.; Rawlinson, E.; Harclerode, Z.; Bennett, J. Developmental Physiology of the Liver, Gastrointestinal Tract, and Renal System. In Gregory’s Pediatric Anesthesia; Wiley: Hoboken, NJ, USA, 2020; pp. 164–190. [Google Scholar]
- King, L.; Syring, J.; Entzie, Y.; Hirchert, M.R.; Crouse, M.S.; Caton, J.; Dahlen, C.R.; Ward, A.K. 180 Supplementation of One-Carbon Metabolites to Beef Heifers During Early Gestation Increases Fetal Organ Weight at Day 63 of Gestation. J. Anim. Sci. 2022, 100 (Suppl. S2), 87. [Google Scholar] [CrossRef]
- Cuezva, J.M.; Ostronoff, L.K.; Ricart, J.; de Heredia, M.L.; Di Liegro, C.M.; Izquierdo, J.M. Mitochondrial biogenesis in the liver during development and oncogenesis. J. Bioenerg. Biomembr. 1997, 29, 365–377. [Google Scholar] [CrossRef]
- Fang, H.; Li, Q.; Wang, H.; Ren, Y.; Zhang, L.; Yang, L. Maternal nutrient metabolism in the liver during pregnancy. Front. Endocrinol. 2024, 15, 1295677. [Google Scholar] [CrossRef]
- Crouse, M.S.; Caton, J.S.; Claycombe-Larson, K.J.; Diniz, W.J.; Lindholm-Perry, A.K.; Reynolds, L.P.; Dahlen, C.R.; Borowicz, P.P.; Ward, A.K. Epigenetic modifier supplementation improves mitochondrial respiration and growth rates and alters DNA methylation of bovine embryonic fibroblast cells cultured in divergent energy supply. Front. Genet. 2022, 13, 812764. [Google Scholar] [CrossRef]
- Robin, E.D.; Wong, R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J. Cell. Physiol. 1988, 136, 507–513. [Google Scholar] [CrossRef]
- Filograna, R.; Mennuni, M.; Alsina, D.; Larsson, N.G. Mitochondrial DNA copy number in human disease: The more the better? FEBS Lett. 2021, 595, 976–1002. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wang, Y.; Ye, K.; Picard, M.; Gu, Z. Independent impacts of aging on mitochondrial DNA quantity and quality in humans. BMC Genom. 2017, 18, 1–14. [Google Scholar] [CrossRef]
- Busnelli, A.; Lattuada, D.; Ferrari, S.; Reschini, M.; Colciaghi, B.; Somigliana, E.; Fedele, L.; Ferrazzi, E. Mitochondrial DNA copy number in peripheral blood in the first trimester of pregnancy and different preeclampsia clinical phenotypes development: A pilot study. Reprod. Sci. 2019, 26, 1054–1061. [Google Scholar] [CrossRef]
- Du, M.; Tong, J.; Zhao, J.; Underwood, K.; Zhu, M.; Ford, S.; Nathanielsz, P. Fetal programming of skeletal muscle development in ruminant animals. J. Anim. Sci. 2010, 88 (Suppl. S13), E51–E60. [Google Scholar] [CrossRef]
- Watkins, P.A.; Ellis, J.M. Peroxisomal acyl-CoA synthetases. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2012, 1822, 1411–1420. [Google Scholar] [CrossRef]
- Rodríguez-Calvo, R.; Girona, J.; Rodríguez, M.; Samino, S.; Barroso, E.; de Gonzalo-Calvo, D.; Guaita-Esteruelas, S.; Heras, M.; van der Meer, R.W.; Lamb, H.J. Fatty acid binding protein 4 (FABP4) as a potential biomarker reflecting myocardial lipid storage in type 2 diabetes. Metabolism 2019, 96, 12–21. [Google Scholar] [CrossRef]
- Nakano, M.; Ogasawara, H.; Shimada, T.; Yamamoto, K.; Ishihama, A. Involvement of cAMP-CRP in transcription activation and repression of the pck gene encoding PEP carboxykinase, the key enzyme of gluconeogenesis. FEMS Microbiol. Lett. 2014, 355, 93–99. [Google Scholar] [CrossRef]
