1. Introduction
The large yellow croaker (
Larimichthys crocea), a member of the Sciaenidae family, is primarily distributed in the southern Yellow, East, and northern South Seas in China [
1,
2,
3,
4]. Valued for its delicate flesh, this species has become a highly sought-after food fish and is one of the most economically important marine fish in China [
5,
6,
7]. Previous studies have classified
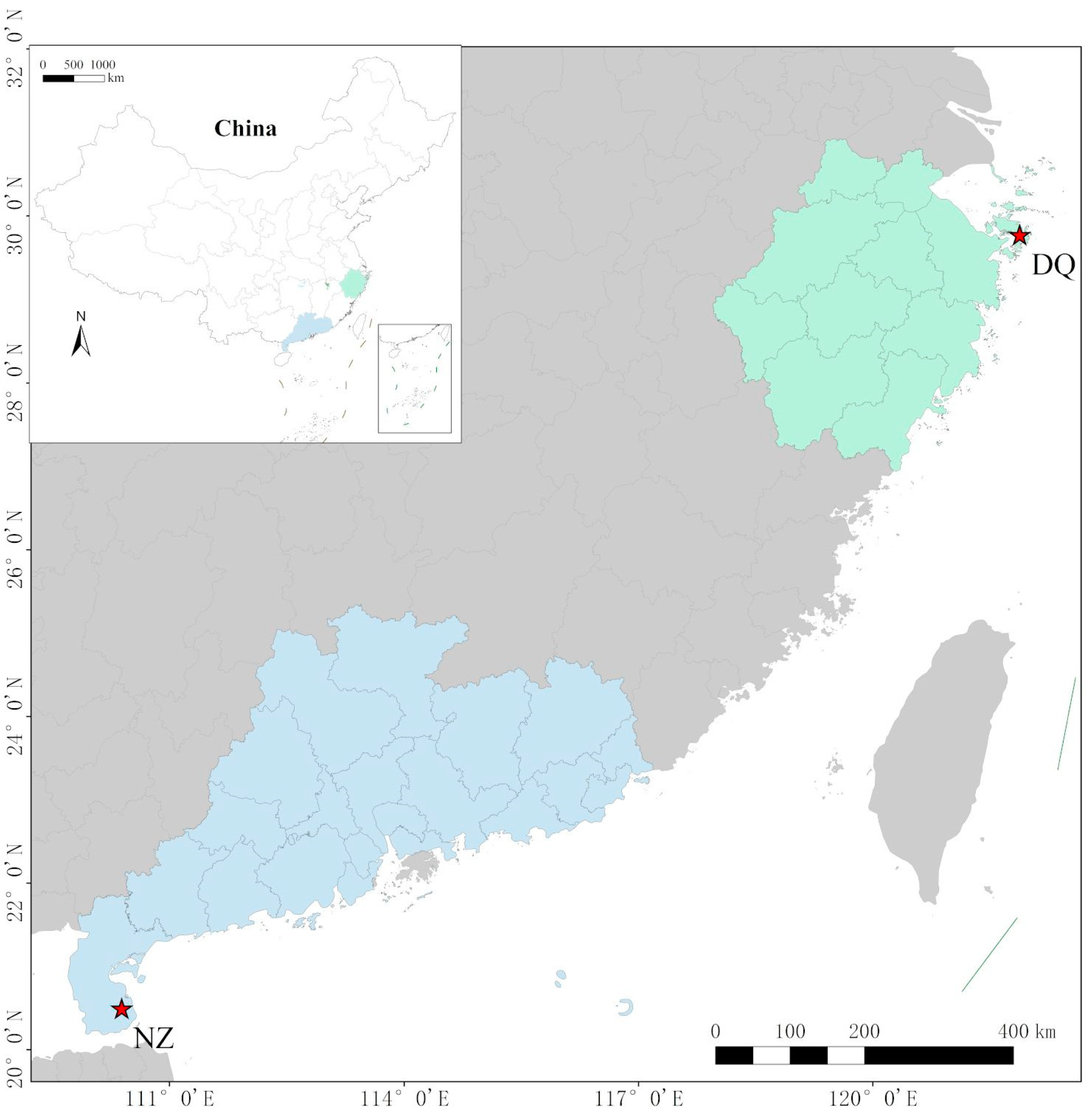
L. crocea into three distinct geographical stocks: the Dai-qu stock (from the southern Yellow Sea to the central East Sea), the Min-Yuedong stock (from the southeastern East Sea to the northern South Sea), and the Naozhou stock (from the west of the Pearl River Estuary to the Qiongzhou Strait in the South Sea) [
8,
9,
10]. The Naozhou stock exhibits distinct phenotypic characteristics that set it apart from the eastern coastal stocks and retains many traits typical of wild populations, demonstrating a richer genetic diversity. Compared with other stocks, individuals from the Naozhou stock show slower yet more stable growth rates, which are advantageous for selective breeding under intensive aquaculture conditions. Their deeper body coloration and brighter skin are highly favored in the consumer market, contributing to increased commercial value. Moreover, this stock demonstrates enhanced tolerance to environmental stressors such as temperature fluctuations and low dissolved oxygen, which is likely attributable to its preservation of wild-type genetic diversity characteristics. In addition, the Naozhou stock possesses thicker muscle fibers and a firmer flesh texture, which improve sensory quality [
1,
2,
3,
7]. Delayed sexual maturation has also been observed, potentially extending the growth period before spawning and enhancing feed conversion efficiency. These characteristics suggest that the Naozhou stock holds valuable characteristics often lost in intensively farmed populations. Its preservation could play a key role in selective breeding programs aimed at enhancing resilience, flesh quality, and adaptability to variable marine environments. However, the Naozhou stock has experienced a marked decline in population size. Therefore, developing a breeding program that enhances traits suitable for deep sea and offshore aquaculture is essential for advancing the large yellow croaker farming industry and expanding it into more complex marine environments.
Microsatellites, also referred to as simple sequence repeats (SSRs) or short tandem repeats (STRs), are DNA sequences composed of tandemly repeated units of 2–6 base pairs (bp), flanked by unique but conserved sequences within populations [
11]. Owing to their abundance, polymorphism, co-dominance, strong repeatability, and widespread distribution throughout the genome, microsatellites have become valuable molecular markers [
