A Markerless Approach for Full-Body Biomechanics of Horses
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
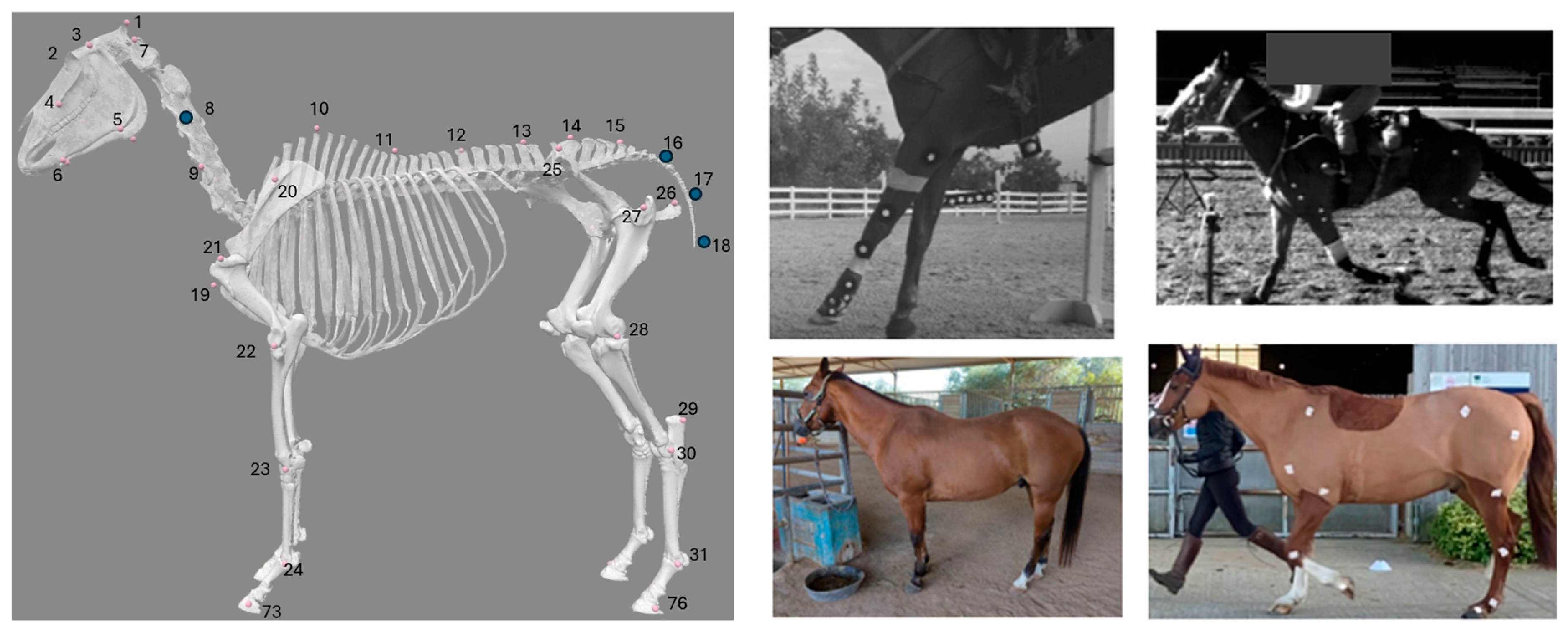
2.1. Equine Datasets
2.2. Skeletal Marker Predictions
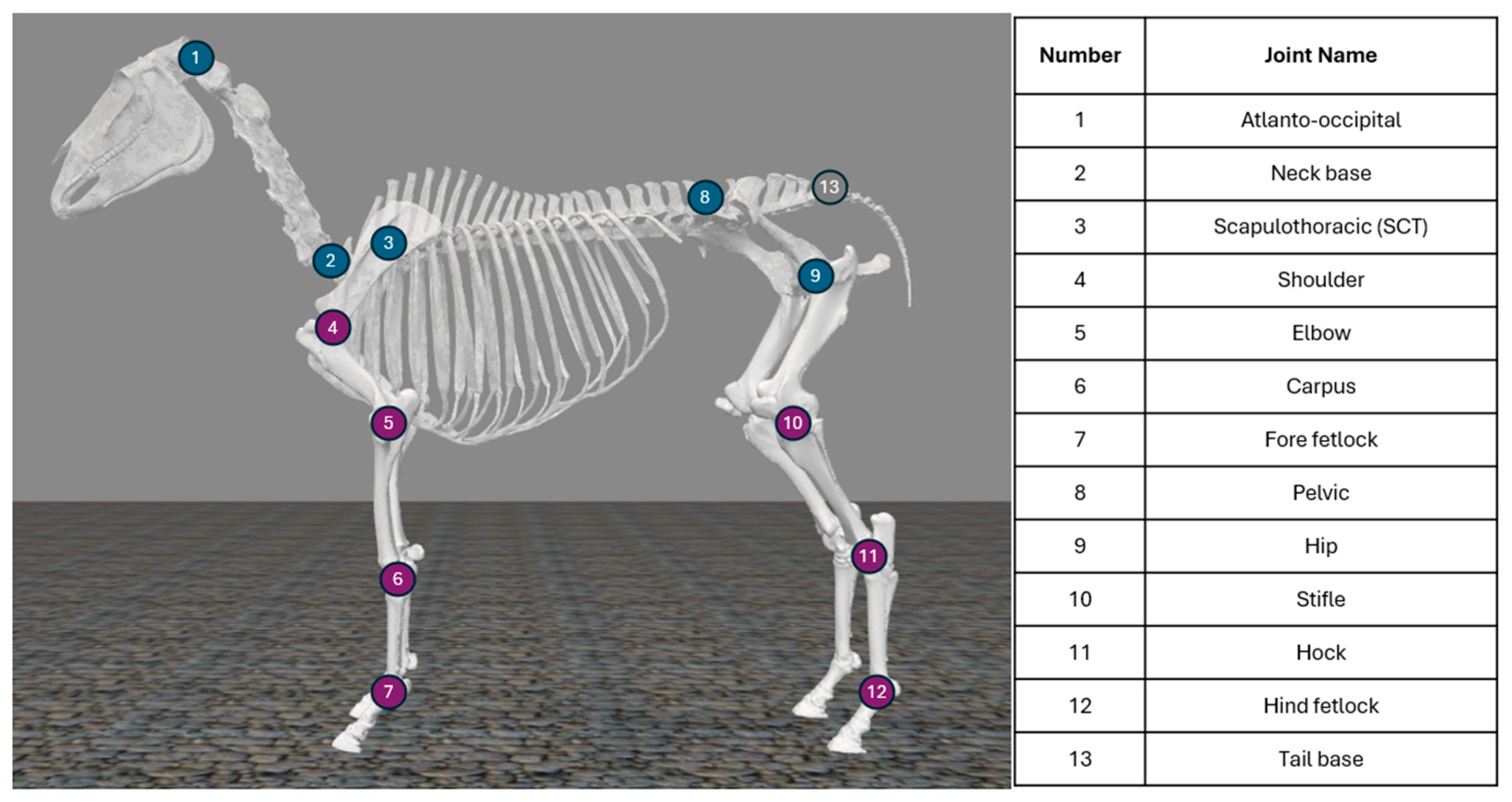
2.3. Equine Biomechanics Model
2.4. Pipeline Validation
3. Results
3.1. Network Training
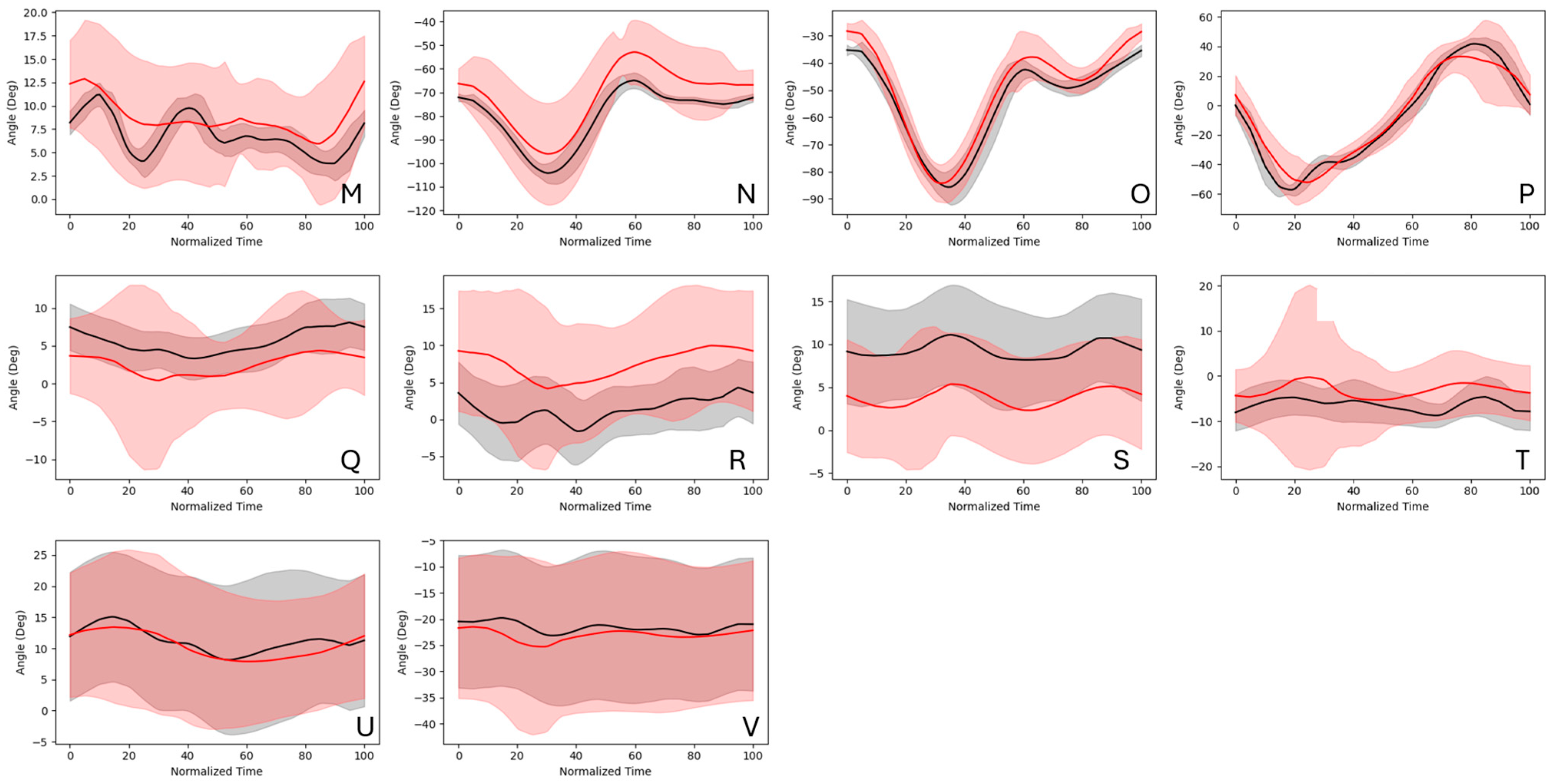
3.2. Kinematics Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| PFERD_Base Prediction Error (cm) Average ± Standard Deviation (Median) | PFERD-SwRI_Horse Prediction Error (cm) Average ± Standard Deviation (Median) | |||
|---|---|---|---|---|
| Axial Markers | Appendicular Markers | Axial Markers | Appendicular Markers | |
| Walk | 10.7 ± 156.1 (4.1) | 9.7 ± 93.7 (5.1) | 4.8 ± 4.4 (3.2) | 3.7 ± 2.4 (3.2) |
| Trot | 16.9 ± 207.5 (4.6) | 6.9 ± 20.7 (5.6) | 6.4 ± 24.8 (3.8) | 4.9 ± 5.4 (3.8) |
| Canter | 17.7 ± 110.8 (4.5) | 6.2 ± 4.1 (5.3) | 5.6 ± 4.6 (4.1) | 4.5 ± 2.7 (3.9) |
| Ground Truth IK Marker Error Average ± Standard Deviation (Median) | PFERD_Base IK Marker Error Average ± Standard Deviation (Median) | PFERD-SwRI_Horse IK Marker Error Average ± Standard Deviation (Median) | ||||
|---|---|---|---|---|---|---|
| Axial Markers | Appendicular Markers | Axial Markers | Appendicular Markers | Axial Markers | Appendicular Markers | |
| Walk | 4.2 ± 2.4 (3.8) | 3.0 ± 2.0 (2.7) | 4.9 ± 3.2 (4.2) | 4.3 ± 2.2 (4.3) | 4.7 ± 3 (4.1) | 3.5 ± 1.9 (3.4) |
| Trot | 4.2 ± 2.3 (3.9) | 3.0 ± 2.0 (2.6) | 4.9 ± 3.1 (4.3) | 4.6 ± 2.4 (4.4) | 5 ± 3.3 (4.3) | 4.1 ± 3.6 (3.8) |
| Canter | 4.1 ± 2.2 (3.8) | 2.9 ± 1.9 (2.6) | 4.7 ± 2.9 (4.0) | 4.3 ± 2.1 (4.2) | 4.9 ± 3.0 (4.2) | 3.6 ± 1.9 (3.5) |
| Angle | Walk | Trot | Canter | |
|---|---|---|---|---|
| Forelimb Joint Angles | SCT; left (Rz) | 4.9 | 3.8 | 3.9 |
| SCT; right (Rz) | 3.9 | 4.6 | 4.4 | |
| SCT; left (Rx) | 2.0 | 3.9 | 3.1 | |
| SCT; right (Rx) | 3.5 | 5.6 | 2.4 | |
