Heterogeneity of Variances in Milk Yield in Murrah Buffaloes
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
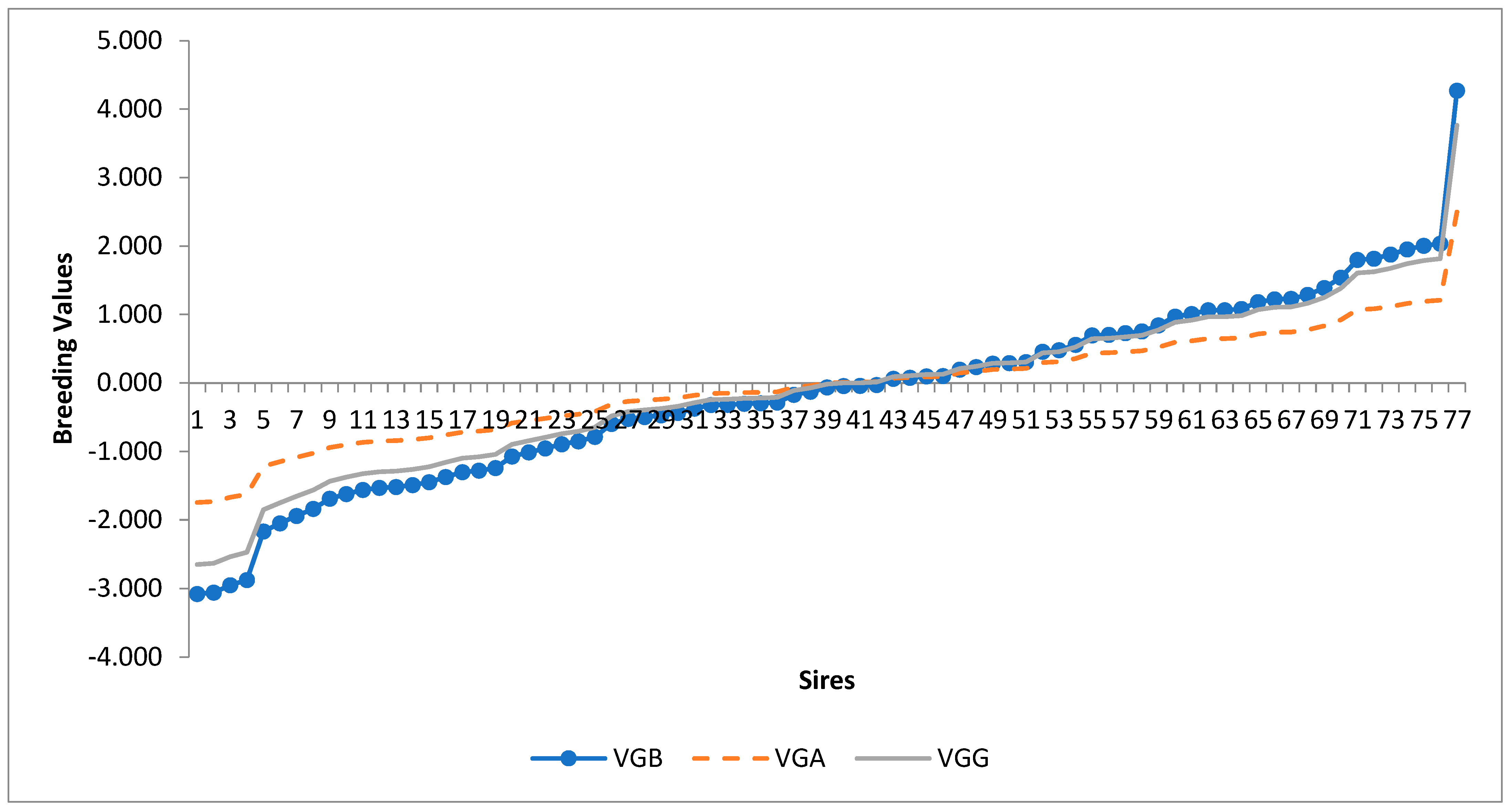
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brito, L.F.; Bedere, N.; Douhard, F.; Oliveira, H.R.; Arnal, M.; Peñagaricano, F.; Schinckel, A.P.; Baes, C.F.; Miglior, F. Review: Genetic selection of high-yield ing dairy cattle toward sustainable farming systems in a rapidly changing world. Animal 2021, 15, 100292. [Google Scholar] [CrossRef] [PubMed]
- Guinan, F.L.; Wiggans, G.R.; Norman, H.D.; Dürr, J.W.; Cole, J.B.; Van Tassell, C.P.; Misztal, I.; Lourenco, D. Changes in genetic trends in US dairy cattle since the implementation of genomic selection. J. Dairy Sci. 2023, 106, 1110–1129. [Google Scholar] [CrossRef]
- Gutierrez-Reinoso, M.A.; Aponte, P.M.; Garcia-Herreros, M. Genomic Analysis, Progress and Future Perspectives in Dairy Cattle Selection: A Review. Animals 2021, 11, 599. [Google Scholar] [CrossRef]
- Pryce, J.E.; Egger-Danner, C.; Simm, G. Strategies and Tools for Genetic Selection in Dairy Cattle and Their Application to Improving Animal Welfare. In Cattle Welfare in Dairy and Beef Systems: A New Approach to Global Issues; Haskell, M., Ed.; Springer International Publishing: Cham, Switzerland, 2023; pp. 323–348. [Google Scholar]
- Wiggans, G.R.; Carrillo, J.A. Genomic selection in United States dairy cattle. Front. Genet. 2022, 13, 994466. [Google Scholar] [CrossRef]
- Consortium, R.D.; Fugeray-Scarbel, A.; Bastien, C.; Dupont-Nivet, M.; Lemarié, S. Why and How to Switch to Genomic Selection: Lessons From Plant and Animal Breeding Experience. Front. Genet. 2021, 12, 629737. [Google Scholar] [CrossRef]
- Knol, E.F.; van der Spek, D.; Zak, L.J. Genetic aspects of piglet survival and related traits: A review. J. Anim. Sci. 2022, 100, skac190. [Google Scholar] [CrossRef] [PubMed]
- Kalds, P.; Zhou, S.; Gao, Y.; Cai, B.; Huang, S.; Chen, Y.; Wang, X. Genetics of the phenotypic evolution in sheep: A molecular look at diversity-driving genes. Genet. Sel. Evol. 2022, 54, 61. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Enciso, M.; Steibel, J.P. Phenomes: The current frontier in animal breeding. Genet. Sel. Evol. 2021, 53, 22. [Google Scholar] [CrossRef]
- Larkina, T.A.; Barkova, O.Y.; Peglivanyan, G.K.; Mitrofanova, O.V.; Dementieva, N.V.; Stanishevskaya, O.I.; Vakhrameev, A.B.; Makarova, A.V.; Shcherbakov, Y.S.; Pozovnikova, M.V.; et al. Evolutionary Subdivision of Domestic Chickens: Implications for Local Breeds as Assessed by Phenotype and Genotype in Comparison to Commercial and Fancy Breeds. Agriculture 2021, 11, 914. [Google Scholar] [CrossRef]
