Transcriptomics Analysis of the Adipogenic Differentiation Mechanism of Bovine Adipose-Derived Neural Crest Stem Cells
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. baNCSCs Isolation, Cell Culture, and Passage Procedure
2.2. Transcriptome Detection of Adipocytes from baNCSCs at Different Differentiation Stages
3. Results
3.1. RNA-seq Quality Control and Reference Genome Alignment Results
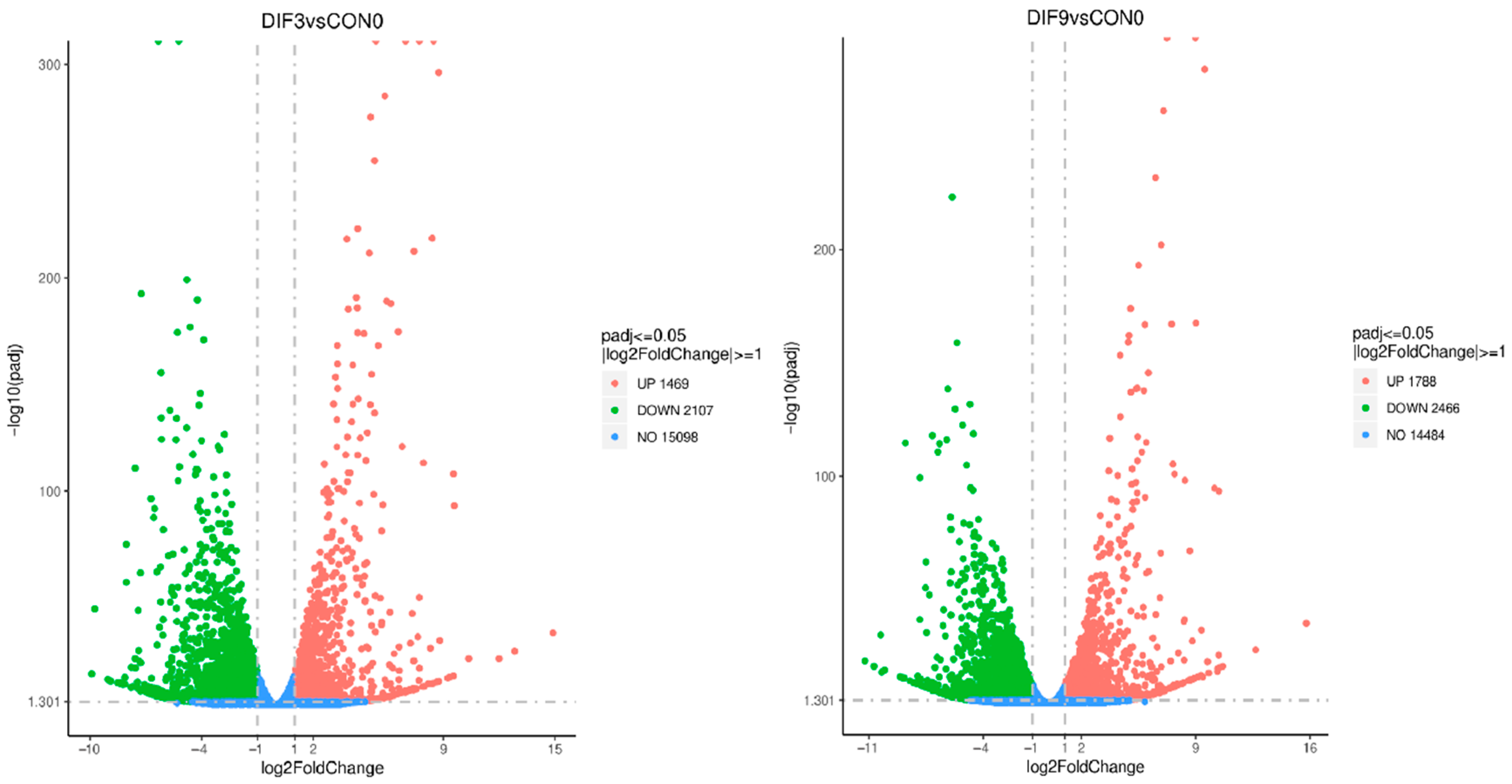
3.2. Analysis of Differentially Expressed Genes
3.3. Gene Ontology Functional Enrichment Analysis of Differentially Expressed Genes
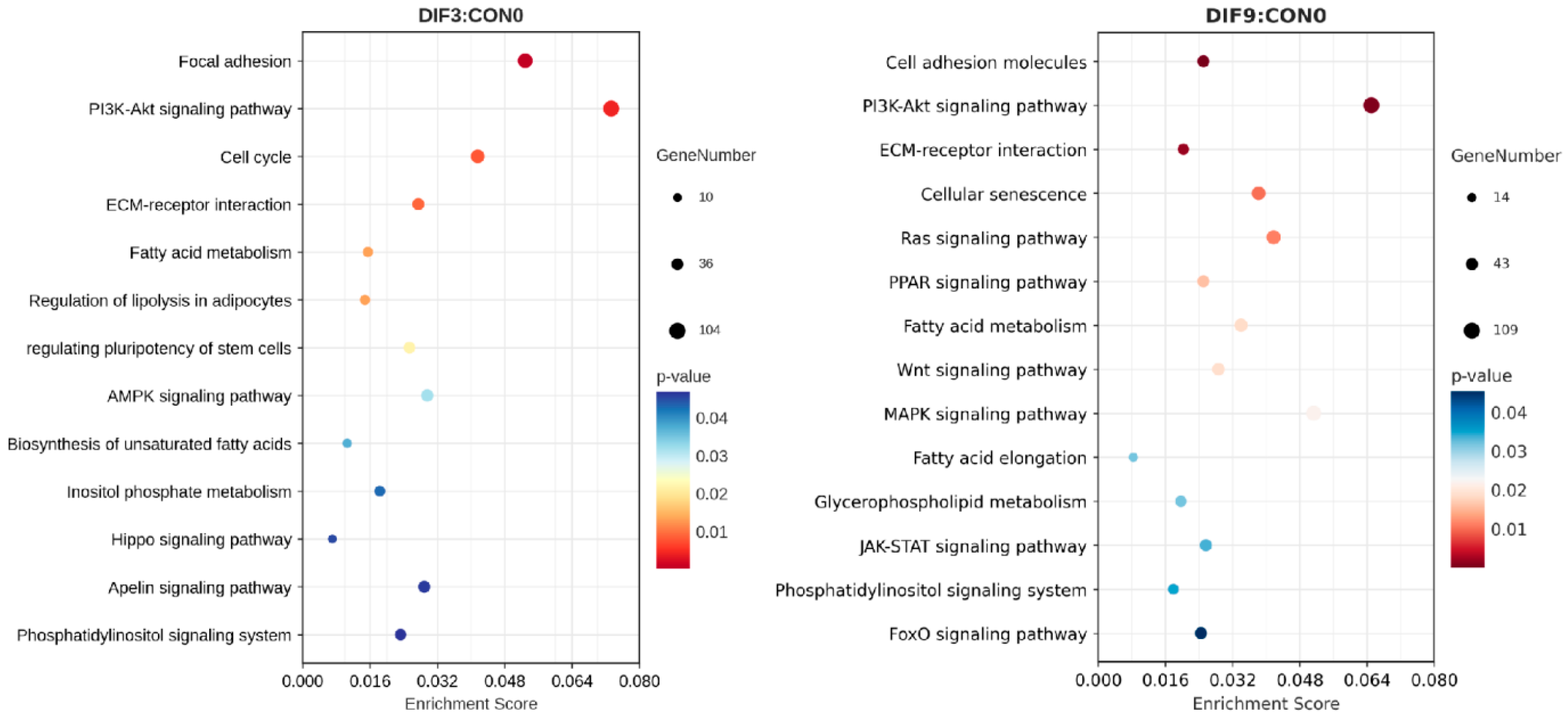
3.4. Enrichment Analysis of KEGG Pathways for Differentially Expressed Genes
3.5. Screening of Core Hub Genes in Adipogenic Metabolism
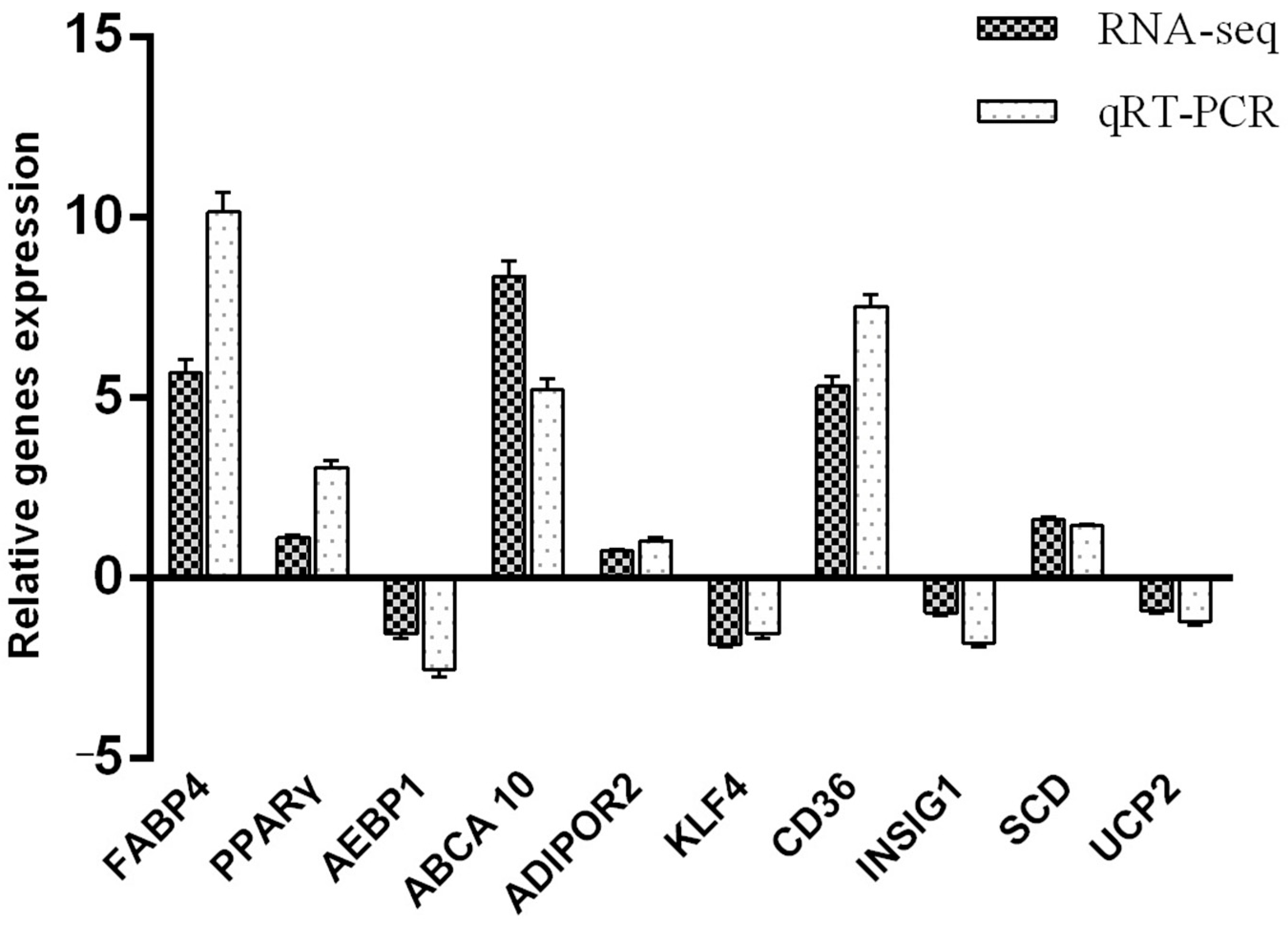
3.6. Quantitative Verification of Differentially Expressed Genes in the Transcriptome
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AKT | serine/threonine kinase |
| baNCSC | bovine adipose-derived neural crest stem cells |
| BP | biological processes |
| CC | cellular components |
| C/EBPα | CCAAT/enhancer binding protein α |
| CHGs | core hub genes |
| DEGs | differentially expressed genes |
| FABP4 | fatty acid binding protein 4 |
| FBS | fetal bovine serum |
| FC | fold change |
| FoxO | forkhead box O |
| FPKM | fragments per kilobase of transcript per million mapped reads |
| GO | gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MAPK | mitogen-activated protein kinase |
| MCC | maximum group centrality |
| MF | molecular functions |
| PI3K | phosphoinositide 3-kinase |
| PPARγ | peroxisome proliferator-activated receptor γ |
| PPI | protein–protein interaction |
References
- Liu, S.S.; Fang, X.; Wen, X.; Liu, J.S.; Alip, M.; Sun, T.; Wang, Y.Y.; Chen, H.W. How mesenchymal stem cells transform into adipocytes: Overview of the current understanding of adipogenic differentiation. World J. Stem Cells 2024, 16, 245–256. [Google Scholar] [CrossRef]
- Wang, G.; Wu, B.; Zhang, L.; Cui, Y.; Zhang, B.; Wang, H. Laquinimod Prevents Adipogenesis and Obesity by Down-Regulating PPAR-γ and C/EBPα through Activating AMPK. ACS Omega 2020, 5, 22958–22965. [Google Scholar] [CrossRef] [PubMed]
- Dou, H.X.; Wang, T.; Su, H.X.; Gao, D.D.; Xu, Y.C.; Li, Y.X.; Wang, H.Y. Exogenous FABP4 Interferes with Differentiation, Promotes Lipolysis and Inflammation in Adipocytes. Endocrine 2019, 67, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Yu, X.; Yin, L. Diazinon Exposure Activated Transcriptional Factors CCAAT-Enhancer-Binding Proteins α (C/EBPα) and Peroxisome Proliferator-Activated Receptor γ (PPARγ) and Induced Adipogenesis in 3T3-L1 Preadipocytes. Pestic. Biochem. Phys. 2018, 150, 48–58. