Clinical and Molecular Characterization of Feline Sporotrichosis in the Brazilian Amazon: PCR-Based Identification of Sporothrix brasiliensis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cats Included in the Study
2.2. Reference Strains of the Sporothrix Genus and Others
2.3. Procedures
2.3.1. Clinical and Epidemiological Characterization
2.3.2. DNA Extraction and PCR Amplification
- (a)
- Silica Column-Based Extraction: DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Briefly, fungal material was incubated with 20 µL of proteinase K and 180 µL of buffer AL at 56 °C for 30 min. After the addition of 200 µL of absolute ethanol, the lysate was transferred to spin columns containing silica membranes. Wash steps were performed with 500 µL each of buffers AW1 and AW2. Genomic DNA was eluted with 100 µL of elution buffer ATE (Tris-EDTA, pH 9.0).
- (b)
- Salt Precipitation-Based Extraction: The protocol employed the MasterPure DNA Purification Kit for Blood (Epicentre, Madison, WI, USA). Fungal cells were lysed with sodium dodecyl sulfate (SDS) and proteinase K, followed by protein precipitation using 3 M sodium acetate. After centrifugation, the DNA-containing supernatant was precipitated with absolute ethanol and washed twice with 70% ethanol. The DNA pellet was air-dried and resuspended in 100 µL of TE buffer (Tris-EDTA).
- (c)
- Phenol–Chloroform Extraction with Mechanical Disruption: This protocol [16] was adapted for Sporothrix spp. to improve cell wall lysis efficiency. Fungal biomass (~200 mg) was transferred to microtubes containing 0.45 mm glass beads and 500 µL of lysis buffer. Samples were subjected to thermal shock (85 °C for 15 min), frozen at −80 °C for 30 min, and then incubated in a boiling water bath at 100 °C for 60 min. Afterwards, the biomass was mechanically disrupted using sterile cotton-free swabs. An additional 500 µL of lysis buffer was added (total volume: 1 mL), and the mixture was vortexed for 30 s. After adding 