- Andersson, L.; Purugganan, M. Molecular genetic variation of animals and plants under domestication. Proc. Natl. Acad. Sci. USA 2022, 119, e2122150119. [Google Scholar] [CrossRef]
- Hidalgo, J.; Tsuruta, S.; Gonzalez, D.; de Oliveira, G.; Sanchez, M.; Kulkarni, A.; Przybyla, C.; Vargas, G.; Vukasinovic, N.; Misztal, I.; et al. Converting estimated breeding values from the observed to probability scale for health traits. J. Dairy Sci. 2024, 107, 9628–9637. [Google Scholar] [CrossRef]
- Mrode, R.A.; Pocrnic, I. Solving Linear Equations. CABI 2023, 19, 314–341. [Google Scholar] [CrossRef]
- Fathoni, A.; Boonkum, W.; Chankitisakul, V.; Duangjinda, M. An Appropriate Genetic Approach for Improving Reproductive Traits in Crossbred Thai–Holstein Cattle under Heat Stress Conditions. Vet. Sci. 2022, 9, 163. [Google Scholar] [CrossRef]
- Tesema, Z.; Deribe, B.; Lakew, M.; Getachew, T.; Tilahun, M.; Belayneh, N.; Kefale, A.; Shibesh, M.; Zegeye, A.; Yizengaw, L.; et al. Genetic and non-genetic parameter estimates for growth traits and Kleiber ratios in Dorper × indigenous sheep. Animal 2022, 16, 100533. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Yin, D.; Fu, Y.; Yuan, X.; Li, X.; Liu, X.; Zhao, S. HIBLUP: An integration of statistical models on the BLUP framework for efficient genetic evaluation using big genomic data. Nucleic Acids Res. 2023, 51, 3501–3512. [Google Scholar] [CrossRef]
- Silva Neto, J.B.; Mota, L.F.M.; Amorim, S.T.; Peripolli, E.; Brito, L.F.; Magnabosco, C.U.; Baldi, F. Genotype-by-environment interactions for feed efficiency traits in Nellore cattle based on bi-trait reaction norm models. Genet. Sel. Evol. 2023, 55, 93. [Google Scholar] [CrossRef] [PubMed]
- Tiezzi, F.; Maltecca, C. Genotype by Environment Interactions in Livestock Farming. In Animal Breeding and Genetics; Springer: Berlin/Heidelberg, Germany, 2023; pp. 77–97. [Google Scholar] [CrossRef]
- Wolf, J.; Zavadilová, L.; Němcová, E. Non-additive effects on milk production in Czech dairy cows. J. Anim. Breed. Genet. 2005, 122, 332–339. [Google Scholar] [CrossRef]
- Diaz, J.R. Aspectos Genéticos da Produção de Leite e Seus Constituíntes em Búfalas Mestiças. Master’s Thesis, Universidade Estadual Paulista Julio de Mesquita Filho, Jaboticabal, São Paulo, 2010. [Google Scholar]
- Silva Neto, J.B.; Mota, L.F.M.; Londoño-Gil, M.; Schmidt, P.I.; Rodrigues, G.R.D.; Ligori, V.A.; Arikawa, L.M.; Magnabosco, C.U.; Brito, L.F.; Baldi, F. Genotype-by-environment interactions in beef and dairy cattle populations: A review of methodologies and perspectives on research and applications. Anim. Genet. 2024, 55, 871–892. [Google Scholar] [CrossRef] [PubMed]