12]. They are widely applied in population genetics, phylogenetic analysis, germplasm identification, genotyping, and the construction of genetic linkage maps, particularly in aquatic species [
13]. Despite their utility, research on microsatellites in large yellow croakers remains limited. The lack of genomic research has impeded the effective management and utilization of this species’ genetic diversity. Consequently, the identification and characterization of additional highly polymorphic and stable microsatellite loci from the
L. crocea genome are urgently needed. Such markers contribute to the genetic improvement and conservation in the large yellow croaker aquaculture industry.
A genome encompasses both functional and non-functional DNA sequences that define an organism’s biological identity [
14]. In recent decades, the advancement of high-throughput sequencing technologies has accelerated genome sequencing efforts across taxa [
15]. As of December 2019, approximately 270 fish genomes had been assembled and made publicly available through the NCBI Genome database, supporting research in comparative genomics, systematics, and aquaculture. With over 34,000 fish species recorded in FishBase, large-scale initiatives like the Earth BioGenome Project are making comprehensive genome sequencing of fish species increasingly feasible, enabling deeper insights into their biology, evolution, and utility in sustainable fisheries and aquaculture [
16,
17,
18,
19,
20].
Genome-wide survey sequencing (GSS), based on high-throughput sequencing technology, offers a rapid and efficient approach for generating a global perspective for high-quality genome assembly. It also serves as a fundamental tool for low-depth sequencing in non-model species that lack reference genomes [
7,
21,
22]. In aquaculture genomics, the identification of genetic determinants of key production and performance traits is central to advancing selective breeding programs. This field has been widely discussed in review papers [
23,
24], conference proceedings [
25,
26], and books [
18,
27]. Whole-genome sequencing (WGS) has been widely applied to a variety of aquatic fish species, including the
Cyprinus carpio [
28],
Platycephalus sp.1 [
29],
Muraenolepis orangiensis [
30],
Paralichthys orbignyanus [
31],
Pampus spp. [
32], and
Chionobathyscus dewitti [
33]. WGS enables the characterization of essential genomic features such as genome size, heterozygosity levels, repeat sequence content, and guanine–cytosine (GC) content. Additionally, the resulting genomic data support the development of genome-wide microsatellite (SSR) markers and the assembly of mitochondrial genome (mtDNA), as demonstrated in
Platycephalus sp.1 and
Acanthocepola indica [
34].