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Y.; Xu, Q.; Yuan, X.; Dai, W.; Shen, X.; Wang, Z.; Chang, G.; Wang, Z.; Chen, G. The Differentiation of Preadipocytes and Gene Expression Related to Adipogenesis in Ducks (Anas Platyrhynchos). PLoS ONE 2018, 13, e0196371. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.A.; Cheung, M. Neural crest stem cells and their potential therapeutic applications. Dev. Biol. 2016, 419, 199–216. [Google Scholar] [CrossRef]
- Méndez-Maldonado, K.; Vega-López, G.A.; Aybar, M.J.; Velasco, I. Neurogenesis From Neural Crest Cells: Molecular Mechanisms in the Formation of Cranial Nerves and Ganglia. Front. Cell Dev. Biol. 2020, 8, 655. [Google Scholar] [CrossRef]
- Ng, T.K.; Yang, Q.; Fortino, V.R.; Lai, N.Y.; Carballosa, C.M.; Greenberg, J.M.; Choy, K.W.; Pelaez, D.; Pang, C.P.; Cheung, H.S. MicroRNA-132 Directs Human Periodontal Ligament-derived Neural Crest Stem Cell Neural Differentiation. J. Tissue Eng. Regen. Med. 2019, 13, 12–24. [Google Scholar] [CrossRef]
- Sowa, Y.; Imura, T.; Numajiri, T.; Takeda, K.; Mabuchi, Y.; Matsuzaki, Y.; Nishino, K. Adipose Stromal Cells Contain Phenotypically Distinct Adipogenic Progenitors Derived from Neural Crest. PLoS ONE 2013, 8, e84206. [Google Scholar] [CrossRef]
- Qi, Y.; Miao, X.; Xu, L.; Fu, M.; Peng, S.; Shi, K.; Li, J.; Ye, M.; Li, R. Isolation, Culture, and Adipogenic Induction of Neural Crest Original Adipose-Derived Stem Cells from Periaortic Adipose Tissue. J. Vis. Exp. 2020, 157, e60691. [Google Scholar] [CrossRef]
- Woonnoi, W.; Suttithumsatid, W.; Muneerungsee, N.; Saetan, J.; Tanasawet, S.; Sukketsiri, W. Sangyod Rice Extract Inhibits Adipocyte Growth and Differentiation via mTOR, Akt, and AMPK Pathways. J. Funct. Foods 2023, 111, 105913. [Google Scholar] [CrossRef]
- Dairi, G.; Al Mahri, S.; Benabdelkamel, H.; Alfadda, A.A.; Alswaji, A.A.; Rashid, M.; Malik, S.S.; Iqbal, J.; Ali, R.; Al Ibrahim, M.; et al. Transcriptomic and Proteomic Analysis Reveals the Potential Role of RBMS1 in Adipogenesis and Adipocyte Metabolism. Int. J. Mol. Sci. 2023, 24, 11300. [Google Scholar] [CrossRef]
- He, D.; Cheng, J.; Xu, F.; Li, B.; Jin, G.; Zhang, X. Detection of selection signatures based on the integrated haplotype score in Chinese Jinnan cattle. Emir. J. Food Agr. 2017, 29, 562–566. [Google Scholar]
- Wang, X.; Zhang, Y.; Zhang, X.; Wang, D.; Jin, G.; Li, B.; Xu, F.; Cheng, J.; Zhang, F.; Wu, S.; et al. The comprehensive liver transcriptome of two cattle breeds with different intramuscular fat content. Biochem. Biophys. Res. Commun. 2017, 490, 1018–1025. [Google Scholar] [CrossRef]