| SCT; left (Ry) | 6.5 | 8.1 | 6.5 | |
| SCT; right (Ry) | 3.7 | 5.7 | 4.2 | |
| Shoulder; left (Rz) | 4.5 | 5.9 | 6.7 | |
| Shoulder; right (Rz) | 4.8 | 8.6 | 6.0 | |
| Elbow; left (Rz) | 7.0 | 7.0 | 7.7 | |
| Elbow; right (Rz) | 6.3 | 8.1 | 6.9 | |
| Carpus; left (Rz) | 12.5 | 13.5 | 12.7 | |
| Carpus; right (Rz) | 12.3 | 11.1 | 11.3 | |
| Fore fetlock; left (Rz) | 36.2 | 38.4 | 41.1 | |
| Fore fetlock; right (Rz) | 32.8 | 36.1 | 27.6 | |
| Hindlimb Joint Angles | Hip; left (Rz) | 7.0 | 6.2 | 4.8 |
| Hip; right (Rz) | 9.8 | 10.2 | 10.4 | |
| Hip; left (Rx) | 3.3 | 5.8 | 4.6 | |
| Hip; right (Rx) | 4.3 | 7.3 | 6.0 | |
| Hip; left (Ry) | 4.8 | 4.9 | 4.1 | |
| Hip; right (Ry) | 5.2 | 6.9 | 8.7 | |
| Stifle; left (Rz) | 10.7 | 9.6 | 8.7 | |
| Stifle; right (Rz) | 13.1 | 14.1 | 13.2 | |
| Hock; left (Rz) | 10.9 | 8.3 | 6.4 | |
| Hock; right (Rz) | 12.4 | 14.9 | 15.1 | |
| Hind fetlock; left (Rz) | 13.6 | 17.6 | 13.4 | |
| Hind fetlock; right (Rz) | 21.9 | 24.9 | 21.6 | |
| Other Joint Angles | Atlanto-occipital (Rx) | 3.9 | 4.5 | 4.5 |
| Atlanto-occipital (Ry) | 4.5 | 7.4 | 9.0 | |
| Atlanto-occipital (Rz) | 2.1 | 3.5 | 3.9 | |
| Neck base (Rx) | 3.1 | 5.9 | 3.8 | |
| Neck base (Ry) | 2.8 | 3.5 | 3.2 | |
| Neck base (Rz) | 2.5 | 2.7 | 2.2 | |
| Pelvic; (Rx) | 3.1 | 5.9 | 3.8 | |
| Pelvic; (Ry) | 2.8 | 3.5 | 3.2 | |
| Pelvic; (Rz) | 2.5 | 2.7 | 2.2 |
| Angle Name | Walk | Trot | Canter | |
|---|---|---|---|---|
| Forelimb Joint Angles | SCT; left (Rz) | 5.5 | 6.6 | 4.2 |
| SCT; right (Rz) | 3.5 | 4.3 | 4.6 | |
| SCT; left (Rx) | 2.1 | 3.3 | 2.6 | |
| SCT; right (Rx) | 2.2 | 5.1 | 2.7 | |
| SCT; left (Ry) | 2.2 | 4.4 | 2.8 | |
| SCT; right (Ry) | 4.1 | 4.7 | 5.0 | |
| Shoulder; left (Rz) | 7.5 | 10.6 | 7.7 | |
| Shoulder; right (Rz) | 8.6 | 10.3 | 11.0 | |
| Elbow; left (Rz) | 4.6 | 6.8 | 7.8 | |
| Elbow; right (Rz) | 8.4 | 10.0 | 9.3 | |
| Carpus; left (Rz) | 10.2 | 9.6 | 9.1 | |