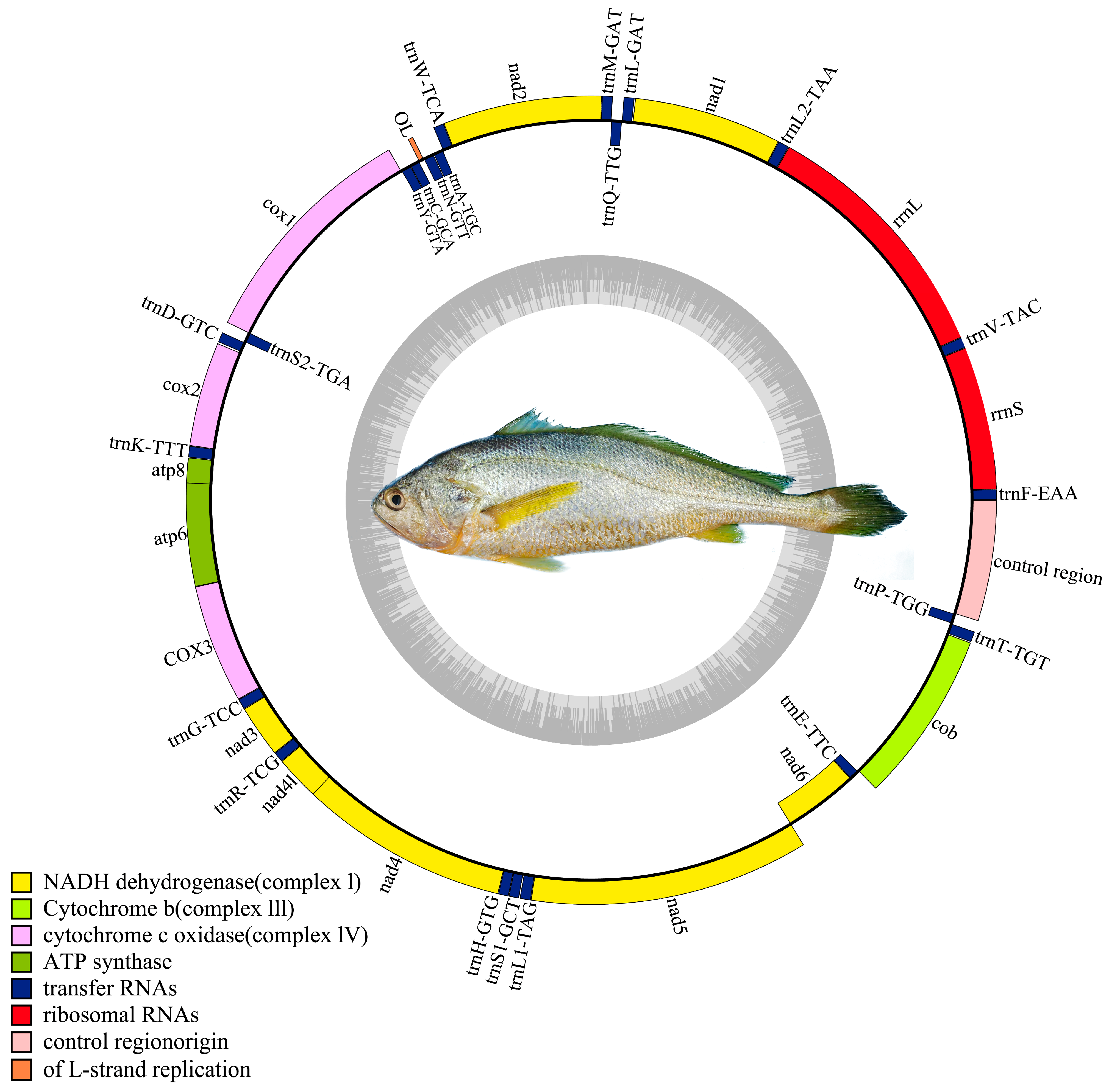
The mtDNA is a circular, double-stranded molecule typically composed of 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a control region [
35,
36,
37]. However, genomic and mtDNA information specific to the Naozhou yellow croaker remains unavailable. To date, only nucleotide sequences from the Dai-qu and Min-Yuedong stocks are available in the GenBank database (
www.ncbi.nlm.nih.gov/genbank/) (accessed on 18 December 2024). This lack of genomic resources limits the implementation of effective genetic breeding strategies and conservation efforts for the Naozhou stock of large yellow croakers. Therefore, a comprehensive genomic investigation of this stock is essential for expanding the genetic resource database of the species, facilitating marker-assisted breeding and optimizing aquaculture.