20 µL of proteinase K, samples were incubated at 56 °C for 1 h. Next, 500 µL of phenol–chloroform–isoamyl alcohol (25:24:1) was added, and the mixture was gently inverted 100 times to homogenize it. Vortexing was avoided to prevent DNA shearing. After centrifugation (13,500 rounds per min (rpm), 15 min), the aqueous phase was transferred to new tubes containing 500 µL of isopropanol and mixed by inversion (50 times). DNA was precipitated by centrifugation (13,500 rpm, 15 min), followed by two washing steps with 500 µL of 70% ethanol. The pellet was air-dried at 65 °C (10 min) and resuspended in 100 µL of TE buffer.
2.3.3. In Silico RFLP Analysis
2.3.4. Phylogenetic Analysis
2.3.5. Antifungal Susceptibility Testing
2.4. Statistical Analysis
3. Results
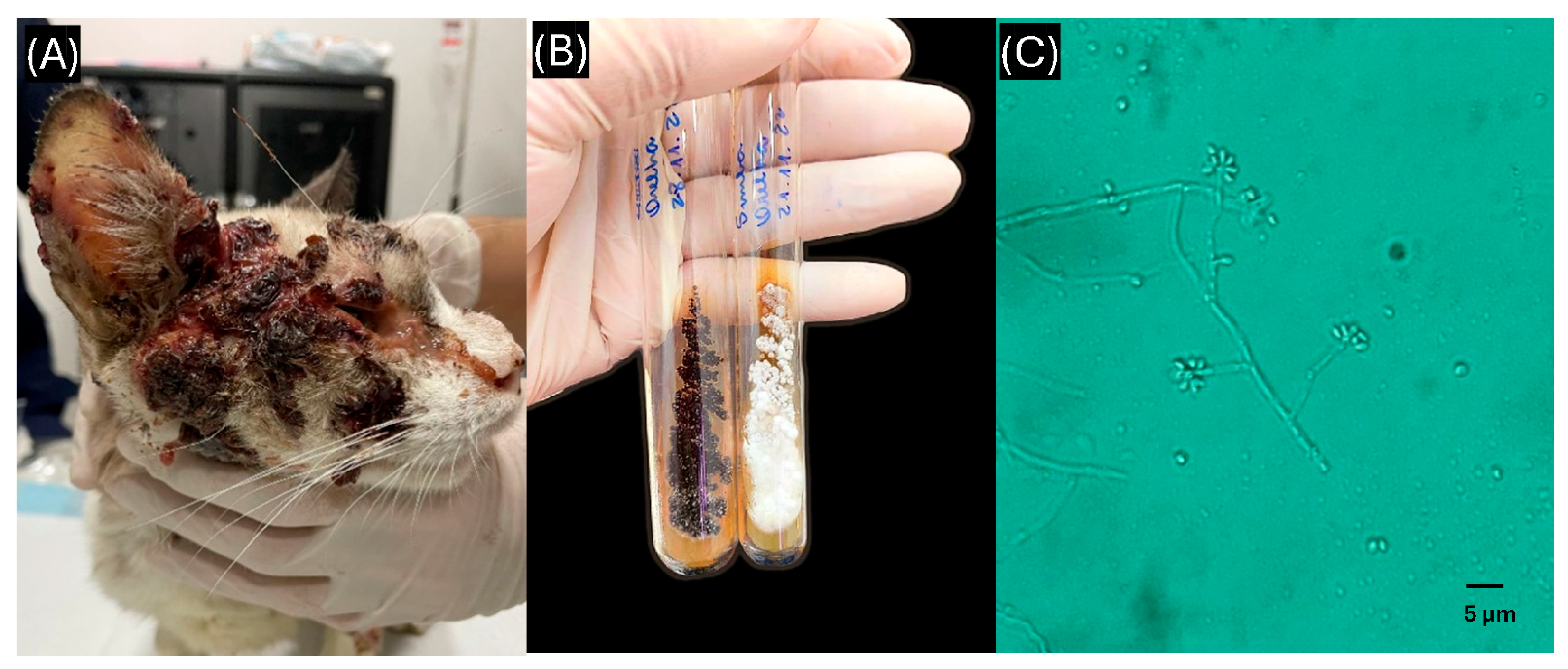
3.1. Clinical and Epidemiological Characteristics
3.2. DNA Extraction Testing and PCR Amplification Efficiency
3.3. In Silico RFLP Analysis
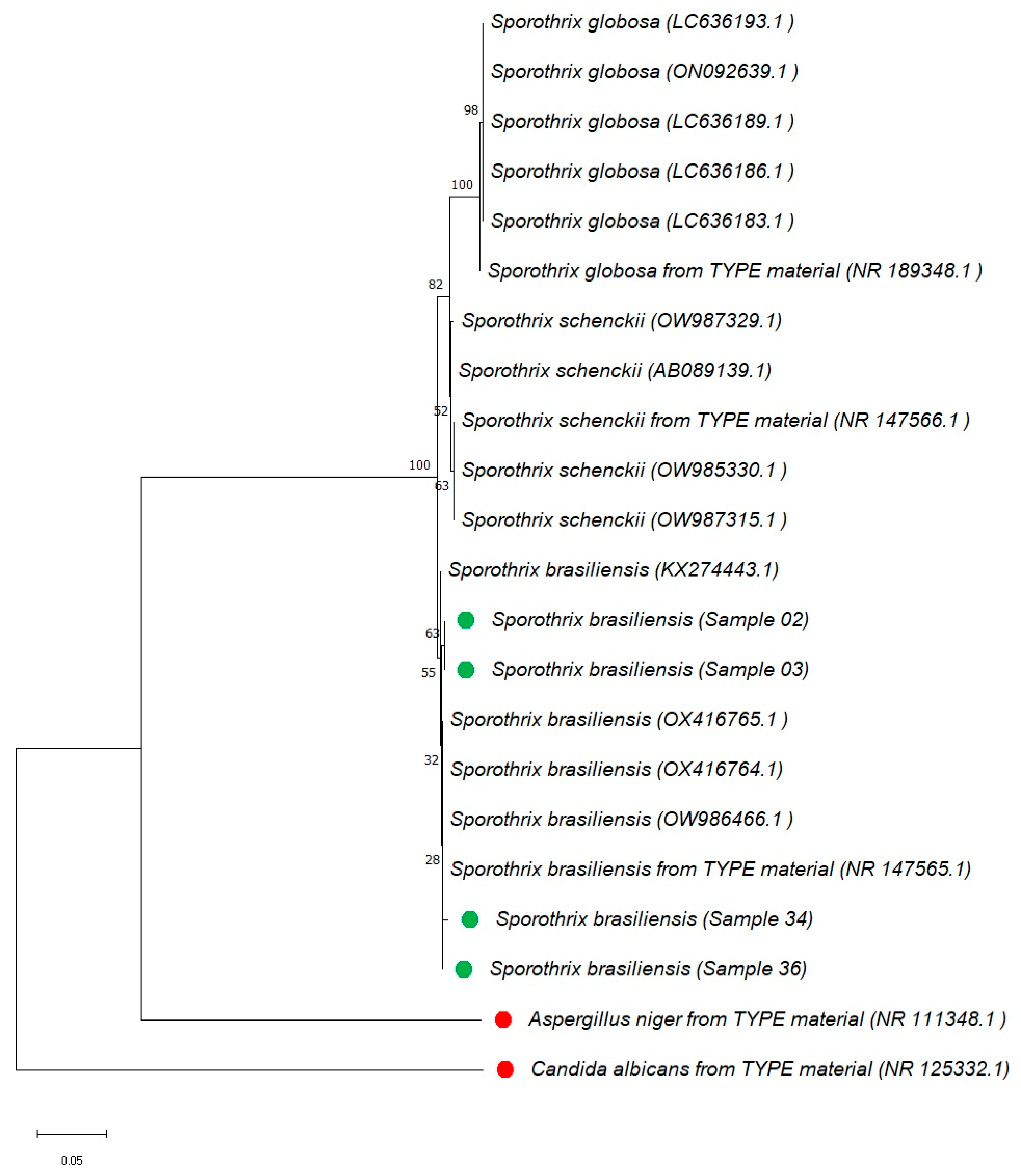
3.4. Phylogenetic Analysis
3.5. Antifungal Susceptibility Profile (MIC Determination)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ITS | Internal Transcribed Spacer |
| SDS | Sodium Dodecyl Sulfate |
| rpm | Revolutions Per Minute |
| AMB | Amphotericin B |
| ITR | Itraconazole |
| CTZ | Clotrimazole |
| RPMI | Roswell Park Memorial Institute (medium) |
| MOPS | 3-(N-morpholino)propanesulfonic acid |
| bp | Base Pairs |
References
- Gremião, I.D.F.; Marques, M.; Oliveira, E.; de Miranda, L.H.M.; Freitas, D.F.S.; Pereira, S.A. Geographic Expansion of Sporotrichosis, Brazil. Emerg. Infect. Dis. 2020, 26, 621. [Google Scholar] [CrossRef]
- Rodrigues, A.M.; Hagen, F.; de Camargo, Z.P. A Spotlight on Sporothrix and Sporotrichosis. Mycopathologia 2022, 187, 407–411. [Google Scholar] [CrossRef]
- Mesquita, V.A.; Talhari, S.; Leturiondo, A.L.; de Souza, G.C.; de Brito, E.M.; de Andrade, S.L.; de Fernandes, D.C.L.; Frota, M.Z.M.; Cruz, R.C.d.S.; Guimarães, J.D.A.R.; et al. Zoonotic Sporotrichosis Outbreak: Emerging Public Health Threat in the Amazon State, Brazil. PLoS Negl. Trop. Dis. 2024, 18, e0012328. [Google Scholar] [CrossRef]