| Carpus; right (Rz) | 4.7 | 9.9 | 7.4 | |
| Fore Fetlock; left (Rz) | 15.0 | 15.3 | 13.6 | |
| Fore Fetlock; right (Rz) | 16.6 | 28.5 | 23.8 | |
| Hindlimb Joint Angles | Hip; left (Rz) | 4.2 | 9.9 | 3.7 |
| Hip; right (Rz) | 2.7 | 4.9 | 3.6 | |
| Hip; left (Rx) | 1.9 | 4.2 | 2.6 | |
| Hip; right (Rx) | 2.3 | 5.0 | 4.5 | |
| Hip; left (Ry) | 4.7 | 6.4 | 3.7 | |
| Hip; right (Ry) | 3.3 | 7.1 | 6.2 | |
| Stifle; left (Rz) | 9.1 | 18.0 | 7.8 | |
| Stifle; right (Rz) | 3.8 | 8.8 | 4.1 | |
| Hock; left (Rz) | 5.9 | 7.8 | 6.5 | |
| Hock; right (Rz) | 6.1 | 7.9 | 7.4 | |
| Hind fetlock; left (Rz) | 8.0 | 16.5 | 15.3 | |
| Hind fetlock; right (Rz) | 7.2 | 14.7 | 12.2 | |
| Other Joint Angle | Atlanto-occipital (Rx) | 3.7 | 7.2 | 3.5 |
| Atlanto-occipital (Ry) | 6.7 | 9.3 | 9.1 | |
| Atlanto-occipital (Rz) | 5.2 | 6.5 | 5.2 | |
| Neck base (Rx) | 3.9 | 10.8 | 5.4 | |
| Neck base (Ry) | 2.8 | 5.0 | 4.5 | |
| Neck base (Rz) | 1.6 | 4.8 | 1.8 | |
| Pelvic; (Rx) | 2.8 | 4.4 | 3.3 | |
| Pelvic; (Ry) | 2.1 | 2.9 | 2.7 | |
| Pelvic; (Rz) | 1.4 | 2.4 | 1.5 |
| Angle | Pearson Correlation Coefficient | Interclass Correlation Coefficient | |||||
|---|---|---|---|---|---|---|---|
| Walk | Trot | Canter | Walk | Trot | Canter | ||
| Forelimb Joint Angles | SCT; left (Rz) | 0.95 [0.91–0.98] | 0.96 [0.88–1.00] | 0.95 [0.87–0.98] | 0.87 [0.75–0.94] | 0.94 [0.80–0.99] | 0.89 [0.77–0.94] |
| SCT; right (Rz) | 0.96 [0.85–0.99] | 0.96 [0.87–1.00] | 0.92 [0.86–0.96] | 0.93 [0.77–0.98] | 0.96 [0.87–0.99] | 0.91 [0.81–0.96] | |
| SCT; left (Rx) | 0.78 [0.02–0.96] | 0.72 [0.13–0.96] | 0.32 [-0.22–0.77] | 0.76 [0.02–0.96] | 0.67 [0.12–0.96] | 0.31 [-0.21–0.77] | |
| SCT; right (Rx) | 0.58 [−0.29–0.92] | 0.64 [0.04–0.91] | 0.18 [−0.26–0.89] | 0.55 [−0.28–0.86] | 0.58 [0.04–0.87] | 0.18 [−0.24–0.86] | |
| SCT; left (Ry) | 0.19 [−0.21–0.80] | 0.30 [−0.80–0.93] | 0.31 [−0.01–0.69] | 0.10 [−0.17–0.52] | 0.25 [−0.76–0.90] | 0.21 [−0.01–0.66] | |