- Romao, J.M.; Jin, W.; He, M.; McAllister, T.; Guan, L.L. Elucidation of Molecular Mechanisms of Physiological Variations between Bovine Subcutaneous and Visceral Fat Depots under Different Nutritional Regimes. PLoS ONE 2013, 8, e83211. [Google Scholar] [CrossRef]
- Tang, X.; Liu, X.; Zhang, K. Effects of Microbial Fermented Feed on Serum Biochemical Profile, Carcass Traits, Meat Amino Acid and Fatty Acid Profile, and Gut Microbiome Composition of Finishing Pigs. Front. Vet. Sci. 2021, 8, 744630. [Google Scholar] [CrossRef]
- Mukai, T.; Kusudo, T. Bidirectional effect of vitamin D on brown adipogenesis of C3H10T1/2 fibroblast-like cells. PeerJ 2023, 11, e14785. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Yoshiike, K.; Watanabe, H.; Watanabe, M. The Marine Factor 3,5-Dihydroxy-4-methoxybenzyl Alcohol Represses Adipogenesis in Mouse 3T3-L1 Adipocytes In Vitro: Regulating Diverse Signaling Pathways. Nutraceuticals 2023, 3, 366–379. [Google Scholar] [CrossRef]
- Brooks, P.T.; Munthe-Fog, L.; Rieneck, K.; Banch Clausen, F.; Rivera, O.B.; Kannik Haastrup, E.; Fischer-Nielsen, A.; Svalgaard, J.D. Application of a deep learning-based image analysis and live-cell imaging system for quantifying adipogenic differentiation kinetics of adipose-derived stem/stromal cells. Adipocyte 2021, 10, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Cui, X.; Zhang, B.; Song, X.; Liu, Q.; Yang, S. Multipotent stem cells with neural crest stem cells characteristics exist in bovine adipose tissue. Biochem. Bioph Res. Commun. 2020, 522, 819–825. [Google Scholar] [CrossRef]
- Guo, W.; Zhang, K.M.; Tu, K.; Li, Y.X.; Zhu, L.; Xiao, H.S.; Yang, Y.; Wu, J.R. Adipogenesis Licensing and Execution Are Disparately Linked to Cell Proliferation. Cell Res. 2008, 19, 216–223. [Google Scholar] [CrossRef]
- Langerman, J.; Lopez, D.; Pellegrini, M.; Smale, S.T. Species-Specific Relationships between DNA and Chromatin Properties of CpG Islands in Embryonic Stem Cells and Differentiated Cells. Stem Cell Rep. 2021, 16, 899–912. [Google Scholar] [CrossRef]
- Musri, M.M.; Gomis, R.; Párrizas, M. A Chromatin Perspective of Adipogenesis. Organogenesis 2010, 6, 15–23. [Google Scholar] [CrossRef]
- Maraldi, T.; Angeloni, C.; Prata, C.; Hrelia, S. NADPH Oxidases: Redox Regulators of Stem Cell Fate and Function. Antioxidants 2021, 10, 973. [Google Scholar] [CrossRef]
- Wang, X.; Li, N.; Zheng, M.; Yu, Y.; Zhang, S. Acetylation and Deacetylation of Histone in Adipocyte Differentiation and the Potential Significance in Cancer. Transl. Oncol. 2024, 39, 101815. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular Matrix Structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef]