- Rodrigues, A.M.; Della Terra, P.P.; Gremião, I.D.; Pereira, S.A.; Orofino-Costa, R.; de Camargo, Z.P. The Threat of Emerging and Re-Emerging Pathogenic Sporothrix Species. Mycopathologia 2020, 185, 813–842. [Google Scholar] [CrossRef]
- Boechat, J.S.; Oliveira, M.M.E.; Gremião, I.D.F.; Almeida-Paes, R.; de Machado, A.C.S.; Zancopé-Oliveira, R.M.; de Oliveira, R.V.C.; Morgado, D.S.; Corrêa, M.L.; Figueiredo, A.B.F.; et al. Sporothrix brasiliensis and Feline Sporotrichosis in the Metropolitan Region of Rio de Janeiro, Brazil (1998–2018). J. Fungi 2022, 8, 749. [Google Scholar] [CrossRef] [PubMed]
- Dib, I.; Gremião, F.; Martins Da Silva Da Rocha, E.; Montenegro, H.; Aroldo, C.; Borges Carneiro, J.; Xavier, M.O.; Rodrigues De Farias, M.; Monti, F.; Mansho, W.; et al. Guideline for the Management of Feline Sporotrichosis Caused by Sporothrix brasiliensis and Literature Revision. Braz. J. Microbiol. 2021, 52, 107–124. [Google Scholar] [CrossRef]
- Maschio-Lima, T.; Domiciano, M.; Marques, R.; Thiago, H.L.; Lemes, H.; Seron Brizzotti-Mazuchi, N.; Maicon, H.C.; Caetano, H.; Gottardo De Almeida, B.; Letícia, M.B.; et al. Clinical and Epidemiological Aspects of Feline Sporotrichosis Caused by Sporothrix brasiliensis and in Vitro Antifungal Susceptibility. Vet. Res. Commun. 2021, 45, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Rodrigues, A.M.; Feng, P.; De Hoog, G.S. Global ITS Diversity in the Sporothrix Schenckii Complex. Fungal Divers. 2014, 66, 153–165. [Google Scholar] [CrossRef]
- Rodrigues, A.M.; De Hoog, G.; Zhang, Y.; De Camargo, Z.P. Emerging Sporotrichosis Is Driven by Clonal and Recombinant Sporothrix Species. Emerg. Microbes Infect. 2014, 3, e32. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J.W.T.J., Eds.; Academic in Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- do Prado, C.M.; Razzolini, E.; Santacruz, G.; Ojeda, L.; Geraldo, M.R.; Segovia, N.; Pereira Brunelli, J.; Vicente, V.A.; Svoboda, W.K.; Queiroz-Telles, F. First Cases of Feline Sporotrichosis Caused by Sporothrix brasiliensis in Paraguay. J. Fungi 2023, 9, 972. [Google Scholar] [CrossRef]