- Wahinya, P.K.; Jeyaruban, G.M.; Swan, A.A.; van der Werf, J.H.J. Optimization of Dairy Cattle Breeding Programs with Genotype by Environment Interaction in Kenya. Agriculture 2022, 12, 1274. [Google Scholar] [CrossRef]
- Berry, D.P.; Hilliard, B.; McCarthy, J.; Kennedy, E. Exploring the presence of genotype-by-environment interactions between dairy cow herds milking once-a-day or twice-a-day for the entire lactation. Ir. J. Agric. Food Res. 2024, 62, 146–155. [Google Scholar] [CrossRef]
- Sampaio Filho, J.S.; Oliveira, I.C.M.; Pastina, M.M.; Campos, M.d.S.; de Oliveira, E.J. Genotype x environment interaction in cassava multi-environment trials via analytic factor. PLoS ONE 2024, 19, e0315370. [Google Scholar] [CrossRef]
- Hasan, M.M.; Thomson, P.C.; Raadsma, H.W.; Khatkar, M.S. A Review and Meta-Analysis of Genotype by Environment Interaction in Commercial Shrimp Breeding. Genes 2024, 15, 1222. [Google Scholar] [CrossRef]
- Martins, R.; Nascimento, B.M.; Valloto, A.A.; Carvalheiro, R.; de Albuquerque, L.G.; de Almeida Teixeira, R.; Dias, L.T. Influence of different environmental challenges on the expression of reproductive traits in Holstein cattle in Southern Brazil. Trop. Anim. Health Prod. 2024, 56, 288. [Google Scholar] [CrossRef]
- Alves, F.C.; Galli, G.; Matias, F.I.; Vidotti, M.S.; Morosini, J.S.; Fritsche-Neto, R. Impact of the complexity of genotype by environment and dominance modeling on the predictive accuracy of maize hybrids in multi-environment prediction models. Euphytica 2021, 217, 37. [Google Scholar] [CrossRef]
- Mumford, M.H.; Forknall, C.R.; Rodriguez, D.; Eyre, J.X.; Kelly, A.M. Incorporating environmental covariates to explore genotype × environment × management (G × E × M) interactions: A one-stage predictive model. Field Crops Res. 2023, 304, 109133. [Google Scholar] [CrossRef]
- Misztal, I.; Tsuruta, S.; Lourenco, D.; Masuda, Y.; Aguilar, I.; Legarra, A.; Vitezica, Z.; Ensat, F. Manual for BLUPF90 Family of Programs; UGA Animal Breeding and Genetics Group: Athens, GA, USA, 2018; p. 142. [Google Scholar]
- Geweke, J.F. Evaluating the Accuracy of Sampling-Based Approaches to the Calculation of Posterior Moments; Federal Reserve Bank of Minneapolis: Oxford, UK, 1991; pp. 169–193. [Google Scholar]
- Lozada-Soto, E.A.; Maltecca, C.; Lu, D.; Miller, S.; Cole, J.B.; Tiezzi, F. Trends in genetic diversity and the effect of inbreeding in American Angus cattle under genomic selection. Genet. Sel. Evol. 2021, 53, 50. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Saharan, P.; Dhaka, S.S.; Patil, C.S.; Sharma, R. Genetic and Phenotypic Trends along with Genetic Parameters of Production Traits in Murrah Buffalo. J. Sci. Res. Rep. 2024, 30, 476–486. [Google Scholar] [CrossRef]
