APOBEC1-Dependent RNA Eiting of TNF Signaling Orchestrates Ileal Villus Morphogenesis in Pigs: Integrative Transcriptomic and Editomic Insights
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Animals and Data Storage
2.3. Alignments and Variant Identification
2.4. Differential Editing Analysis of RNA Editing Events
2.5. RNA Differential Expression Genes Analysis
2.6. Pathway Enrichment
2.7. Correlation Analysis Between Genes RNA Editing and Expression
2.8. Statistical Analysis
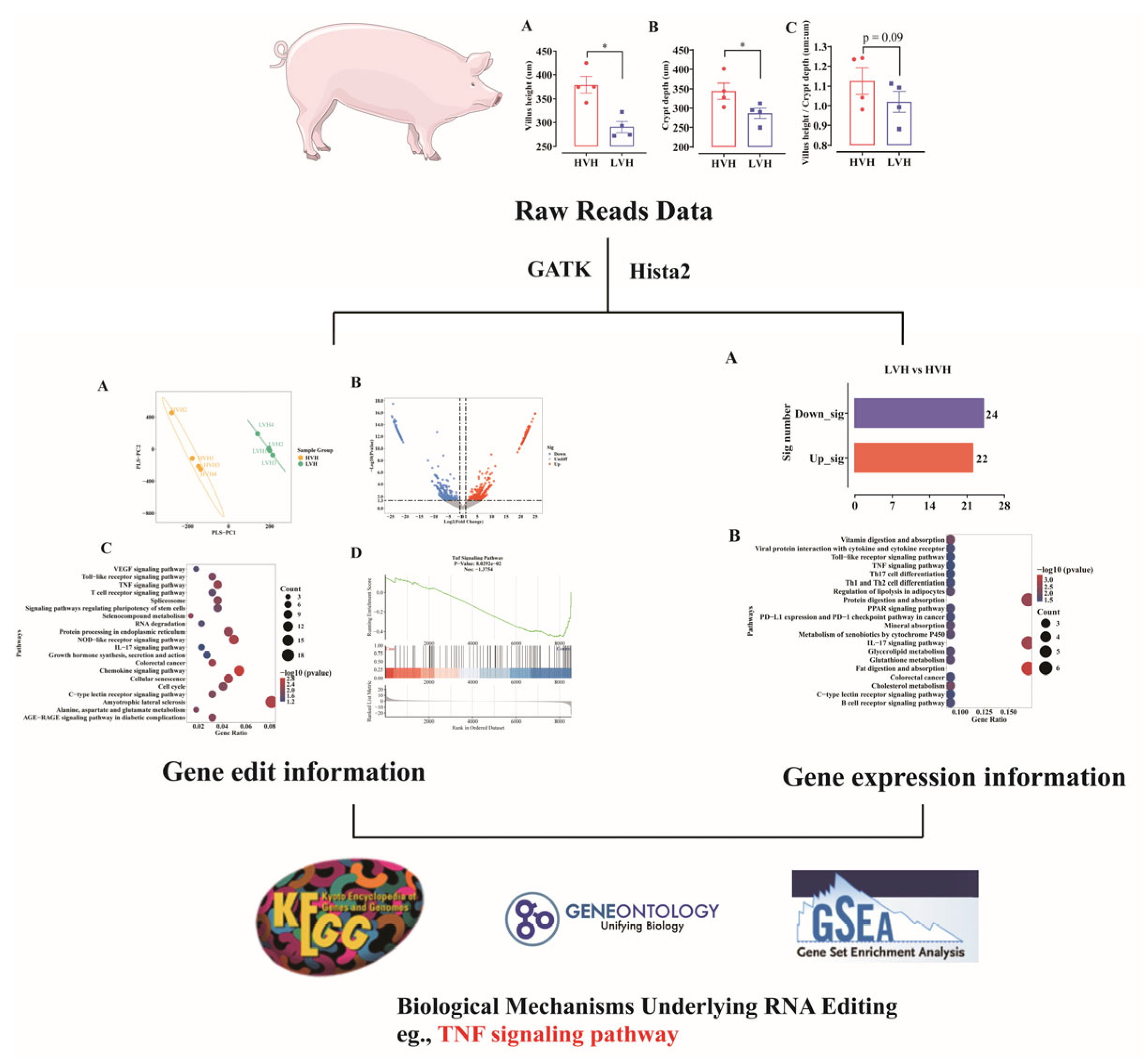
3. Results
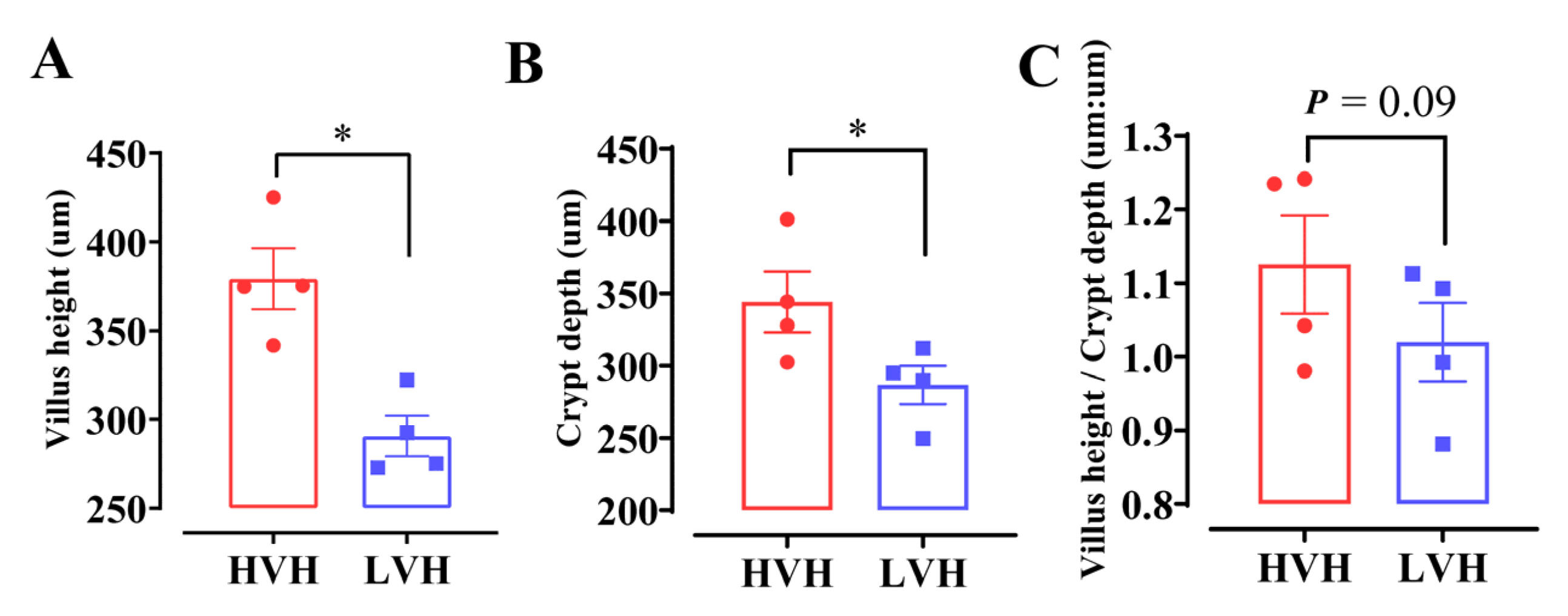
3.1. General Characteristics of RNA Editing Events in HVH and LVH
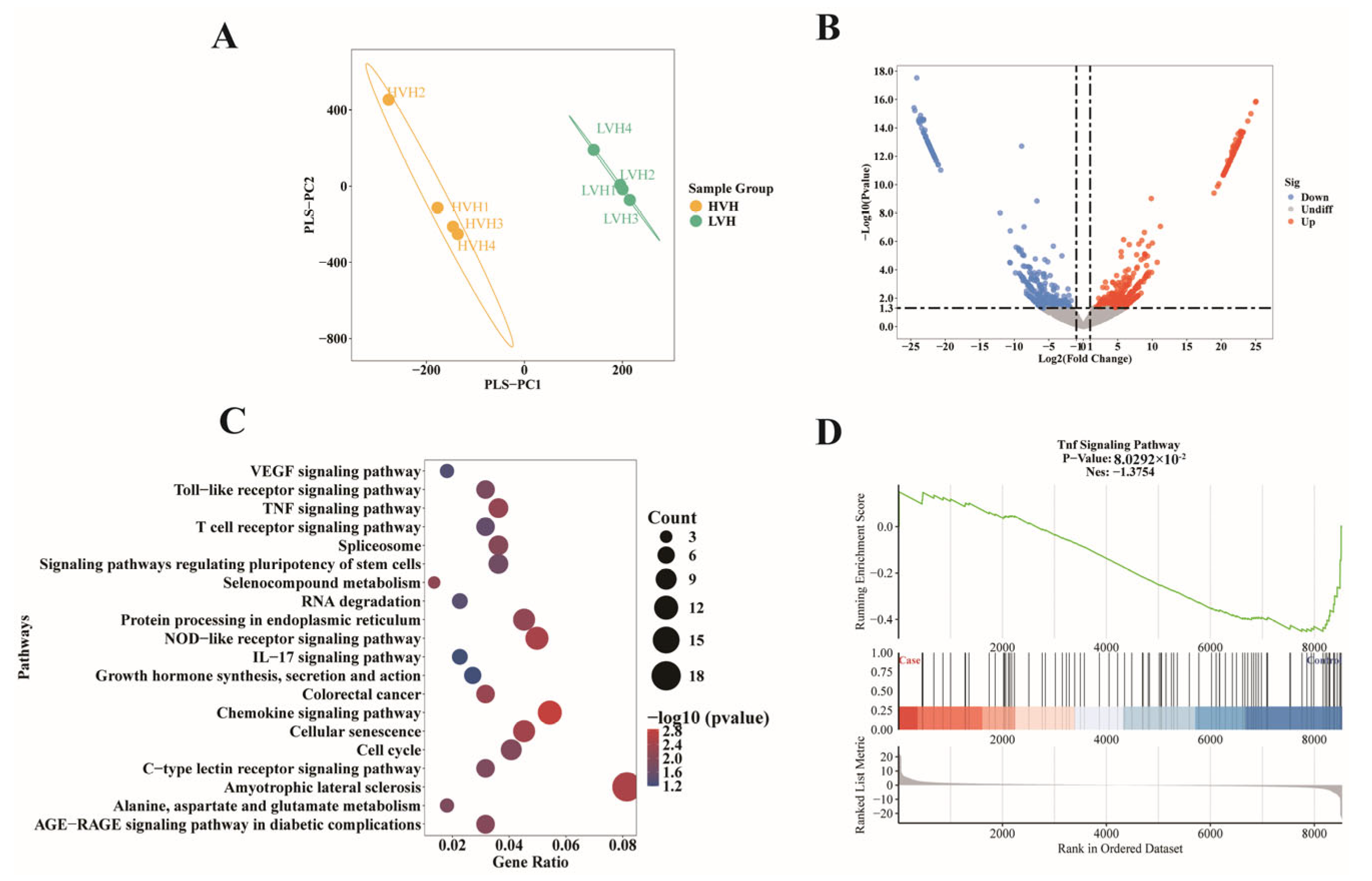
3.2. RNA Editing Genes Are Significantly Enriched in the TNF Signaling Pathway in HVH
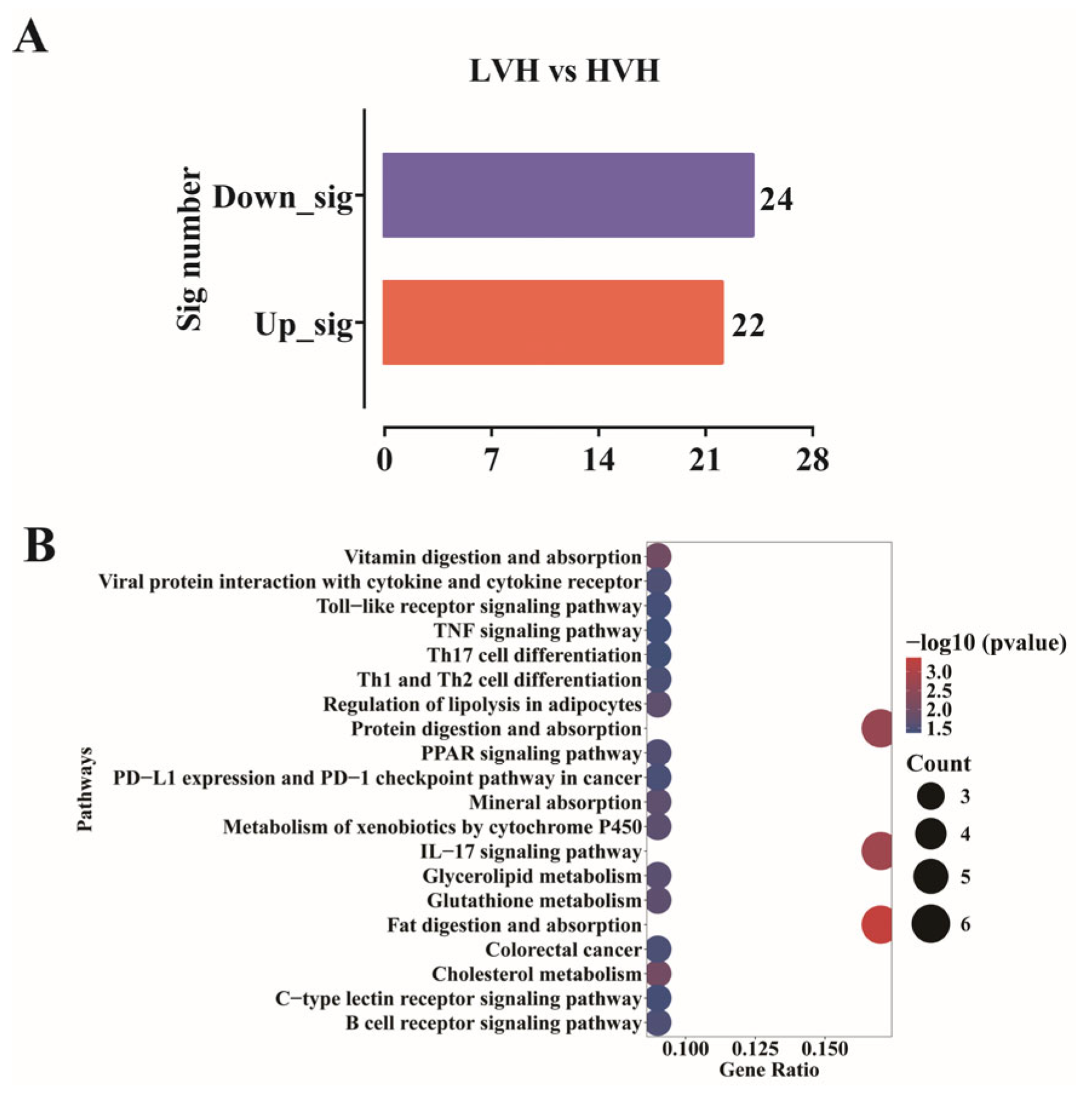
3.3. RNA Expression Genes Are Significantly Enriched in the TNF Signaling Pathway in LVH
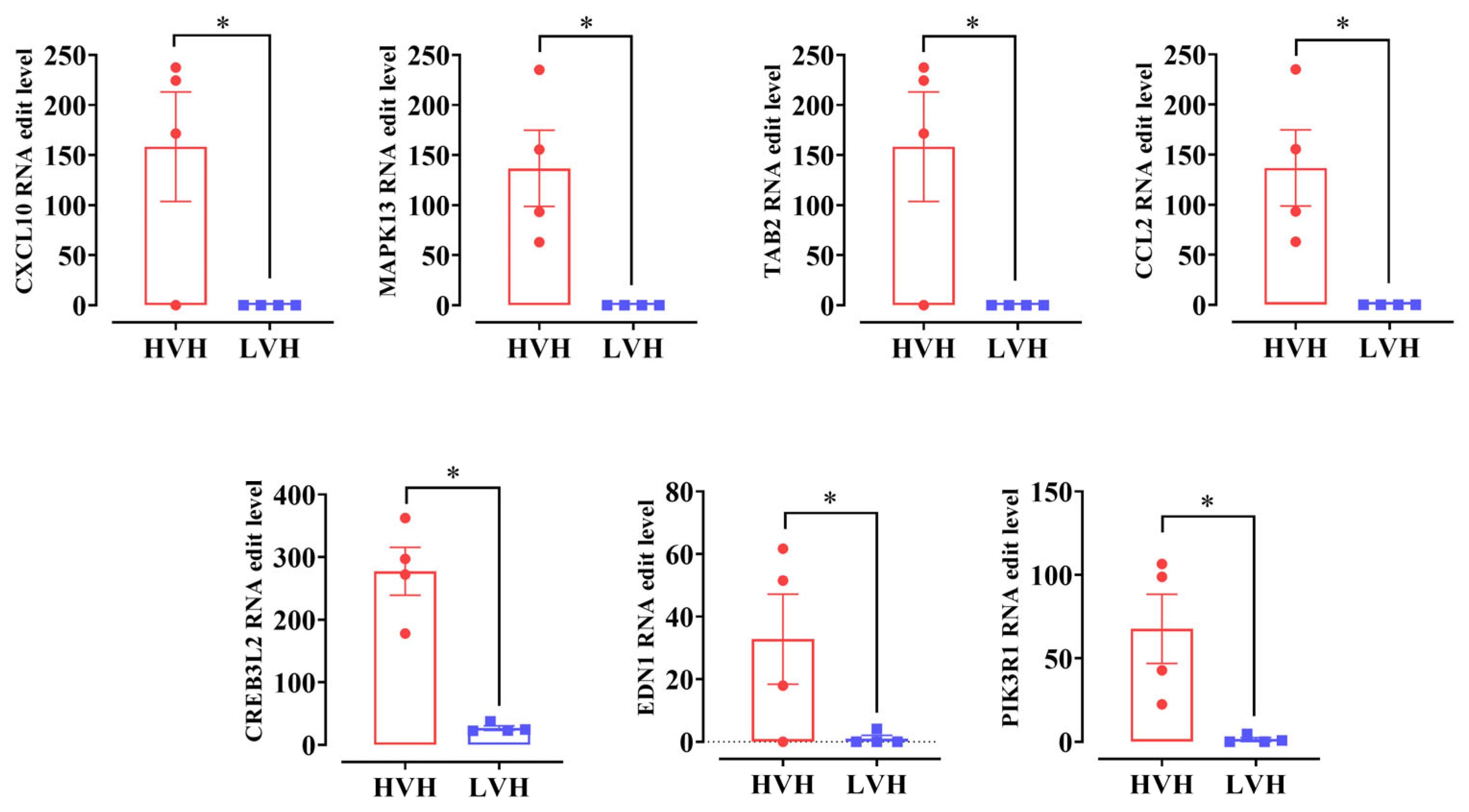
3.4. High RNA Editing Attenuates Ileum Inflammation: Integrated Analysis of RNA Editing and Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bai, M.; Wang, L.; Liu, H.; Xu, K.; Deng, J.; Huang, R.; Yin, Y. Imbalanced dietary methionine-to-sulfur amino acid ratio can affect amino acid profiles, antioxidant capacity, and intestinal morphology of piglets. Anim. Nutr. 2020, 6, 447–456. [Google Scholar] [CrossRef]
- Wang, X.; Yin, L.; Geng, C.; Zhang, J.; Li, J.; Huang, P.; Li, Y.; Wang, Q.; Yang, H. Impact of different feed intake levels on intestinal morphology and epithelial cell differentiation in piglets. J. Anim. Sci. 2025, 103, skae262. [Google Scholar] [CrossRef]