| SCT; right (Ry) | 0.49 [−0.30–0.83] | 0.23 [−0.47–0.83] | 0.33 [−0.18–0.68] | 0.46 [−0.30–0.82] | 0.22 [−0.42–0.83] | 0.31 [−0.17–0.68] | |
| Shoulder; left (Rz) | 0.83 [0.69–0.96] | 0.80 [0.09–0.98] | 0.61 [0.22–0.88] | 0.76 [0.49–0.92] | 0.77 [0.09–0.93] | 0.53 [0.21–0.82] | |
| Shoulder; right (Rz) | 0.80 [0.56–0.91] | 0.69 [−0.57–0.92] | 0.63 [0.39–0.92] | 0.77 [0.51–0.91] | 0.65 [−0.52–0.90] | 0.61 [0.39–0.90] | |
| Elbow; left (Rz) | 0.98 [0.96–0.99] | 0.96 [0.92–0.98] | 0.95 [0.93–0.98] | 0.95 [0.90–0.98] | 0.95 [0.84–0.97] | 0.93 [0.88–0.97] | |
| Elbow; right (Rz) | 0.93 [0.74–0.98] | 0.93 [0.79–0.99] | 0.92 [0.80–0.96] | 0.93 [0.73–0.98] | 0.92 [0.74–0.99] | 0.92 [0.76–0.96] | |
| Carpus; left (Rz) | 0.84 [0.62–0.98] | 0.91 [0.70–0.99] | 0.90 [0.80–0.98] | 0.83 [0.60–0.98] | 0.89 [0.62–0.99] | 0.90 [0.77–0.98] | |
| Carpus; right (Rz) | 0.93 [0.77–0.99] | 0.95 [0.75–1.00] | 0.97 [0.96–0.98] | 0.92 [0.77–0.99] | 0.94 [0.70–1.00] | 0.95 [0.89–0.97] | |
| Fore fetlock; left (Rz) | 0.82 [0.67–0.89] | 0.82 [-0.75–0.98] | 0.65 [0.26–0.92] | 0.68 [0.55–0.79] | 0.77 [-0.63–0.95] | 0.62 [0.24–0.92] | |
| Fore fetlock; right (Rz) | 0.64 [0.09–0.96] | 0.45 [-0.69–0.97] | 0.82 [0.63–0.95] | 0.60 [0.09–0.94] | 0.42 [-0.69–0.95] | 0.78 [0.61–0.95] | |
| Hindlimb Joint Angles | Hip; left (Rz) | 0.95 [0.71–0.99] | 0.97 [0.86–1.00] | 0.98 [0.98–0.99] | 0.93 [0.60–0.99] | 0.97 [0.86–1.00] | 0.98 [0.98–0.99] |
| Hip; right (Rz) | 0.97 [0.90–0.99] | 0.96 [0.88–1.00] | 0.92 [0.85–0.98] | 0.93 [0.81–0.99] | 0.95 [0.88–0.99] | 0.91 [0.85–0.98] | |
| Hip; left (Rx) | 0.93 [0.83–0.98] | 0.80 [-0.31–0.99] | 0.83 [0.58–0.94] | 0.92 [0.82–0.98] | 0.77 [−0.19–0.99] | 0.77 [0.56–0.92] | |
| Hip; right (Rx) | 0.81 [0.47–0.96] | 0.53 [-0.67–0.88] | 0.53 [0.16–0.79] | 0.79 [0.46–0.95] | 0.51 [−0.47–0.84] | 0.46 [0.16–0.79] | |