- Suhardi, S.; Summpunn, P.; Duangjinda, M.; Wuthisuthimethavee, S. Buffalo (Bubalus bubalis) phenotypic diversity characterization reveals the need for improved performance based on quantitative and qualitative characteristics: A comparison of Kalang buffalo in Kalimantan, Indonesia and Thale Noi buffalo in Phatthalung, Thailand. Buffalo Bull. 2022, 41, 373–390. [Google Scholar] [CrossRef]
- Medrado, B.D.; Pedrosa, V.B.; Pinto, L.F.B. Meta-analysis of genetic parameters for economic traits in buffaloes. Livest. Sci. 2021, 251, 104614. [Google Scholar] [CrossRef]
- Mota-Rojas, D.; Bragaglio, A.; Braghieri, A.; Napolitano, F.; Domínguez-Oliva, A.; Mora-Medina, P.; Álvarez-Macías, A.; De Rosa, G.; Pacelli, C.; José, N.; et al. Dairy Buffalo Behavior: Calving, Imprinting and Allosuckling. Animals 2022, 12, 2899. [Google Scholar] [CrossRef]
- Al Kalaldeh, M.; Swaminathan, M.; Gaundare, Y.; Joshi, S.; Aliloo, H.; Strucken, E.M.; Ducrocq, V.; Gibson, J.P. Genomic evaluation of milk yield in a smallholder crossbred dairy production system in India. Genet. Sel. Evol. 2021, 53, 73. [Google Scholar] [CrossRef]
- Mancin, E.; Tuliozi, B.; Sartori, C.; Guzzo, N.; Mantovani, R. Genomic Prediction in Local Breeds: The Rendena Cattle as a Case Study. Animals 2021, 11, 1815. [Google Scholar] [CrossRef]
- El Sabry, M.I.; Almasri, O. Space allowance: A tool for improving behavior, milk and meat production, and reproduction performance of buffalo in different housing systems—A review. Trop. Anim. Health Prod. 2022, 54, 266. [Google Scholar] [CrossRef] [PubMed]
- Arana, Á.J.; Sánchez, L. Knockout, Knockdown, and the Schrödinger Paradox: Genetic Immunity to Phenotypic Recapitulation in Zebrafish. Genes 2024, 15, 1164. [Google Scholar] [CrossRef]
- Shao, B.; Sun, H.; Ahmad, M.J.; Ghanem, N.; Abdel-Shafy, H.; Du, C.; Deng, T.; Mansoor, S.; Zhou, Y.; Yang, Y.; et al. Genetic Features of Reproductive Traits in Bovine and Buffalo: Lessons From Bovine to Buffalo. Front. Genet. 2021, 23, 617128. [Google Scholar] [CrossRef]
- Rehman, S.u.; Hassan, F.-u.; Luo, X.; Li, Z.; Liu, Q. Whole-Genome Sequencing and Characterization of Buffalo Genetic Resources: Recent Advances and Future Challenges. Animals 2021, 11, 904. [Google Scholar] [CrossRef]
- Gómez, M.; Rossi, D.; Cimmino, R.; Zullo, G.; Gombia, Y.; Altieri, D.; Di Palo, R.; Biffani, S. Accounting for Genetic Differences Among Unknown Parents in Bubalus bubalis: A Case Study From the Italian Mediterranean Buffalo. Front. Genet. 2021, 12, 625335. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, V.B.; Boerman, J.P.; Gloria, L.S.; Chen, S.-Y.; Montes, M.E.; Doucette, J.S.; Brito, L.F. Genomic-based genetic parameters for milkability traits derived from automatic milking systems in North American Holstein cattle. J. Dairy Sci. 2023, 106, 2613–2629. [Google Scholar] [CrossRef] [PubMed]