- Li, W.; Zeng, X.; Wang, L.; Yin, L.; Wang, Q.; Yang, H. Comparative Analysis of Gut Microbiota Diversity Across Different Digestive Tract Sites in Ningxiang Pigs. Animals 2025, 15, 936. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Li, Z.; Zhang, Y. Effects of the Partial Substitution of Corn with Wheat or Barley on the Growth Performance, Blood Antioxidant Capacity, Intestinal Health and Fecal Microbial Composition of Growing Pigs. Antioxidants 2022, 11, 1614. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wu, L.; Han, H.; Li, Y.; Wang, L.; Yin, J.; Fan, W.; Bai, M.; Yao, J.; Huang, X.; et al. Reduced dietary nitrogen with a high Lys:CP ratio restricted dietary N excretion without negatively affecting weaned piglets. Anim. Nutr. 2019, 5, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y. Fatty acids, inflammation and intestinal health in pigs. J. Anim. Sci. Biotechnol. 2015, 6, 41. [Google Scholar] [CrossRef]
- Huang, S.M.; Wu, Z.H.; Li, T.T.; Liu, C.; Han, D.D.; Tao, S.Y.; Pi, Y.; Li, N.; Wang, J.J. Perturbation of the lipid metabolism and intestinal inflammation in growing pigs with low birth weight is associated with the alterations of gut microbiota. Sci. Total Environ. 2020, 719, 137382. [Google Scholar] [CrossRef]
- Lima, M.S.R.; de Lima, V.C.O.; Piuvezam, G.; de Azevedo, K.P.M.; Maciel, B.L.L.; Morais, A.H.A. Mechanisms of action of anti-inflammatory proteins and peptides with anti-TNF-alpha activity and their effects on the intestinal barrier: A systematic review. PLoS ONE 2022, 17, e0270749. [Google Scholar] [CrossRef]