| Hip; left (Ry) | 0.57 [−0.11–0.90] | 0.39 [−0.48–0.77] | 0.61 [−0.45–0.92] | 0.55 [−0.11–0.90] | 0.35 [−0.36–0.74] | 0.59 [−0.45–0.91] | |
| Hip; right (Ry) | 0.22 [−0.35–0.54] | 0.46 [−0.20–0.89] | 0.52 [−0.15–0.86] | 0.20 [−0.32–0.53] | 0.41 [−0.20–0.88] | 0.43 [−0.12–0.71] | |
| Stifle; left (Rz) | 0.78 [0.38–0.94] | 0.92 [0.60–0.99] | 0.92 [0.73–0.98] | 0.74 [0.35–0.93] | 0.88 [0.59–0.99] | 0.85 [0.61–0.94] | |
| Stifle; right (Rz) | 0.91 [0.80–0.98] | 0.85 [0.64–0.98] | 0.60 [−0.08–0.89] | 0.90 [0.78–0.97] | 0.81 [0.56–0.98] | 0.57 [−0.07–0.88] | |
| Hock; left (Rz) | 0.68 [0.30–0.93] | 0.94 [0.76–0.98] | 0.91 [0.81–0.98] | 0.66 [0.30–0.92] | 0.93 [0.76–0.98] | 0.90 [0.76–0.96] | |
| Hock; right (Rz) | 0.71 [0.38–0.98] | 0.74 [0.24–0.98] | 0.63 [−0.28–0.97] | 0.69 [0.38–0.97] | 0.71 [0.23–0.97] | 0.60 [−0.28–0.97] | |
| Hind fetlock; left (Rz) | 0.90 [0.72–0.97] | 0.91 [0.74–0.98] | 0.96 [0.93–0.98] | 0.89 [0.72–0.97] | 0.90 [0.73–0.98] | 0.95 [0.92–0.97] | |
| Hind fetlock; right (Rz) | 0.77 [0.61–0.98] | 0.78 [0.22–0.97] | 0.82 [0.08–0.99] | 0.76 [0.61–0.98] | 0.74 [0.22–0.96] | 0.81 [0.08–0.99] | |
| Other Joint Angles | Atlanto-occipital (Rx) | 0.37 [−0.44–0.72] | 0.43 [−0.47–0.94] | 0.55 [−0.37–0.96] | 0.33 [−0.44–0.71] | 0.38 [−0.37–0.88] | 0.44 [−0.33–0.73] |
| Atlanto-occipital (Ry) | 0.48 [−0.07–0.95] | 0.41 [−0.62–0.92] | 0.28 [−0.38–0.71] | 0.41 [−0.07–0.92] | 0.36 [−0.49–0.91] | 0.22 [−0.22–0.51] | |
| Atlanto-occipital (Rz) | 0.85 [0.68–0.99] | 0.72 [0.19–0.99] | 0.72 [−0.40–0.99] | 0.82 [0.68–0.99] | 0.66 [0.16–0.98] | 0.71 [−0.33–0.97] | |
| Neck base (Rx) | 0.64 [0.13–0.91] | 0.31 −0.42–0.89] | 0.58 [−0.06–0.91] | 0.59 [0.11–0.89] | 0.30 [−0.33–0.89] | 0.53 [−0.04–0.90] | |
| Neck base (Ry) | 0.83 [0.01–0.99] | 0.56 [−0.86–1.00] | 0.61 [−0.46–0.95] | 0.81 [0.01–0.99] | 0.53 [−0.83–0.99] | 0.56 [−0.36–0.81] | |