- Mulim, H.A.; Brito, L.F.; Pinto, L.F.B.; Ferraz, J.B.S.; Grigoletto, L.; Silva, M.R.; Pedrosa, V.B. Characterization of runs of homozygosity, heterozygosity-enriched regions, and population structure in cattle populations selected for different breeding goals. BMC Genom. 2022, 23, 209. [Google Scholar] [CrossRef]
- Macciotta, N.P.P.; Colli, L.; Cesarani, A.; Ajmone-Marsan, P.; Low, W.Y.; Tearle, R.; Williams, J.L. The distribution of runs of homozygosity in the genome of river and swamp buffaloes reveals a history of adaptation, migration and crossbred events. Genet. Sel. Evol. 2021, 53, 20. [Google Scholar] [CrossRef]
- Lázaro, S.F.; Tonhati, H.; Oliveira, H.R.; Silva, A.A.; Nascimento, A.V.; Santos, D.J.A.; Stefani, G.; Brito, L.F. Genomic studies of milk-related traits in water buffalo (Bubalus bubalis) based on single-step genomic best linear unbiased prediction and random regression models. J. Dairy Sci. 2021, 104, 5768–5793. [Google Scholar] [CrossRef]
- Colangelo, P.; Di Civita, M.; Bento, C.M.; Franchini, P.; Meyer, A.; Orel, N.; das Neves, L.C.B.G.; Mulandane, F.C.; Almeida, J.S.; Senczuk, G.; et al. Genome-wide diversity, population structure and signatures of inbreeding in the African buffalo in Mozambique. BMC Ecol. Evol. 2024, 24, 29. [Google Scholar] [CrossRef]
- Petrocchi Jasinski, F.; Evangelista, C.; Basiricò, L.; Bernabucci, U. Responses of Dairy Buffalo to Heat Stress Conditions and Mitigation Strategies: A Review. Animals 2023, 13, 1260. [Google Scholar] [CrossRef] [PubMed]
- Siraj, M.; Ibrahim, M.; Sabiha, B.; Ahmad, S. Evaluation of Lipin 1 polymorphisms for genetic markers in association with performance traits in Azikheli buffalo. Reprod. Breed. 2024, 4, 61–72. [Google Scholar] [CrossRef]
- Chen, S.Y.; Boerman, J.P.; Gloria, L.S.; Pedrosa, V.B.; Doucette, J.; Brito, L.F. Genomic-based genetic parameters for resilience across lactations in North American Holstein cattle based on variability in daily milk yield records. J. Dairy Sci. 2023, 106, 4133–4146. [Google Scholar] [CrossRef]
- Gómez-Carpio, M.; Cesarani, A.; Zullo, G.; Cimmino, R.; Neglia, G.; Campanile, G.; Biffani, S. Genetic parameters for reproductive traits in the Italian Mediterranean buffalo using milk yield as a correlated trait. J. Dairy Sci. 2023, 106, 9016–9025. [Google Scholar] [CrossRef]
- Ozturk, N.; Kocak, O.; Peker, A.; Serva, L.; Kaygisiz, F.; Kecici, P.D.; Yalcintan, H.; Kilic, H.I.; Magrin, L. Characteristics of Buffalo Farming Systems in Turkey Based on a Multivariate Aggregation of Indicators: A Survey Study. Animals 2022, 12, 3056. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, F.B.; Malhado, C.H.M.; Carneiro, P.L.S.; Ambrosini, D.P.; Rezende, M.P.G.; Bozzi, R.; Song, J. Genotype by environment interactions for body weight in Mediterranean buffaloes using reaction norm models. Rev. Colomb. Cienc. Pecu. 2021, 34, 166–176. [Google Scholar] [CrossRef]