In this study, we conducted the first GSS of the Naozhou yellow croaker using DNBseq technology. Key genomic features, including genome size, GC content, and heterozygosity, were estimated and analyzed. Additionally, genome-wide SSRs were identified and applied to assess the population structure across two L. crocea stocks. The mtDNA of the Naozhou stock was also assembled, and its PCGs were analyzed. These genomic resources provide valuable data for future studies on the genetic breeding and population genetics of the Naozhou stock of L. crocea.
4. Discussion
The advancement of next-generation sequencing technologies has made it more accessible for researchers to explore a wide range of genome-related biological questions, particularly in non-model species [
44,
58]. WGS data enable the estimation of key genomic characteristics, including genome size, heterozygosity ratio, and repeat ratio, using bioinformatics approaches without requiring prior knowledge [
59]. The comprehensive whole-genome survey and analysis of the Naozhou stock of large yellow croakers have provided valuable insights into its genomic characteristics, genetic diversity, and potential applications in aquaculture optimization and genetic conservation. This study successfully identified genome-wide SSR markers and assembled the mtDNA, which are critical for genetic evaluation, selective breeding, and conservation. In recent years, microsatellite markers have gained widespread application across various fields, including studies of genetic diversity [
60,
61], marker-assisted breeding [
62], gene mapping, and quantitative trait loci (QTL) analysis [