| Neck base (Rz) | 0.95 [0.81–1.00] | 0.90 [0.73–1.00] | 0.93 [0.79–0.98] | 0.95 [0.81–1.00] | 0.87 [0.44–0.99] | 0.92 [0.76–0.98] | |
| Pelvic; (Rx) | 0.86 [0.74–0.96] | 0.64 [−0.48–0.94] | 0.44 [0.09–0.74] | 0.84 [0.72–0.96] | 0.58 [-0.47–0.94] | 0.41 [0.08–0.71] | |
| Pelvic; (Ry) | 0.72 [−0.01–0.91] | 0.47 [−0.40–0.95] | 0.45 [−0.05–0.87] | 0.64 [−0.01–0.89] | 0.43 [−0.40–0.85] | 0.42 [−0.04–0.81] | |
| Pelvic; (Rz) | 0.63 [0.01–0.79] | 0.67 [−0.14–0.92] | 0.97 [0.93–0.99] | 0.59 [0.01–0.77] | 0.58 [−0.14–0.86] | 0.95 [0.88–0.98] | |
| Angle | Pearson Correlation Coefficient | Interclass Correlation Coefficient | |||||
|---|---|---|---|---|---|---|---|
| Walk | Trot | Canter | Walk | Trot | Canter | ||
| Forelimb Joint Angles | SCT; left (Rz) | 0.97 [0.86–0.99] | 0.95 [0.65–0.99] | 0.97 [0.90–0.99] | 0.96 [0.83–0.99] | 0.94 [0.63–0.99] | 0.96 [0.89–0.99] |
| SCT; right (Rz) | 0.97 [0.93–0.99] | 0.95 [0.67–0.99] | 0.97 [0.90–0.98] | 0.97 [0.92–0.99] | 0.93 [0.67–0.99] | 0.95 [0.89–0.97] | |
| SCT; left (Rx) | 0.77 [0.36–0.97] | 0.73 [0.06–0.96] | 0.43 [−0.20–0.70] | 0.75 [0.33–0.97] | 0.68 [0.06–0.96] | 0.38 [−0.18–0.70] | |
| SCT; right (Rx) | 0.82 [0.61–0.95] | 0.57 [−0.40–0.95] | −0.03 [−0.50–0.63] | 0.79 [0.50–0.93] | 0.51 [−0.39–0.94] | −0.01 [−0.42–0.50] | |
| SCT; left (Ry) | 0.46 [−0.22–0.96] | 0.44 [−0.64–0.94] | 0.28 [−0.06–0.80] | 0.32 [−0.22–0.96] | 0.43 [−0.41–0.93] | 0.23 [−0.06–0.79] | |
| SCT; right (Ry) | 0.78 [0.51–0.93] | 0.68 [0.14–0.96] | 0.39 [−0.26–0.93] | 0.74 [0.48–0.93] | 0.62 [0.13–0.95] | 0.35 [−0.26–0.90] | |
| Shoulder; left (Rz) | 0.91 [0.66–0.97] | 0.77 [0.04–0.97] | 0.64 [0.32–0.78] | 0.88 [0.54–0.96] | 0.75 [0.04–0.96] | 0.57 [0.23–0.75] | |
| Shoulder; right (Rz) | 0.80 [0.60–0.91] | 0.59 [−0.09–0.95] | 0.66 [−0.04–0.88] | 0.78 [0.51–0.91] | 0.56 [v0.09–0.94] | 0.65 [−0.04–0.87] | |
| Elbow; left (Rz) | 0.98 [0.95–1.00] | 0.96 [0.91–0.98] | 0.94 [0.79–0.97] | 0.97 [0.92–0.99] | 0.95 [0.91–0.97] | 0.92 [0.74–0.97] | |