- Silva, A.A.; Brito, L.F.; Silva, D.A.; Lazaro, S.F.; Silveira, K.R.; Stefani, G.; Tonhati, H. Random regression models using B-splines functions provide more accurate genomic breeding values for milk yield and lactation persistence in Murrah buffaloes. J. Anim. Breed. Genet. 2023, 140, 167–184. [Google Scholar] [CrossRef] [PubMed]
- Bayssa, M.; Yigrem, S.; Betsha, S.; Tolera, A. Production, reproduction and some adaptation characteristics of Boran cattle breed under changing climate: A systematic review and meta-analysis. PLoS ONE 2021, 16, e0244836. [Google Scholar] [CrossRef]
- Valente, G.F.; Ferraz, G.A.e.S.; Santana, L.S.; Ferraz, P.F.P.; Mariano, D.d.C.; dos Santos, C.M.; Okumura, R.S.; Simonini, S.; Barbari, M.; Rossi, G. Mapping Soil and Pasture Attributes for Buffalo Management through Remote Sensing and Geostatistics in Amazon Biome. Animals 2022, 12, 2374. [Google Scholar] [CrossRef]
- Ünal, E.Ö.; Işık, R.; Şen, A.; Geyik Kuş, E.; Soysal, M.İ. Evaluation of Genetic Diversity and Structure of Turkish Water Buffalo Population by Using 20 Microsatellite Markers. Animals 2021, 11, 1067. [Google Scholar] [CrossRef]
- Freese, J. Genetics and the social science explanation of individual outcomes. Am. J. Sociol. 2008, 114, S1–S35. [Google Scholar] [CrossRef]
- Burns, J.G.; Glenk, K.; Eory, V.; Simm, G.; Wall, E. Preferences of European dairy stakeholders in breeding for resilient and efficient cattle: A best-worst scaling approach. J. Dairy Sci. 2022, 105, 1265–1280. [Google Scholar] [CrossRef]
- Sahana, G.; Cai, Z.; Sanchez, M.P.; Bouwman, A.C.; Boichard, D. Invited review: Good practices in genome-wide association studies to identify candidate sequence variants in dairy cattle. J. Dairy Sci. 2023, 106, 5218–5241. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.-L.; Wiggans, G.R.; Norman, H.D.; Caputo, M.J.; Miles, A.M.; Van Tassell, C.P.; Baldwin, R.L.; Sievert, S.; Mattison, J.; Burchard, J.; et al. Updating test-day milk yield factors for use in genetic evaluations and dairy production systems: A comprehensive review. Front. Genet. 2023, 14, 1298114. [Google Scholar] [CrossRef] [PubMed]
- Dahiyta, S.; Kumar, M.; Ratwan, P.; Dhillod, S. Genetic analysis of biometric traits in Murrah buffaloes using Bayesian approach. Turk. J. Vet. Anim. Sci. 2022, 46, 285–292. [Google Scholar] [CrossRef]
- Salem, M.M.I.; Amin, A.M.S.; Ashour, A.F.; El Nagar, A.G. Estimation of genetic parameters for semen traits in Egyptian buffalo bulls. Trop. Anim. Health Prod. 2023, 55, 264. [Google Scholar] [CrossRef]