- Crouse, M.S.; Caton, J.S.; Cushman, R.A.; McLean, K.J.; Dahlen, C.R.; Borowicz, P.P.; Reynolds, L.P.; Ward, A.K. Moderate nutrient restriction of beef heifers alters expression of genes associated with tissue metabolism, accretion, and function in fetal liver, muscle, and cerebrum by day 50 of gestation. Transl. Anim. Sci. 2019, 3, 855–866. [Google Scholar] [CrossRef]
- Imami, K.; Milek, M.; Bogdanow, B.; Yasuda, T.; Kastelic, N.; Zauber, H.; Ishihama, Y.; Landthaler, M.; Selbach, M. Phosphorylation of the ribosomal protein RPL12/uL11 affects translation during mitosis. Mol. Cell 2018, 72, 84–98.e9. [Google Scholar] [CrossRef] [PubMed]
- Stram, A.R.; Payne, R.M. Post-translational modifications in mitochondria: Protein signaling in the powerhouse. Cell. Mol. Life Sci. 2016, 73, 4063–4073. [Google Scholar] [CrossRef] [PubMed]
- Adelizzi, A.; Giri, A.; Di Donfrancesco, A.; Boito, S.; Prigione, A.; Bottani, E.; Bollati, V.; Tiranti, V.; Persico, N.; Brunetti, D. Fetal and obstetrics manifestations of mitochondrial diseases. J. Transl. Med. 2024, 22, 853. [Google Scholar] [CrossRef] [PubMed]



| Ingredient, % of Dietary DM | |
| Ground corn | 27.0 |
| Corn silage | 9.0 |
| Alfalfa | 15.0 |
| Alfalfa/grass hay | 45.0 |
| Top-dressed supplement 1 | 4.0 |
| Average Estimated Dietary Composition | |
| Dry matter, % | 83.00 |
| Crude protein, % | 13.50 |
| Dietary metabolizable energy, Mcal/kg 2 | 2.50 |
| Dietary net energy of maintenance, Mcal/kg 2 | 2.00 |
| Dietary net energy of gain, Mcal/kg 2 | 1.00 |
| Supplement Ingredients on a g/d Basis | |
| Trouw dairy VTM w/Optimins 3 | 10.0 |
| Rumen protected methionine 4 | 7.5 |
| Rumen protected choline 5 | 44.5 |
| Chemical Composition of Trouw VTM Premix 6 | |
| Ca, % | 10.00 to 12.00 |
| Mg, % | 5.00 |
| K, % | 5.00 |
| Co, mg/kg | 180.00 |
| Cu, mg/kg | 5100.00 |
| I, mg/k | 375.00 |
| Fe, % | 1.00 |
| Mg, % | 3.00 |
| Se, mg/kg | 132.00 |
| Zn, % | 3.00 |
| Vitamin A, IU/kg | 2,755,775.00 |
| Vitamin D3, IU/kg | 771,617.00 |
| Vitamin E, IU/kg | 13,228.00 |
| Restricted | Control | SEM | p value | |||||
|---|---|---|---|---|---|---|---|---|
| −OCM | +OCM | −OCM | +OCM | Gain | OCM | Gain × OCM | ||
| State 3 respiration | 586 ab | 482 a | 437 a | 646 b | 52.2 | 0.88 | 0.30 | 0.004 |
| State 4o respiration | 72.9 b | 57.2 a | 64.6 ab | 78.1 ab | 7.07 | 0.87 | 0.35 | 0.04 |
| * RCR | 8.04 | 8.43 | 6.76 | 8.27 | 0.99 | 0.85 | 0.08 | 0.37 |
| Restricted | Control | SEM | p value | |||||
|---|---|---|---|---|---|---|---|---|
| −OCM | +OCM | −OCM | +OCM | Gain | OCM | Gain × OCM | ||
| State 3 respiration | 608 | 582 | 585 | 714 | 133.6 | 0.66 | 0.68 | 0.54 |
| State 4o respiration | 83.8 | 85.5 | 120 | 96.0 | 33.37 | 0.46 | 0.73 | 0.68 |
| * RCR | 7.25 | 6.81 | 4.88 | 7.44 | 1.259 | 0.61 | 0.99 | 0.76 |
| Treatment Group | Name | Ensembl ID | Fold Change | FDR p Value | p Value | Biotype |
|---|---|---|---|---|---|---|
| CON − OCM vs. CON + OCM | RPL12 | ENSBTAG00000006963 | 2484.8215 | <0.001 | <0.001 | protein_coding |
| CON − OCM vs. RES + OCM | RPL12 | ENSBTAG00000006963 | 1104.8939 | <0.001 | <0.001 | protein_coding |
| CYP3A5_2 | ENSBTAG00000053645 | −8.1855 | 0.007 | <0.001 | protein_coding | |
| RES − OCM vs. RES + OCM | CYP3A5_2 | ENSBTAG00000053645 | −8.5048 | 0.005 | <0.001 | protein_coding |
| CON − OCM vs. RES − OCM | RPL12 | ENSBTAG00000006963 | 1535.9445 | <0.001 | <0.001 | protein_coding |