- Zhang, M.; Li, F.; Li, R.; Gong, J.; Zhao, F. Fast Diagnosis of Sporotrichosis Caused by Sporothrix Globosa, Sporothrix Schenckii, and Sporothrix brasiliensis Based on Multiplex Real-Time PCR. PLoS Negl. Trop. Dis. 2019, 13, e0007219. [Google Scholar] [CrossRef]
- Amadei, S.; Campos, J.; Bertão-Santos, A.; Frentzel, A.; Ávila, H.; Monti, F.S.; Farias, M.R. A Novel Approach for Feline Sporotrichosis Pathogen Detection Based on Loop-Mediated Isothermal Amplification. Vet. Dermatol. 2025, 36, 474–484. [Google Scholar] [CrossRef]
- Lacaz, C.; Porto, E.; Costa, J.; Heins-Vaccari, E.; Melo, N. Tratado de Micologia Médica; Sarvier: São Paulo, Brazil, 2002. [Google Scholar]
- Sidrim, J.; Rocha, M. Micologia Médica à Luz de Autores Conteporâneos; Guanaba Koogan: Rio de Janeiro, Brazil, 2004. [Google Scholar]
- Sambrook, J.; Russell, D.W. Purification of Nucleic Acids by Extraction with Phenol: Chloroform. Cold Spring Harb. Protoc. 2006, 2006, pdb-prot4455. [Google Scholar] [CrossRef]
- Vincze, T.; Posfai, J.; Roberts, R.J. NEBcutter: A Program to Cleave DNA with Restriction Enzymes. Nucleic Acids Res. 2003, 31, 3688. [Google Scholar] [CrossRef]
- Espinel-Ingroff, A.; Chowdhary, A.; Gonzalez, G.M.; Guinea, J.; Hagen, F.; Meis, J.F.; Iii, G.R.T.; Turnidgeh, J. Multicenter Study of Isavuconazole Mic Distributions and Epidemiological Cutoff Values for the Cryptococcus Neoformans-Cryptococcus Gattii Species Complex Using the Clsi M27-A3 Broth Microdilution Method. Antimicrob. Agents Chemother. 2015, 59, 666–668. [Google Scholar] [CrossRef]
- CLSI M27M44S; Performance Standards for Antifungal Susceptibility Testing of Yeasts. Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2022.