| Elbow; right (Rz) | 0.98 [0.95–0.99] | 0.93 [0.78–0.99] | 0.93 [0.89–0.97] | 0.98 [0.92–0.99] | 0.93 [0.75–0.99] | 0.92 [0.88–0.97] | |
| Carpus; left (Rz) | 0.96 [0.82–1.00] | 0.98 [0.85–1.00] | 0.98 [0.98–0.99] | 0.96 [0.82–1.00] | 0.97 [0.85–0.99] | 0.97 [0.89–0.98] | |
| Carpus; right (Rz) | 0.99 [0.98–1.00] | 0.95 [0.34–1.00] | 0.98 [0.96–0.99] | 0.99 [0.98–1.00] | 0.95 [0.33–0.99] | 0.98 [0.96–0.99] | |
| Fore fetlock; left (Rz) | 0.96 [0.86–0.98] | 0.97 [0.86–0.99] | 0.97 [0.95–0.98] | 0.96 [0.86–0.98] | 0.96 [0.83–0.99] | 0.96 [0.95–0.98] | |
| Fore fetlock; right (Rz) | 0.92 [0.66–0.97] | 0.67 [-0.65–0.98] | 0.78 [0.37–0.97] | 0.90 [0.65–0.96] | 0.65 [−0.65–0.96] | 0.72 [0.30–0.93] | |
| Hindlimb Joint Angles | Hip; left (Rz) | 0.98 [0.95–0.99] | 0.94 [0.51–1.00] | 0.98 [0.97–0.99] | 0.97 [0.95–0.99] | 0.93 [0.40–0.99] | 0.98 [0.94–0.99] |
| Hip; right (Rz) | 0.98 [0.94–1.00] | 0.98 [0.95–1.00] | 0.97 [0.91–0.99] | 0.98 [0.90–0.99] | 0.97 [0.72–1.00] | 0.97 [0.91–0.98] | |
| Hip; left (Rx) | 0.97 [0.94–0.99] | 0.90 [0.06–1.00] | 0.91 [0.77–0.96] | 0.97 [0.94–0.99] | 0.89 [0.06–0.99] | 0.89 [0.74–0.96] | |
| Hip; right (Rx) | 0.95 [0.88–0.98] | 0.73 [−0.40–0.98] | 0.76 [0.22–0.96] | 0.94 [0.85–0.98] | 0.71 [−0.35–0.98] | 0.72 [0.20–0.95] | |
| Hip; left (Ry) | 0.62 [0.07–0.84] | 0.47 [−0.59–0.80] | 0.77 [0.52–0.91] | 0.59 [0.07–0.84] | 0.43 [−0.43–0.75] | 0.75 [0.52–0.89] | |
| Hip; right (Ry) | 0.60 [0.18–0.81] | 0.65 [−0.04–0.96] | 0.64 [−0.34–0.86] | 0.59 [0.17–0.81] | 0.62 [−0.03–0.96] | 0.64 [−0.22–0.86] | |
| Stifle; left (Rz) | 0.96 [0.89–0.98] | 0.90 [−0.71–0.99] | 0.95 [0.93–0.99] | 0.92 [0.85–0.98] | 0.87 [−0.46–0.99] | 0.94 [0.91–0.99] | |
| Stifle; right (Rz) | 0.97 [0.91–0.99] | 0.94 [0.41–0.99] | 0.95 [0.89–0.99] | 0.96 [0.91–0.99] | 0.92 [0.23–0.99] | 0.93 [0.84–0.98] | |