3,
28,
63,
64].
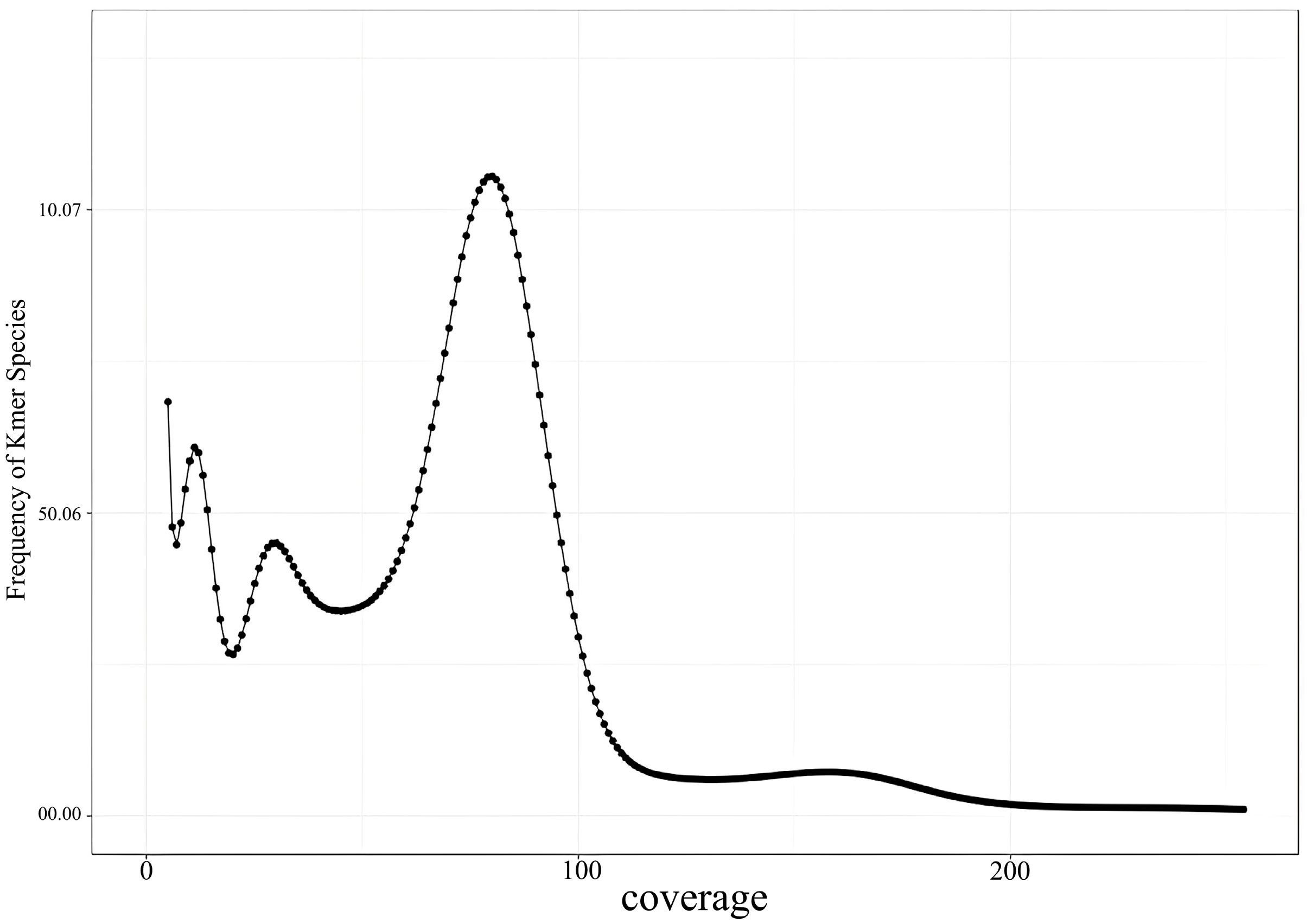
The K-mer analysis conducted in this study revealed the genome size of the Naozhou large yellow croaker to be approximately 677.78 Mb, which is smaller than that of other marine fish species, including the
C. dewitti (880 Mb) [
33],
Morone saxatilis (797 Mb) [
65]
Anthias nicholsi (815 Mb) (Liu et al., 2024) [
66], and
Clupea harengus (850 Mb) [
67] but similar to that of
Sardina pilchardus (625–637 Mb) [
68] and
Trachinotus ovatus (642.68 Mb) [
36] The size and variability of eukaryotic genomes are influenced by various factors, including mutation pressure, transposon activity, genome ploidy, biological life history traits, and environmental conditions [
69,
70]. Larger genomes are generally associated with longer evolutionary histories and a higher risk of extinction [
71]. The repeat sequence proportion in the genome of the Naozhou large yellow croaker was 22.181%, which was considered medium-low, lower than that of the
Chiloscyllium plagiosum (63.53%) [
72],
Hemitripterus villosus (38.61%) [
73], and
Trachinotus carolinus (30.19%) [
74], but similar to the
Ameiurus nebulosus (39.65%) [
44],
M. saxatilis (39.22%) [
65], and
A. nicholsi (39.69%) [
66]. The observed genome heterozygosity and repeated content suggest that the Naozhou large yellow croaker retains genetic traits that may confer advantages for adaptation and resilience in natural and aquaculture environments. These results suggest that the Naozhou stock exhibits moderate genome complexity, with a relatively lower repeat sequence proportion compared to other marine fish species.