- von Erlach, T.C.; Bertazzo, S.; Wozniak, M.A.; Horejs, C.-M.; Maynard, S.A.; Attwood, S.; Robinson, B.K.; Autefage, H.; Kallepitis, C.; del Río Hernández, A.; et al. Cell-Geometry-Dependent Changes in Plasma Membrane Order Direct Stem Cell Signalling and Fate. Nat. Mat. 2018, 17, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Spiegelman, B.M.; Ginty, C.A. Fibronectin Modulation of Cell Shape and Lipogenic Gene Expression in 3t3-Adipocytes. Cell 1983, 35, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Paganelli, A.; Benassi, L.; Rossi, E.; Magnoni, C. Extracellular Matrix Deposition by Adipose-Derived Stem Cells and Fibroblasts: A Comparative Study. Arch. Dermatol. Res. 2019, 312, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, B.; Serpell, C.J.; Fong, I.L.; Wong, E.H. Molecular Mechanisms of Adipogenesis: The Anti-Adipogenic Role of AMP-Activated Protein Kinase. Front. Mol. Biosci. 2020, 7, 76. [Google Scholar] [CrossRef]
- Liu, D.D.; Han, C.C.; Wan, H.F.; He, F.; Xu, H.Y.; Wei, S.H.; Du, X.H.; Xu, F. Effects of Inhibiting PI3K-Akt-mTOR Pathway on Lipid Metabolism Homeostasis in Goose Primary Hepatocytes. Animal 2016, 10, 1319–1327. [Google Scholar] [CrossRef]
- Peng, H.; Lin, X.; Wang, Y.; Chen, J.; Zhao, Q.; Chen, S.; Cheng, Q.; Chen, C.; Sang, T.; Zhou, H.; et al. Epigallocatechin Gallate Suppresses Mitotic Clonal Expansion and Adipogenic Differentiation of Preadipocytes through Impeding JAK2/STAT3-Mediated Transcriptional Cascades. Phytomedicine 2024, 129, 155563. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Wei, D.; Tang, L.; Wang, S.; Pan, C.; Ma, Y.F.; Ma, Y. SGK1 Affects the Phosphorylation of FOXO1/FOXO3 Promoting Bovine Fat Deposition via the PI3K/Akt Signaling Pathway. Research Square. 2022. preprint. [Google Scholar] [CrossRef]
- Huang, X.; Liu, G.; Guo, J.; Su, Z. The PI3K/AKT pathway in obesity and type 2 diabetes. Int. J. Biol. Sci. 2018, 14, 1483–1496. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, J.; Raza, S.H.A.; Deng, J.; Ma, J.; Qu, X.; Yu, S.; Zhang, D.; Alshammari, A.M.; Almohaimeed, H.M.; et al. Identification of the hub genes related to adipose tissue metabolism of bovine. Front. Vet. Sci. 2022, 9, 1014286. [Google Scholar] [CrossRef]
- Cao, Y.; Chen, Y.; Miao, K.; Zhang, S.; Deng, F.; Zhu, M.; Wang, C.; Gu, W.; Huang, Y.; Shao, Z.; et al. PPARγ As a Potential Target for Adipogenesis Induced by Fine Particulate Matter in 3T3-L1 Preadipocytes. Environ. Sci. Technol. 2023, 57, 7684–7697. [Google Scholar] [CrossRef]