- Wang, S.; Li, H.; Du, C.; Liu, Q.; Yang, D.; Chen, L.; Zhu, Q.; Wang, Z. Effects of dietary supplementation with Lactobacillus acidophilus on the performance, intestinal physical barrier function, and the expression of NOD-like receptors in weaned piglets. PeerJ 2018, 6, e6060. [Google Scholar] [CrossRef]
- Liu, B.; Jiang, X.; Cai, L.; Zhao, X.; Dai, Z.; Wu, G.; Li, X. Putrescine mitigates intestinal atrophy through suppressing inflammatory response in weanling piglets. J. Anim. Sci. Biotechnol. 2019, 10, 69. [Google Scholar] [CrossRef]
- Zhang, S.; Xiong, L.; Cui, C.; Zhao, H.; Zhang, Y.; Tian, Z.; Guan, W.; Chen, F. Maternal supplementation with Artemisia annua L. ameliorates intestinal inflammation via inhibiting the TLR4/NF-kappaB and MAPK pathways and improves the oxidative stability of offspring. Food Funct. 2022, 13, 9311–9323. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, M.; Zhao, X.; Feng, J. Ammonia exposure induced intestinal inflammation injury mediated by intestinal microbiota in broiler chickens via TLR4/TNF-alpha signaling pathway. Ecotoxicol. Environ. Saf. 2021, 226, 112832. [Google Scholar] [CrossRef] [PubMed]
- Anandam, K.Y.; Alwan, O.A.; Subramanian, V.S.; Srinivasan, P.; Kapadia, R.; Said, H.M. Effect of the proinflammatory cytokine TNF-alpha on intestinal riboflavin uptake: Inhibition mediated via transcriptional mechanism(s). Am. J. Physiol. Cell Physiol. 2018, 315, C653–C663. [Google Scholar] [CrossRef] [PubMed]
- Porath, H.T.; Knisbacher, B.A.; Eisenberg, E.; Levanon, E.Y. Massive A-to-I RNA editing is common across the Metazoa and correlates with dsRNA abundance. Genome Biol. 2017, 18, 185. [Google Scholar] [CrossRef]