The proportion of repeat sequences in a genome is crucial for designing genome sequencing strategies, as it facilitates the selection of appropriate genome assembly methods. The GC content in most fish species typically ranges from 40% to 46% [
75,
76,
77]. For the large yellow croaker, the heterozygosity rate was 0.839%, with a GC content of 41.47%. This value is lower than that of the
Hemitripterus villosus (43.13%) [
73] and the icefish
C. dewitti (49.9%) [
33], but comparable to that of the
sebastiscus marmoratus (41.3%) [
44],
Acanthogobius omaturus (40.88%) [
76], and
Acanthopagrus latus (42.07%) [
78]. The GC content of 41.47% falls within the typical range for fish species (40–46%), indicating a stable genome composition. Additionally, the absence of contamination and high sequence quality (Q20: 98.14%; Q30: 93.96%) ensures the reliability of subsequent analyses.
This study identifies a high-density panel of 195,263 SSR markers in the Naozhou stock genome, providing a valuable resource for genetic applications. The relative abundance (288 loci/Mb) and high polymorphism rate, particularly of AC/GT dinucleotide repeats, are comparable to or exceed those reported in related species such as
Harpadon nehereus,
Synbranchus marmoratus,
Gadus macrocephalus,
Pogonophryne albipinna,
Siganus oramin, and
Acanthogobius ommaturus [
23,
29,
75,
76,
79,
80,
81]. These SSRs, especially the 28 primer pairs with a high amplification success rate (93.3%), provide a solid foundation for constructing high-resolution genetic maps, which are essential for marker-assisted selection (MAS) in aquaculture [
23]. This genome-wide SSR development marks a considerable improvement over earlier studies that employed traditional methods for SSR identification, typically relying on expressed sequence tag (EST) libraries or limited genomic libraries. For example, previous works such as Zhang et al. [
3] employed mitochondrial COI sequences to study population structure, which, while useful for phylogeographic inference, lacked the resolution and co-dominant inheritance pattern of SSRs. Additionally, the Naozhou stock exhibited high genetic diversity (Na = 4–22; He up to 0.9238), exceeding that of the Dai-qu stock. This suggests strong adaptive potential and resilience, which are vital for selective breeding under diverse aquaculture conditions. The PIC values, averaging 0.718, indicate a robust capacity to discriminate between individuals, a prerequisite for effective parentage analysis, QTL mapping, and population structure assessments [
45,
82]. In Chen’s study on the heat resistance of large yellow croakers (
L. crocea), all three microsatellite markers associated with thermal tolerance contained AC as their repeat motifs, which is consistent with the findings of the present study (thermal tolerance evaluation and related microsatellite marker screening and identification in the large yellow croaker (
L. crocea)). With increasing fishing pressure, Wang et al. conducted a microsatellite analysis of both wild and cultured populations of
L. crocea and found that the cultured populations exhibited lower genetic diversity compared to wild populations, further underscoring the importance of conserving wild genetic resources (loss of genetic diversity in the cultured stocks of the large yellow croaker,
L. crocea, revealed by microsate).
The high-frequency AC/GT motifs are not merely statistical artifacts; these motifs are associated with regulatory regions involved in gene expression and chromatin structure [
38]. Their prevalence may indicate genomic regions of evolutionary and functional importance in
L. crocea, particularly under environmental pressures such as salinity and temperature fluctuations common in offshore farming. In comparison to other marine teleosts, the SSR composition of the Naozhou genome aligns with broader trends in fish genomics but also reveals stock-specific signatures. For example, while trinucleotide repeats dominate in species such as
Dicentrarchus labrax,
Salmo salar, and
Takifugu rubripes, the Naozhou stock is characterized by a predominance of dinucleotide SSRs. This divergence may reflect distinct evolutionary histories and selection pressures, potentially linked to ecological niches or demographic events [
83]. By establishing a dense SSR marker panel for the previously under-studied Naozhou stock, this study addresses a critical gap in marine aquaculture. These markers not only function as genetic barcodes but also facilitate lineage tracking, inbreeding monitoring, and adaptive capacity assessment. This directly supports long-term sustainability in breeding programs and biodiversity conservation [
25,
84].