- Fayyad, A.M.; Khan, A.A.; Abdallah, S.H.; Alomran, S.S.; Bajou, K.; Khattak, M.N.K. Rosiglitazone Enhances Browning Adipocytes in Association with MAPK and PI3-K Pathways During the Differentiation of Telomerase-Transformed Mesenchymal Stromal Cells into Adipocytes. Int. J. Mol. Sci. 2019, 20, 1618. [Google Scholar] [CrossRef]
- Huang, W.; Guo, Y.; Du, W.; Zhang, X.; Li, A.; Miao, X. Global transcriptome analysis identifies differentially expressed genes related to lipid metabolism in Wagyu and Holstein cattle. Sci. Rep. 2017, 7, 5278. [Google Scholar] [CrossRef]
- Du, L.; Li, K.; Chang, T.; An, B.; Liang, M.; Deng, T.; Cao, S.; Du, Y.; Cai, W.; Gao, X.; et al. Integrating genomics and transcriptomics to identify candidate genes for subcutaneous fat deposition in beef cattle. Genomics 2022, 114, 110406. [Google Scholar] [CrossRef]
- Vitali, M.; Dimauro, C.; Sirri, R.; Zappaterra, M.; Zambonelli, P.; Manca, E.; Sami, D.; Lo Fiego, D.P.; Davoli, R. Effect of dietary polyunsaturated fatty acid and antioxidant supplementation on the transcriptional level of genes involved in lipid and energy metabolism in swine. PLoS ONE 2018, 13, e0204869. [Google Scholar] [CrossRef] [PubMed]
- Latorre, J.; Martínez, C.; Ortega, F.; Oliveras-Cañellas, N.; Díaz-Sáez, F.; Aragonés, J.; Camps, M.; Gumà, A.; Ricart, W.; Fernández-Real, J.M.; et al. The relevance of EGFR, ErbB receptors and neuregulins in human adipocytes and adipose tissue in obesity. Biomed. Pharmacother. 2022, 156, 113972. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J. Emerging roles of Myc in stem cell biology and novel tumor therapies. J. Exp. Clin. Cancer Res. 2018, 37, 173. [Google Scholar] [CrossRef]
- Gouw, A.M.; Margulis, K.; Liu, N.S.; Raman, S.J.; Mancuso, A.; Toal, G.G.; Tong, L.; Mosley, A.; Hsieh, A.L.; Sullivan, D.K.; et al. The MYC Oncogene Cooperates with Sterol-Regulated Element-Binding Protein to Regulate Lipogenesis Essential for Neoplastic Growth. Cell Metab. 2019, 30, 556–572.e5. [Google Scholar] [CrossRef] [PubMed]
- Yuzuriha, A.; Nakamura, S.; Sugimoto, N.; Kihara, S.; Nakagawa, M.; Yamamoto, T.; Sekiguchi, K.; Eto, K. Extracellular laminin regulates hematopoietic potential of pluripotent stem cells through integrin β1-ILK-β-catenin-JUN axis. Stem Cell Res. 2021, 53, 102287. [Google Scholar] [CrossRef]
- Nakamura, K.; Nakano, S.; Miyoshi, T.; Yamanouchi, K.; Matsuwaki, T.; Nishihara, M. Age-related resistance of skeletal muscle-derived progenitor cells to SPARC may explain a shift from myogenesis to adipogenesis. Aging 2012, 4, 40–48. [Google Scholar] [CrossRef]