- Eisenberg, E.; Levanon, E.Y. A-to-I RNA editing—Immune protector and transcriptome diversifier. Nat. Rev. Genet. 2018, 19, 473–490. [Google Scholar] [CrossRef]
- Erdmann, E.A.; Mahapatra, A.; Mukherjee, P.; Yang, B.; Hundley, H.A. To protect and modify double-stranded RNA—The critical roles of ADARs in development, immunity and oncogenesis. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 54–87. [Google Scholar] [CrossRef]
- Schmauss, C.; Howe, J.R. RNA editing of neurotransmitter receptors in the mammalian brain. Sci. STKE 2002, 2002, pe26. [Google Scholar] [CrossRef]
- Zhang, D.; Zhu, L.; Gao, Y.; Wang, Y.; Li, P. RNA editing enzymes: Structure, biological functions and applications. Cell Biosci. 2024, 14, 34. [Google Scholar] [CrossRef]
- Blanc, V.; Xie, Y.; Kennedy, S.; Riordan, J.D.; Rubin, D.C.; Madison, B.B.; Mills, J.C.; Nadeau, J.H.; Davidson, N.O. Apobec1 complementation factor (A1CF) and RBM47 interact in tissue-specific regulation of C to U RNA editing in mouse intestine and liver. RNA 2019, 25, 70–81. [Google Scholar] [CrossRef]
- Song, J.; Zhuang, Y.; Yi, C. Programmable RNA base editing via targeted modifications. Nat. Chem. Biol. 2024, 20, 277–290. [Google Scholar] [CrossRef]
- Medina-Munoz, H.C.; Kofman, E.; Jagannatha, P.; Boyle, E.A.; Yu, T.; Jones, K.L.; Mueller, J.R.; Lykins, G.D.; Doudna, A.T.; Park, S.S.; et al. Expanded palette of RNA base editors for comprehensive RBP-RNA interactome studies. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cao, D.; Shi, M.; Yang, X. BIRC3 RNA Editing Modulates Lipopolysaccharide-Induced Liver Inflammation: Potential Implications for Animal Health. Int. J. Mol. Sci. 2025, 26, 2941. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Tessier, L.; Cote, O.; Bienzle, D. Sequence variant analysis of RNA sequences in severe equine asthma. PeerJ 2018, 6, e5759. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Lu, Y.; Rosenfeld, R.; Simon, I.; Nau, G.J.; Bar-Joseph, Z. A probabilistic generative model for GO enrichment analysis. Nucleic Acids Res. 2008, 36, e109. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Ekstrom, C.T.; Carstensen, B. Statistical models for assessing agreement for quantitative data with heterogeneous random raters and replicate measurements. Int. J. Biostat. 2024, 20, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, M.; Wang, Y.; Chen, J.; Zhang, Y.; Wang, Q.; Yang, H. The relationship of N-glycosylation and porcine duodenal morphology and function. J. Anim. Sci. 2025, 103, skaf087. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, Y.; Wen, M.; Zhong, F.; Shu, X.; Xu, R.; Xiong, P.; Zhou, Z.; He, X.; Tang, X.; et al. Supplementary Hesperidin Alleviated CPT-11-Induced Diarrhea by Modulating Gut Microbiota Inhibiting the IL-17 Signaling Pathway. J. Agric. Food Chem. 2025, 73, 5915–5930. [Google Scholar] [CrossRef]
- Zhao, X.; Ma, L.; Dai, L.; Zuo, D.; Li, X.; Zhu, H.; Xu, F. TNF-alpha promotes the malignant transformation of intestinal stem cells through the NF-kappaB and Wnt/beta-catenin signaling pathways. Oncol. Rep. 2020, 44, 577–588. [Google Scholar] [CrossRef]
- Johnson, J.S.; Sapkota, A.; Lay, D.C., Jr. Rapid cooling after acute hyperthermia alters intestinal morphology and increases the systemic inflammatory response in pigs. J. Appl. Physiol. 2016, 120, 1249–1259. [Google Scholar] [CrossRef]