Moreover, microsatellite loci from earlier studies were often limited in number and polymorphism, with reported allele numbers typically ranging from 3 to 8 [
3,
85]. Furthermore, while genome-wide approaches have been used to examine stress adaptation in
L. crocea, such as in Ao et al. [
86], who explored the molecular responses to hypoxia and thermal stress, these studies did not focus on the systematic development of polymorphic SSRs. Similarly, Xu et al. [
87] characterized the hsp70 gene family under cold and heat stress conditions but did not provide transferable genetic markers for population studies or breeding programs. The SSR markers developed in this study are directly representative of this wild-type diversity and thus have high relevance for selective breeding, conservation, and population structure monitoring in both hatchery and natural settings. Additionally, the newly developed SSRs are positioned to overcome the limitations of earlier markers. For instance, SSRs used in population differentiation studies like those by Zhang et al. [
3] and Kon et al. [
85] often suffered from poor genome coverage and limited polymorphism, which constrained their ability to capture fine-scale genetic structure or support genome-wide association studies (GWASs). In contrast, the current study provides a genome-wide inventory with comprehensive coverage, higher repeat motif diversity (dominated by AC dinucleotides), and validated primer sets that exhibit strong amplification and polymorphic potential. To further enhance the contribution of this study, the newly developed SSR loci should be compared against existing published SSR datasets in future study. Metrics such as polymorphism information content (PIC), expected heterozygosity (He), and allelic diversity should be evaluated to determine the relative informativeness and transferability across other geographical stocks, including the Dai-qu and Min-Yuedong populations [
3,
85,
88]. The genome-wide SSR development for the Naozhou stock of
L. crocea provides a rich molecular resource with higher resolution and broader applicability than prior microsatellite datasets. It represents a critical step toward the genetic improvement, conservation, and management of this economically significant species. Comparing these new loci with existing SSR panels will further consolidate their utility in nationwide aquaculture genetics and breeding strategies.
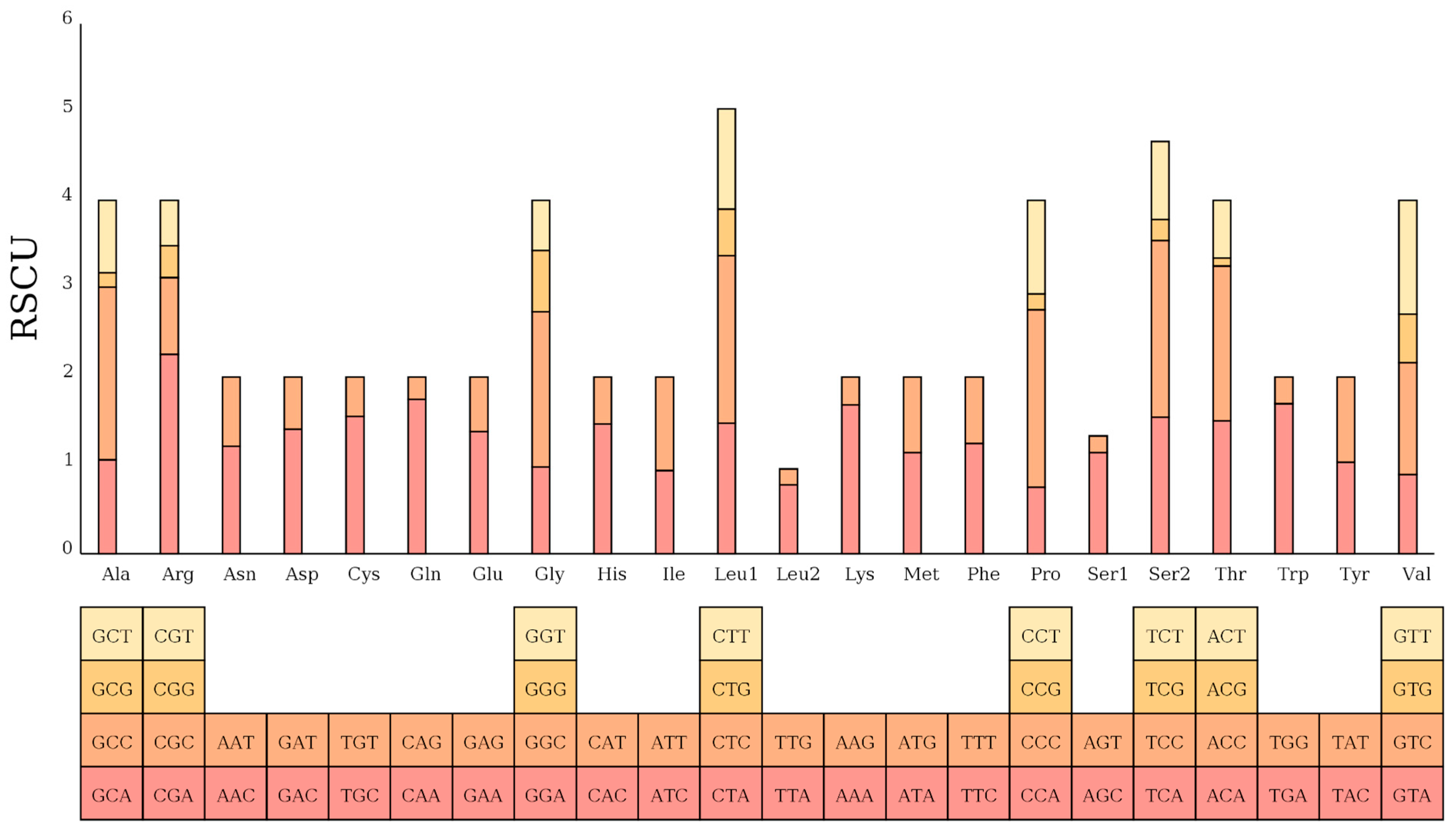