- Etchecopaz, A.; Toscanini, M.A.; Gisbert, A.; Mas, J.; Scarpa, M.; Iovannitti, C.A.; Bendezú, K.; Nusblat, A.D.; Iachini, R.; Cuestas, M.L. Sporothrix brasiliensis: A Review of an Emerging South American Fungal Pathogen, Its Related Disease, Presentation and Spread in Argentina. J. Fungi 2021, 7, 170. [Google Scholar] [CrossRef]
- Sampaio, I.D.L.; Freire, A.K.L.; Ogusko, M.M.; Salem, J.I.; De Souza, J.V.B. Selection and Optimization of PCR-Based Methods for the Detection of Histoplasma Capsulatum Var. Capsulatum. Rev. Iberoam. Micol. 2012, 29, 34–39. [Google Scholar] [CrossRef]
- Rodrigues, A.M.; de Hoog, G.S.; de Camargo, Z.P. Sporothrix Species Causing Outbreaks in Animals and Humans Driven by Animal–Animal Transmission. PLoS Pathog. 2016, 12, e1005638. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, D.R.T.; da Silva Santos, C.S.; Wanke, B.; da Silva, R.M.; dos Santos, M.C.; Cruz, K.S.; Monte, R.L.; Nocker, A.; de Souza, J.V.B. PCR-RFLP as a Useful Tool for Diagnosis of Invasivemycoses in a Healthcare Facility in the North of Brazil. Electron. J. Biotechnol. 2015, 18, 231–235. [Google Scholar] [CrossRef]
- Rodrigues, A.M.; de Hoog, G.S.; de Camargo, Z.P. Molecular Diagnosis of Pathogenic Sporothrix Species. PLoS Negl. Trop. Dis. 2015, 9, e0004190. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, J.A.; Monteiro, R.C.; Hagen, F.; de Camargo, Z.P.; Rodrigues, A.M. Trends in Molecular Diagnostics and Genotyping Tools Applied for Emerging Sporothrix Species. J. Fungi 2022, 8, 809. [Google Scholar] [CrossRef] [PubMed]
- Gremião, I.D.F.; Miranda, L.H.M.; Reis, E.G.; Rodrigues, A.M.; Pereira, S.A. Zoonotic Epidemic of Sporotrichosis: Cat to Human Transmission. PLoS Pathog. 2017, 13, e1006077. [Google Scholar] [CrossRef] [PubMed]
- Naiff, R.D.; Barrett, T.V.; Naiff, M.D.F.; Ferreira, L.C.D.L.; Arias, J.R. New Records of Histoplasma Capsulatum from Wild Animals in the Brazilian Amazon. Rev. Inst. Med. Trop. São Paulo 1996, 38, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Waller, S.B.; Dalla Lana, D.F.; Quatrin, P.M.; Ferreira, M.R.A.; Fuentefria, A.M.; Mezzari, A. Antifungal Resistance on Sporothrix Species: An Overview. Clin. Microbiol. Rev. 2021, 52, 73–80. [Google Scholar] [CrossRef]
- Valente, R.M.; de Verçosa, J.V.M.; de Souza, E.R.; Gordiano, N.P.; Barroso, L.d.C.; Carvalho, S.M.d.S.; Alves, M.J.; Fonseca, F.R.; Grisolia, M.E.; de Almeida, M.E.M.; et al. Expansion of Human and Animal Sporotrichosis in Manaus, Amazonas State, Brazil. Cad. Saude Publica 2025, 41, e00180024. [Google Scholar] [CrossRef]

| Epidemiological and Clinical Characteristics | Sporotrichosis Positive Isolates (%) |
|---|---|
| Age | |
| 6 months | 1/29 (3.4%) |
| 8 months | 1/29 (3.4%) |
| 1 year | 6/29 (20.7%) |
| 2 years | 14/29 (48.3%) |
| 3 years | 5/29 (17.2%) |
| 4 years | 1/29 (3.4%) |
| 7 years | 1/29 (3.4%) |
| Sex | |
| Male | 24/29 (82.7%) |
| Female | 5/29 (17.2%) |
| Behavioral features | |
| Indoor | 3/29 (10.3%) |
| Semi-indoor | 13/29 (44.8%) |
| Free-roaming | 13/29 (44.8%) |
| Clinical Manifestation | |
| Localized lesions (lesions confined to a single anatomical region) | 16/29 (55.2%) |