- Miguel, M.G.; da Silva, C.I.; Farah, L.; Castro Braga, F.; Figueiredo, A.C. Effect of Essential Oils on the Release of TNF-alpha and CCL2 by LPS-Stimulated THP-1 Cells. Plants 2020, 10, 50. [Google Scholar] [CrossRef]
- Giri, J.; Das, R.; Nylen, E.; Chinnadurai, R.; Galipeau, J. CCL2 and CXCL12 Derived from Mesenchymal Stromal Cells Cooperatively Polarize IL-10+ Tissue Macrophages to Mitigate Gut Injury. Cell Rep. 2020, 30, 1923–1934. [Google Scholar] [CrossRef]
- Sato, N.; Garcia-Castillo, V.; Yuzawa, M.; Islam, M.A.; Albarracin, L.; Tomokiyo, M.; Ikeda-Ohtsubo, W.; Garcia-Cancino, A.; Takahashi, H.; Villena, J.; et al. Immunobiotic Lactobacillus jensenii TL2937 Alleviates Dextran Sodium Sulfate-Induced Colitis by Differentially Modulating the Transcriptomic Response of Intestinal Epithelial Cells. Front. Immunol. 2020, 11, 2174. [Google Scholar] [CrossRef]
- He, J.; Song, Y.; Li, G.; Xiao, P.; Liu, Y.; Xue, Y.; Cao, Q.; Tu, X.; Pan, T.; Jiang, Z.; et al. Fbxw7 increases CCL2/7 in CX3CR1hi macrophages to promote intestinal inflammation. J. Clin. Investig. 2019, 129, 3877–3893. [Google Scholar] [CrossRef]
- Kawano, Y.; Nakae, J.; Watanabe, N.; Kikuchi, T.; Tateya, S.; Tamori, Y.; Kaneko, M.; Abe, T.; Onodera, M.; Itoh, H. Colonic Pro-inflammatory Macrophages Cause Insulin Resistance in an Intestinal Ccl2/Ccr2-Dependent Manner. Cell Metab. 2016, 24, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Szymczak, F.; Cohen-Fultheim, R.; Thomaidou, S.; de Brachene, A.C.; Castela, A.; Colli, M.; Marchetti, P.; Levanon, E.; Eizirik, D.; Zaldumbide, A. ADAR1-dependent editing regulates human beta cell transcriptome diversity during inflammation. Front. Endocrinol. 2022, 13, 1058345. [Google Scholar] [CrossRef] [PubMed]