| Name | Ensembl ID | Fold Change | FDR p Value | p Value | Biotype |
|---|---|---|---|---|---|
| ACLY | ENSBTAG00000016740 | −2.8592 | 0.0391 | 0.000111 | protein_coding |
| ACSM1 | ENSBTAG00000001417 | −130.5345 | 0.0001 | <0.001 | protein_coding |
| AGPAT2 | ENSBTAG00000025161 | −4.4916 | 0.0163 | <0.001 | protein_coding |
| BDH1 | ENSBTAG00000000448 | −4.9496 | 0.0509 | <0.001 | protein_coding |
| FABP4 | ENSBTAG00000037526 | −176.6854 | 0.0000 | <0.001 | protein_coding |
| FASN | ENSBTAG00000015980 | −4.7550 | 0.0078 | <0.001 | protein_coding |
| GLYAT | ENSBTAG00000038323 | −45.5870 | 0.0032 | <0.001 | protein_coding |
| GPAM | ENSBTAG00000011917 | −5.0158 | 0.0177 | <0.001 | protein_coding |
| GPD1 | ENSBTAG00000016296 | −3.0421 | 0.0875 | <0.001 | protein_coding |
| IDH1 | ENSBTAG00000020527 | −3.3821 | 0.0479 | <0.001 | protein_coding |
| LRRC39 | ENSBTAG00000015786 | 2.2008 | 0.0447 | <0.001 | protein_coding |
| MGST1 | ENSBTAG00000008541 | −4.7780 | 0.0600 | <0.001 | protein_coding |
| PCK2 | ENSBTAG00000011934 | −5.4512 | 0.0096 | <0.001 | protein_coding |
| PCK1 | ENSBTAG00000001936 | −15.8713 | 0.0001 | <0.001 | protein_coding |
| PPARG | ENSBTAG00000001333 | −9.8412 | 0.0002 | <0.001 | protein_coding |
| RPL12 | ENSBTAG00000006963 | 295.4703 | 0.0000 | <0.001 | protein_coding |
| SLC16A11 | ENSBTAG00000020852 | −4.6394 | 0.0215 | <0.001 | protein_coding |
| TKT | ENSBTAG00000003758 | −3.1795 | 0.0244 | <0.001 | protein_coding |
| Name | Ensembl ID | Fold Change | FDR p Value | p Value | Biotype |
|---|---|---|---|---|---|
| ACSM1 | ENSBTAG00000001417 | −134.9288 | 0.0004 | <0.001 | protein_coding |
| AGPAT2 | ENSBTAG00000025161 | −4.6326 | 0.0172 | <0.001 | protein_coding |
| FABP4 | ENSBTAG00000037526 | −68.8895 | 0.0000 | <0.001 | protein_coding |
| FASN | ENSBTAG00000015980 | −4.2455 | 0.0421 | <0.001 | protein_coding |
| GLYAT | ENSBTAG00000038323 | −29.9011 | 0.0168 | <0.001 | protein_coding |
| GPAM | ENSBTAG00000011917 | −4.7862 | 0.0413 | <0.001 | protein_coding |
| PCK2 | ENSBTAG00000011934 | −5.0033 | 0.0384 | <0.001 | protein_coding |
| PCK1 | ENSBTAG00000001936 | −22.7321 | 0.0000 | <0.001 | protein_coding |
| PPARG | ENSBTAG00000001333 | −6.0840 | 0.0855 | 0.000226 | protein_coding |
| RPL12 | ENSBTAG00000006963 | 171.7712 | 0.0000 | <0.001 | protein_coding |
| SLC16A11 | ENSBTAG00000020852 | −3.7431 | 0.0914 | 0.000251 | protein_coding |
| TKT | ENSBTAG00000003758 | −3.2251 | 0.0521 | 0.000135 | protein_coding |
| UCP1 | ENSBTAG00000004647 | −23.3073 | 0.0878 | 0.000237 | protein_coding |
| Name | Ensembl ID | Fold Change | FDR p Value | p Value | Biotype |
|---|---|---|---|---|---|
| ACSM1 | ENSBTAG00000001417 | −42.8149 | 0.0038 | <0.001 | protein_coding |
| AGPAT2 | ENSBTAG00000025161 | −4.5288 | 0.0247 | <0.001 | protein_coding |
| FABP4 | ENSBTAG00000037526 | −30.3606 | 0.0027 | <0.001 | protein_coding |
| BDH1 | ENSBTAG00000000448 | −4.6942 | 0.0681 | 0.000165 | protein_coding |
| FASN | ENSBTAG00000015980 | −4.2605 | 0.0589 | 0.000131 | protein_coding |
| GPD1 | ENSBTAG00000016296 | −3.2988 | 0.0954 | 0.000258 | protein_coding |
| PCK2 | ENSBTAG00000011934 | −6.6713 | 0.0033 | <0.001 | protein_coding |
| RPL12 | ENSBTAG00000006963 | 169.0700 | 0.0000 | <0.001 | protein_coding |