The assembled mtDNA sequence of the Naozhou large yellow croaker was 16,467 bp in length, consistent with the Dai-qu stock of large yellow croakers (PRJNA927338). The genome adheres to the canonical gene order observed in teleosts, comprising 13 PCGs, 22 tRNAs, and two rRNAs, providing confidence in the completeness and integrity of the assembly [
29,
61,
89]. Most PCGs initiate with the ATG codon, and several utilize incomplete stop codons (e.g., T) that are post-transcriptionally completed, consistent with mechanisms seen in other
L. crocea stocks [
8] and marine fishes generally [
90]. The overall mtDNA structure closely mirrors that of the Dai-qu stock, with minor variations in the control region, a recognized hotspot for polymorphism and regulatory evolution [
35]. Codon usage exhibits a bias toward UAA and codons encoding Proline, Threonine, and Leucine, suggesting functional constraints and possible translational optimization. Codon bias and AT richness (overall GC content of 46.93%) in the mtDNA may influence mitochondrial gene expression, thereby impacting traits like energy metabolism, stress tolerance, and growth performance. These characteristics are critical for aquaculture productivity, particularly under variable environmental conditions [
91]. This study’s dual contributions, namely the development of high-density SSR marker and full mtDNA assembly, substantially enhance genomic resources for
L. crocea, especially for the genetically distinct and underutilized Naozhou stock. The genetic differentiation from other stocks highlights the necessity of implementing stock-specific breeding programs to maintain genetic integrity and avoid homogenization [
7]. These results enable fine-scale genetic mapping and MAS for traits such as disease resistance and growth. Additionally, they support population genomic studies to monitor stock integrity and inbreeding, enable comparative genomic studies with other marine species for evolutionary and functional analyses, and contribute to conservation strategies for declining stocks. By integrating both nuclear (SSR) and mtDNA genomic insights, this study provides a comprehensive genomic toolkit that can guide future transcriptomic, epigenetic, and functional validation studies [
14,
18]. The development of genome-wide SSR markers and mtDNA assembly in this study significantly advances current knowledge of the Naozhou stock of
L. crocea. It marks a transition from mere genomic description to providing actionable insights for functional studies, evolutionary biology, and sustainable aquaculture. These foundational resources not only facilitate immediate applications in breeding and conservation but also open avenues for integrative omics approaches aimed at exploring genotype–phenotype–environment relationships.