- Blanc, V.; Park, E.; Schaefer, S.; Miller, M.; Lin, Y.; Kennedy, S.; Billing, A.M.; Ben Hamidane, H.; Graumann, J.; Mortazavi, A.; et al. Genome-wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver. Genome Biol. 2014, 15, R79. [Google Scholar] [CrossRef] [PubMed]
- Katrekar, D.; Chen, G.; Meluzzi, D.; Ganesh, A.; Worlikar, A.; Shih, Y.R.; Varghese, S.; Mali, P. In vivo RNA editing of point mutations via RNA-guided adenosine deaminases. Nat. Methods 2019, 16, 239–242. [Google Scholar] [CrossRef]
- Reautschnig, P.; Wahn, N.; Wettengel, J.; Schulz, A.E.; Latifi, N.; Vogel, P.; Kang, T.W.; Pfeiffer, L.S.; Zarges, C.; Naumann, U.; et al. CLUSTER guide RNAs enable precise and efficient RNA editing with endogenous ADAR enzymes in vivo. Nat. Biotechnol. 2022, 40, 759–768. [Google Scholar] [CrossRef]
- Stroppel, A.S.; Latifi, N.; Hanswillemenke, A.; Tasakis, R.N.; Papavasiliou, F.N.; Stafforst, T. Harnessing self-labeling enzymes for selective and concurrent A-to-I and C-to-U RNA base editing. Nucleic Acids Res. 2021, 49, e95. [Google Scholar] [CrossRef]
- Blanc, V.; Xie, Y.; Luo, J.; Kennedy, S.; Davidson, N.O. Intestine-specific expression of Apobec-1 rescues apolipoprotein BRNAediting alters chylomicron production in Apobec1−/− mice. J. Lipid Res. 2012, 53, 2643–2655. [Google Scholar] [CrossRef]
- Teng, B.; Ishida, B.; Forte, T.M.; Blumenthal, S.; Song, L.Z.; Gotto, A.M., Jr.; Chan, L. Effective lowering of plasma, LDL, and esterified cholesterol in LDL receptor-knockout mice by adenovirus-mediated gene delivery of ApoB mRNA editing enzyme (Apobec1). Arter. Thromb. Vasc. Biol. 1997, 17, 889–897. [Google Scholar] [CrossRef]