| UCP1 | ENSBTAG00000004647 | −34.6716 | 0.0792 | 0.000209 | protein_coding |
| Name | Ensembl ID | Fold Change | FDR p Value | p Value | Biotype |
|---|---|---|---|---|---|
| ACSM1 | ENSBTAG00000001417 | −66.7699 | 0.0000 | <0.001 | protein_coding |
| AGPAT2 | ENSBTAG00000025161 | −2.7690 | 0.0459 | 0.000107 | protein_coding |
| FABP4 | ENSBTAG00000037526 | −52.2625 | 0.0000 | <0.001 | protein_coding |
| FASN | ENSBTAG00000015980 | −2.7656 | 0.0361 | <0.001 | protein_coding |
| GLYAT | ENSBTAG00000038323 | −20.3391 | 0.0014 | <0.001 | protein_coding |
| GPAM | ENSBTAG00000011917 | −3.0415 | 0.0391 | <0.001 | protein_coding |
| MGST1 | ENSBTAG00000008541 | −3.0183 | 0.0564 | 0.000135 | protein_coding |
| PCK2 | ENSBTAG00000011934 | −2.9884 | 0.1086 | 0.000289 | protein_coding |
| PCK1 | ENSBTAG00000001936 | −9.6135 | 0.0001 | <0.001 | protein_coding |
| PPARG | ENSBTAG00000001333 | −4.5198 | 0.0089 | <0.001 | protein_coding |
| TKT | ENSBTAG00000003758 | −2.1670 | 0.0707 | 0.000173 | protein_coding |
| Name | Ensembl ID | Fold Change | FDR p Value | p Value | Biotype |
|---|---|---|---|---|---|
| ACSM1 | ENSBTAG00000001417 | −29.7676 | 0.0025 | <0.001 | protein_coding |
| FABP4 | ENSBTAG00000037526 | −19.2508 | 0.0025 | <0.001 | protein_coding |
| PCK2 | ENSBTAG00000011934 | −3.1443 | 0.0891 | 0.00014 | protein_coding |
| PCK1 | ENSBTAG00000001936 | −10.4104 | 0.0001 | <0.001 | protein_coding |
| UCP1 | ENSBTAG00000004647 | −15.0831 | 0.0621 | <0.001 | protein_coding |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Safain, K.S.; Crouse, M.S.; Hirchert, M.R.; Entzie, Y.L.; Syring, J.G.; Daneshi, M.; Anas, M.; King, L.E.; Reynolds, L.P.; Borowicz, P.P.; et al. Tissue-Specific Mitochondrial Functionality and Mitochondrial-Related Gene Profiles in Response to Maternal Nutrition and One-Carbon Metabolite Supplementation During Early Pregnancy in Heifers. Animals 2025, 15, 2689. https://doi.org/10.3390/ani15182689
Safain KS, Crouse MS, Hirchert MR, Entzie YL, Syring JG, Daneshi M, Anas M, King LE, Reynolds LP, Borowicz PP, et al. Tissue-Specific Mitochondrial Functionality and Mitochondrial-Related Gene Profiles in Response to Maternal Nutrition and One-Carbon Metabolite Supplementation During Early Pregnancy in Heifers. Animals. 2025; 15(18):2689. https://doi.org/10.3390/ani15182689
Chicago/Turabian StyleSafain, Kazi Sarjana, Matthew S. Crouse, Mara R. Hirchert, Yssi L. Entzie, Jessica G. Syring, Mojtaba Daneshi, Muhammad Anas, Layla E. King, Lawrence P. Reynolds, Pawel P. Borowicz, and et al. 2025. "Tissue-Specific Mitochondrial Functionality and Mitochondrial-Related Gene Profiles in Response to Maternal Nutrition and One-Carbon Metabolite Supplementation During Early Pregnancy in Heifers" Animals 15, no. 18: 2689. https://doi.org/10.3390/ani15182689
APA StyleSafain, K. S., Crouse, M. S., Hirchert, M. R., Entzie, Y. L., Syring, J. G., Daneshi, M., Anas, M., King, L. E., Reynolds, L. P., Borowicz, P. P., Dahlen, C. R., Ward, A. K., Caton, J. S., & Swanson, K. C. (2025). Tissue-Specific Mitochondrial Functionality and Mitochondrial-Related Gene Profiles in Response to Maternal Nutrition and One-Carbon Metabolite Supplementation During Early Pregnancy in Heifers. Animals, 15(18), 2689. https://doi.org/10.3390/ani15182689