| Class of SD | NCG | N | Mean | SD | CV(%) | Confidence Interval (0.95) |
|---|---|---|---|---|---|---|
| Low | 91 | 1562 | 2071.54 | 449.79 | 21.71 | [1977.91; 2165.17] |
| High | 70 | 830 | 2642.69 | 594.28 | 22.48 | [2501.04; 2784.34] |
| General | 161 | 2392 | 2269.72 | 573.13 | 25.25 | [2180.56; 2358.88] |
| Class of Standard Deviation | ||||
|---|---|---|---|---|
| Parameters | Low | High | General | |
| σ2a | Mean ± SD | 353.51 ± 0.57 | 1134.22 ±1.59 | 472.47 ± 0.54 |
| Median | 347.70 | 1119 | 467.60 | |
| CI | 178 a 543.30 | 637.30 a 1656 | 303.30 a 651.80 | |
| Geweke | 0.01 | −0.01 | 0.01 | |
| σ2e | Mean ± SD | 1480.62 ± 0.45 | 2180.15 ± 0.97 | 1804.00 ± 0.41 |
| Median | 1478 | 2174 | 1803 | |
| CI | 1342 a 1633 | 1880 a 2503 | 1670 a 1940 | |
| Geweke | 0.001 | 0.01 | −0.01 | |
| h2 | 0.19 ± 0.0004 | 0.34 ± 0.0003 | 0.21 ± 0.0002 | |
| rg | 0.61 ± 0.001 | |||
| N = 77 (100%) | ||||
|---|---|---|---|---|
| Class of Standard Deviation | ||||
| Low | High | General | ||
| N = 33 (43%) | Low | 1 | 0.87 | 0.95 |
| High | 0.78 | 1.00 | 0.73 | |
| General | 0.87 | 0.51 | 1.00 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Camargo Júnior, R.N.C.; de Araújo, C.V.; Marques, J.R.F.; Bonin Gomes, M.d.N.; da Silva, W.C.; Belo, T.S.; Sousa, C.E.L.; da Silva, É.B.R.; Marques, L.C.; da Silva, M.M.; et al. Heterogeneity of Variances in Milk Yield in Murrah Buffaloes. Animals 2025, 15, 2686. https://doi.org/10.3390/ani15182686
Camargo Júnior RNC, de Araújo CV, Marques JRF, Bonin Gomes MdN, da Silva WC, Belo TS, Sousa CEL, da Silva ÉBR, Marques LC, da Silva MM, et al. Heterogeneity of Variances in Milk Yield in Murrah Buffaloes. Animals. 2025; 15(18):2686. https://doi.org/10.3390/ani15182686
Chicago/Turabian StyleCamargo Júnior, Raimundo Nonato Colares, Cláudio Vieira de Araújo, José Ribamar Felipe Marques, Marina de Nadai Bonin Gomes, Welligton Conceição da Silva, Tatiane Silva Belo, Carlos Eduardo Lima Sousa, Éder Bruno Rebelo da Silva, Larissa Coelho Marques, Mauro Marinho da Silva, and et al. 2025. "Heterogeneity of Variances in Milk Yield in Murrah Buffaloes" Animals 15, no. 18: 2686. https://doi.org/10.3390/ani15182686
APA StyleCamargo Júnior, R. N. C., de Araújo, C. V., Marques, J. R. F., Bonin Gomes, M. d. N., da Silva, W. C., Belo, T. S., Sousa, C. E. L., da Silva, É. B. R., Marques, L. C., da Silva, M. M., Picanço, M. L. R., Lourenço-Júnior, J. d. B., Santos, A. M., de Oliveira, A. S., Cara, J. R. F., & Silva, A. G. M. e. (2025). Heterogeneity of Variances in Milk Yield in Murrah Buffaloes. Animals, 15(18), 2686. https://doi.org/10.3390/ani15182686