- Cole, D.C.; Chung, Y.; Gagnidze, K.; Hajdarovic, K.H.; Rayon-Estrada, V.; Harjanto, D.; Bigio, B.; Gal-Toth, J.; Milner, T.A.; McEwen, B.S.; et al. Loss of APOBEC1 RNA-editing function in microglia exacerbates age-related CNS pathophysiology. Proc. Natl. Acad. Sci. USA 2017, 114, 13272–13277. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Chen, W.; Wang, Y.; Wang, Q.; Yang, H.; Wang, Q.; Wang, B. APOBEC1-Dependent RNA Eiting of TNF Signaling Orchestrates Ileal Villus Morphogenesis in Pigs: Integrative Transcriptomic and Editomic Insights. Animals 2025, 15, 2419. https://doi.org/10.3390/ani15162419
Li W, Chen W, Wang Y, Wang Q, Yang H, Wang Q, Wang B. APOBEC1-Dependent RNA Eiting of TNF Signaling Orchestrates Ileal Villus Morphogenesis in Pigs: Integrative Transcriptomic and Editomic Insights. Animals. 2025; 15(16):2419. https://doi.org/10.3390/ani15162419
Chicago/Turabian StyleLi, Wangchang, Wenxin Chen, Yancan Wang, Qianqian Wang, Huansheng Yang, Qiye Wang, and Bin Wang. 2025. "APOBEC1-Dependent RNA Eiting of TNF Signaling Orchestrates Ileal Villus Morphogenesis in Pigs: Integrative Transcriptomic and Editomic Insights" Animals 15, no. 16: 2419. https://doi.org/10.3390/ani15162419
APA StyleLi, W., Chen, W., Wang, Y., Wang, Q., Yang, H., Wang, Q., & Wang, B. (2025). APOBEC1-Dependent RNA Eiting of TNF Signaling Orchestrates Ileal Villus Morphogenesis in Pigs: Integrative Transcriptomic and Editomic Insights. Animals, 15(16), 2419. https://doi.org/10.3390/ani15162419