| Disseminated lesions | 13/29 (44.8%) |
| Anatomical distribution of all reported lesions 1 | |
| Nasal planum | 10/43 (23.3%) |
| Face (excluding nasal planum, eyes and ears) | 8/43 (18.6%) |
| Paws (including forelimbs and hindlimbs) | 7/43 (16.3%) |
| Ribs and thoracic flank | 4/43 (9.3%) |
| Ears (auricle) | 3/43 (7.0%) |
| Tail | 3/43 (7.0%) |
| Eye and periocular region | 1/43 (2.3%) |
| Penile region | 1/43 (2.3%) |
| Post-surgical abdominal site | 1/43 (2.3%) |
| Dorsum (thoracolumbar) | 1/43 (2.3%) |
| Paw pads | 2/43 (4.7%) |
| Total number of lesion sites identified | 43/29 cats 2 |
| Enzyme | S. brasiliensis CBS 120339 | S. schenckii CBS 359.36 | S. globosa CBS 120340 | A. niger ATCC 16888 | C. albicans CBS 562 |
|---|---|---|---|---|---|
| HaeIII (GG/CC) | 82, 33, 276, 21, 116 | 81, 33, 276, 21, 114 | 97, 33, 277, 21, 113 | 47, 45, 15, 29, 291, 73, 76 | 69, 426 |
| MspI (C/CGG) | 463, 65 | 462, 63 | 479, 62 | 111, 21, 15, 93, 101, 42, 32, 86, 75 | 276, 219 |
| HinfI (G/ANTC) | 130, 60, 16, 64, 258 | 129, 61, 16, 64, 255 | 145, 63, 16, 64, 253 | 19, 208, 16, 56, 248, 29 | 162, 16, 56, 8, 137, 116 |
| DdeI (C/TNAG) | 154, 374 | 153, 372 | 169, 372 | 187, 389 | 397, 98 |
| BsaI (G/TAC) | 22, 415, 91 | 22, 414, 89 | 41, 412, 88 | 73, 503 | 495 |
| Antifungal Agent | MIC Range (µg/mL) | Geometric Mean (µg/mL) |
|---|---|---|
| Ketoconazole | 0.125–0.5 | 0.25 |
| Itraconazole | 0.25–1 | 0.57 |
| Amphotericin B | 4–8 | 7.27 |
| Fluconazole | 64 | 64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lazameth-Diniz, N.F.; Almeida, D.B.d.; Fernandes, F.d.S.; Queiroz, A.O.d.S.; Souza, É.S.d.; Cruz, K.S.; Matsuura, A.B.J.; Frickmann, H.; Souza, J.V.B.d. Clinical and Molecular Characterization of Feline Sporotrichosis in the Brazilian Amazon: PCR-Based Identification of Sporothrix brasiliensis. Animals 2025, 15, 2318. https://doi.org/10.3390/ani15152318
Lazameth-Diniz NF, Almeida DBd, Fernandes FdS, Queiroz AOdS, Souza ÉSd, Cruz KS, Matsuura ABJ, Frickmann H, Souza JVBd. Clinical and Molecular Characterization of Feline Sporotrichosis in the Brazilian Amazon: PCR-Based Identification of Sporothrix brasiliensis. Animals. 2025; 15(15):2318. https://doi.org/10.3390/ani15152318
Chicago/Turabian StyleLazameth-Diniz, Nayara Fátima, Danielle Barreto de Almeida, Flávia da Silva Fernandes, Adriana Oliveira da Silva Queiroz, Érica Simplicio de Souza, Kátia Santana Cruz, Ani Beatriz Jackisch Matsuura, Hagen Frickmann, and João Vicente Braga de Souza. 2025. "Clinical and Molecular Characterization of Feline Sporotrichosis in the Brazilian Amazon: PCR-Based Identification of Sporothrix brasiliensis" Animals 15, no. 15: 2318. https://doi.org/10.3390/ani15152318
APA StyleLazameth-Diniz, N. F., Almeida, D. B. d., Fernandes, F. d. S., Queiroz, A. O. d. S., Souza, É. S. d., Cruz, K. S., Matsuura, A. B. J., Frickmann, H., & Souza, J. V. B. d. (2025). Clinical and Molecular Characterization of Feline Sporotrichosis in the Brazilian Amazon: PCR-Based Identification of Sporothrix brasiliensis. Animals, 15(15), 2318. https://doi.org/10.3390/ani15152318