- Zhu, B.; Ferry, C.H.; Blazanin, N.; Bility, M.T.; Khozoie, C.; Kang, B.H.; Glick, A.B.; Gonzalez, F.J.; Peters, J.M. PPARβ/δ promotes HRAS-induced senescence and tumor suppression by potentiating p-ERK and repressing p-AKT signaling. Oncogene 2014, 33, 5348–5359. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, F.; Xu, D.; Hou, K.; Fang, W.; Li, Y. The Function of RAS Mutation in Cancer and Advances in its Drug Research. Curr. Pharm. Des. 2019, 25, 1105–1114. [Google Scholar] [CrossRef]
- Sivoňová, M.K.; Híveš, M.; Kliment, J.; Dušenka, R.; Grendár, M.; Evin, D.; Kaplán, P.; Brožová, M.K.; Vilčková, M.; Vondrák, A.; et al. Genetic Alterations of Cyclin D-CDK4/6-INK4-RB Pathway in Prostate Cancer. Mol. Biol. Rep. 2025, 52, 439. [Google Scholar] [CrossRef]
- Mathieu, P.S.; Loboa, E.G. Cytoskeletal and focal adhesion influences on mesenchymal stem cell shape, mechanical properties, and differentiation down osteogenic, adipogenic, and chondrogenic pathways. Tissue Eng. Part. B Rev. 2012, 18, 436–444. [Google Scholar] [CrossRef]
- Peach, C.J.; Mignone, V.W.; Arruda, M.A.; Alcobia, D.C.; Hill, S.J.; Kilpatrick, L.E.; Woolard, J. Molecular Pharmacology of VEGF-A Isoforms: Binding and Signalling at VEGFR2. Int. J. Mol. Sci. 2018, 19, 1264. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, X.; Yang, L.; Eckel-Mahan, K.; Tong, Q.; Gu, X.; Kolonin, M.G.; Sun, K. Transient Overexpression of Vascular Endothelial Growth Factor A in Adipose Tissue Promotes Energy Expenditure via Activation of the Sympathetic Nervous System. Mol. Cell Biol. 2018, 38, e00242-18. [Google Scholar] [CrossRef]
- Dai, C.; Xie, Y.; Zhuang, X.; Yuan, Z. MiR-206 inhibits epithelial ovarian cancer cells growth and invasion via blocking c-Met/AKT/mTOR signaling pathway. Biomed. Pharmacother. 2018, 104, 763–770. [Google Scholar] [CrossRef]
- Zeng, M.; Liu, C.; Gong, H.; Tang, Z.; Wen, J.; Wang, S.; Xiao, S. Therapeutic Potential of Tyrosine-Protein Kinase MET in Osteosarcoma. Front. Mol. Biosci. 2024, 11, 1367331. [Google Scholar] [CrossRef] [PubMed]
- Baliakas, P.; Soussi, T. The TP53 Tumor Suppressor Gene: From Molecular Biology to Clinical Investigations. J. Intern. Med. 2025, 298, 78–96. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Xiong, K.; Liu, J.; Li, M. Lipopolysaccharide promotes apoptosis and oxidative injury of porcine small intestinal epithelial cells by down-regulating the expression of glutamine transporter ASCT2. J. Anim. Sci. 2023, 101, skad229. [Google Scholar] [CrossRef]
- Lin, W.H.; Chang, Y.W.; Hong, M.X.; Hsu, T.C.; Lee, K.C.; Lin, C.; Lee, J.L. STAT3 phosphorylation at Ser727 and Tyr705 differentially regulates the EMT-MET switch and cancer metastasis. Oncogene 2021, 40, 791–805. [Google Scholar] [CrossRef]
- Qin, A.; Chen, S.; Wang, P.; Huang, X.; Zhang, Y.; Liang, L.; Du, L.R.; Lai, D.H.; Ding, L.; Yu, X.; et al. Knockout of NOS2 Promotes Adipogenic Differentiation of Rat MSCs by Enhancing Activation of JAK/STAT3 Signaling. Front. Cell Dev. Biol. 2021, 9, 638518. [Google Scholar] [CrossRef]
- Li, K.; Huang, W.; Wang, Z.; Nie, Q. m6A demethylase FTO regulate CTNNB1 to promote adipogenesis of chicken preadipocyte. J. Anim. Sci. Biotechnol. 2022, 13, 147. [Google Scholar] [CrossRef] [PubMed]
- Janson, N.D.; Jehanathan, N.; Jung, S.; Priyathilaka, T.T.; Nam, B.H.; Kim, M.J.; Lee, J. Insight into the molecular function and transcriptional regulation of activator protein 1 (AP-1) components c-Jun/c-Fos ortholog in red lip mullet (Liza haematocheila). Fish. Shellfish. Immunol. 2019, 93, 597–611. [Google Scholar] [CrossRef]
- Kineman, R.D.; del Rio-Moreno, M.; Sarmento-Cabral, A. 40 YEARS of IGF1: Understanding the Tissue-Specific Roles of IGF1/IGF1R in Regulating Metabolism Using the Cre/loxP System. J. Mol. Endocrinol. 2018, 61, T187–T198. [Google Scholar] [CrossRef]
- Wang, Y.; He, G.; Wang, F.; Zhang, C.; Ge, Z.; Zheng, X.; Deng, H.; Yuan, C.; Zhou, B.; Tao, X.; et al. Aspirin inhibits adipogenesis of tendon stem cells and lipids accumulation in rat injury tendon through regulating PTEN/PI3K/AKT signalling. J. Cell Mol. Med. 2019, 23, 7535–7544. [Google Scholar] [CrossRef] [PubMed]



| Gene | Primer Sequence (5′-3′) | Product Length (bp) | Accession No. |
|---|---|---|---|
| FABP4 | forward: AGGTACCTGGAAACTTGTCTCCA | 92 | NM_174314.2 |
| reverse: CCATGCCAGCCACTTTCCTG | |||
| PPARγ | forward: AGTGGAGCCTGTATCCCCAC | 125 | NM_181024.2 |
| reverse: ACCCTGACGCTTTATCCCCA | |||
| ADIPOR2 | forward: CCCGGCAAGTGTGACATCT | 92 | NM_001040499.2 |
| reverse: TTCGAGACCCCGTGGAAGT | |||
| CD36 | forward: GCATTCTGAAAGTGCGTTGA | 179 | NM_001278621.1 |
| reverse: CGGGTCTGATGAAAGTGGTT | |||
| SCD1 | forward: TTATTCCGTTATGCCCTTGG | 151 | OP920982.1 |
| reverse: GGTAGTTGTGGAAGCCCTCA | |||
| ABCA10 | forward: CGCCCAAGAAACGACTC | 193 | XM_070774182.1 |
| reverse: GAAAAGCCACAAACCCG | |||
| AEBP1 | forward: GGAGTGGGCTCCAGTAGAGA | 175 | XM_024991120.2 |
| reverse: CACGCCCCATCGTAGTAGTC | |||
| INSIG1 | forward: AGAGCCACACAAGTTCAAGC | 288 | NM_001077909.1 |
| reverse: AGCCAGGAGCGGATGTAGAG | |||
| KLF4 | forward: GGAGACGGAGGAGTTCAATGAT | 118 | XM_005210496.5 |
| reverse: GGACGAGGATGAGGCTGATG | |||
| UCP2 | forward: GTTCTACACCAAGGGCTCTGA | 117 | NM_001033611.2 |
| reverse: AACCGGACCTTCACCACAT | |||
| GAPDH | forward: TGAACCACGAGAAGTATAACAACAC | 125 | NM_001034034.2 |
| reverse: GGTCATAAGTCCCTCCACGAT |
| Sample | Clean Reads | Q30 (%) | GC Content (%) | Total Map (%) | Unique Map (%) |
|---|---|---|---|---|---|
| CON0-1 | 42,247,574 | 95.33 | 47.60 | 93.71 | 91.44 |
| CON0-2 | 44,361,380 | 95.51 | 52.19 | 94.36 | 92.14 |
| CON0-3 | 45,886,196 | 95.37 | 52.34 | 94.43 | 92.20 |
| DIF3-1 | 50,051,578 | 95.43 | 50.87 | 92.42 | 90.47 |
| DIF3-2 | 41,690,784 | 95.04 | 49.29 | 91.24 | 89.37 |
| DIF3-3 | 41,965,278 | 94.53 | 49.37 | 90.67 | 88.76 |
| DIF9-1 | 43,073,620 | 95.43 | 49.47 | 92.33 | 90.37 |
| DIF9-2 | 41,494,356 | 95.14 | 45.55 | 93.64 | 91.70 |
| DIF9-3 | 39,759,198 | 95.24 | 44.96 | 94.05 | 92.01 |
| Genes | DIF3 vs. CON0 | DIF9 vs. CON0 | Significantly Enriched Lipid Metabolism Pathways |
|---|---|---|---|
| ITGB1 | down | normal | PI3K-AKT signaling pathway |
| KRAS | down | normal | PI3K-AKT and apelin signaling pathway |
| CCND1 | down | down | PI3K-AKT, hippo and apelin signaling pathway |
| ACTB | down | down | Hippo signaling pathway |
| VEGFA | down | down | PI3K-AKT signaling pathway |
| MET | down | down | PI3K-AKT signaling pathway |
| ERBB2 | up | normal | PI3K-AKT signaling pathway |
| HRAS | down | down | PI3K-AKT, Apelin, FoxO, Ras, and MAPK signaling pathway |
| EGFR | up | up | PI3K-AKT, FoxO, Ras, and MAPK signaling pathway |
| MYC | up | up | PI3K-AKT, Hippo, MAPK, and Wnt signaling pathway |
| IGF1R | down | down | PI3K-AKT, FoxO, Ras, and MAPK signaling pathway |
| PTEN | normal | down | PI3K-AKT and FoxO signaling pathway |
| TP53 | up | up | PI3K-AKT, MAPK, and Wnt signaling pathway |
| STAT3 | up | up | FoxO signaling pathway |
| JUN | up | up | Wnt and MAPK signaling pathway |
| CASP3 | up | up | MAPK signaling pathway |
| CTNNB1 | normal | up | Wnt signaling pathway |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.; Tang, X.; Zhao, R.; Yan, Y.; Song, X. Transcriptomics Analysis of the Adipogenic Differentiation Mechanism of Bovine Adipose-Derived Neural Crest Stem Cells. Animals 2025, 15, 2353. https://doi.org/10.3390/ani15162353
Zhang K, Tang X, Zhao R, Yan Y, Song X. Transcriptomics Analysis of the Adipogenic Differentiation Mechanism of Bovine Adipose-Derived Neural Crest Stem Cells. Animals. 2025; 15(16):2353. https://doi.org/10.3390/ani15162353
Chicago/Turabian StyleZhang, Kai, Xiaopeng Tang, Rui Zhao, Yibo Yan, and Xianyi Song. 2025. "Transcriptomics Analysis of the Adipogenic Differentiation Mechanism of Bovine Adipose-Derived Neural Crest Stem Cells" Animals 15, no. 16: 2353. https://doi.org/10.3390/ani15162353
APA StyleZhang, K., Tang, X., Zhao, R., Yan, Y., & Song, X. (2025). Transcriptomics Analysis of the Adipogenic Differentiation Mechanism of Bovine Adipose-Derived Neural Crest Stem Cells. Animals, 15(16), 2353. https://doi.org/10.3390/ani15162353