| Hock; left (Rz) | 0.95 [0.91–0.99] | 0.96 [0.63–0.99] | 0.94 [0.91–0.97] | 0.94 [0.90–0.99] | 0.95 [0.62–0.99] | 0.94 [0.91–0.96] | |
| Hock; right (Rz) | 0.97 [0.93–0.99] | 0.96 [0.84–0.99] | 0.96 [0.94–0.98] | 0.97 [0.93–0.99] | 0.96 [0.72–0.99] | 0.95 [0.93–0.98] | |
| Hind fetlock; left (Rz) | 0.97 [0.84–0.99] | 0.91 [0.04–0.99] | 0.94 [0.77–0.98] | 0.97 [0.84–0.99] | 0.90 [0.04–0.99] | 0.93 [0.75–0.98] | |
| Hind fetlock; right (Rz) | 0.98 [0.95–0.99] | 0.95 [0.75–0.99] | 0.95 [0.85–0.99] | 0.97 [0.94–0.99] | 0.94 [0.75–0.99] | 0.94 [0.84–0.99] | |
| Other Joint Angles | Atlanto-occipital (Rx) | 0.24 [−0.70–0.90] | 0.48 [−0.56–0.97] | 0.69 [0.04–0.99] | 0.22 [-0.59–0.85] | 0.47 [−0.25–0.97] | 0.64 [0.03–0.96] |
| Atlanto-occipital (Ry) | 0.45 [−0.24–0.96] | 0.27 [−0.71–0.97] | 0.38 [−0.19–0.62] | 0.37 [−0.17–0.94] | 0.24 [−0.60–0.92] | 0.32 [−0.16–0.56] | |
| Atlanto-occipital (Rz) | 0.83 [0.61–0.98] | 0.74 [0.09–0.97] | 0.72 [−0.30–0.99] | 0.81 [0.60–0.97] | 0.68 [0.09–0.97] | 0.71 [−0.29–0.99] | |
| Neck base (Rx) | 0.53 [0.11–0.93] | 0.16 [−0.63–0.92] | 0.52 [−0.59–0.94] | 0.43 [0.11–0.84] | 0.15 [−0.63–0.92] | 0.47 [−0.55–0.88] | |
| Neck base (Ry) | 0.81 [−0.33–0.99] | 0.68 [−0.05–0.99] | 0.63 [−0.05–0.93] | 0.79 [−0.33–0.99] | 0.62 [−0.03–0.98] | 0.51 [−0.05–0.86] | |
| Neck base (Rz) | 0.97 [0.91–1.00] | 0.85 [−0.37–1.00] | 0.92 [0.77–0.98] | 0.96 [0.89–1.00] | 0.81 [−0.13–0.99] | 0.90 [0.75–0.97] | |
| Pelvic; (Rx) | 0.86 [0.69–0.95] | 0.73 [−0.53–0.98] | 0.63 [0.19–0.93] | 0.83 [0.66–0.94] | 0.68 [−0.39–0.98] | 0.61 [0.18–0.92] | |
| Pelvic; (Ry) | 0.86 [0.56–0.97] | 0.77 [0.21–0.98] | 0.38 [−0.19–0.95] | 0.83 [0.55–0.96] | 0.70 [0.17–0.96] | 0.36 [−0.17–0.87] | |
| Pelvic; (Rz) | 0.78 [0.64–0.93] | 0.80 [−0.44–0.97] | 0.97 [0.94–0.99] | 0.76 [0.61–0.91] | 0.77 [−0.31–0.96] | 0.96 [0.93–0.99] | |
References
- Kane, A.; Traub-Dargatz, J.; Losinger, W.; Garber, L. The Occurrence and Causes of Lameness and Laminitis in the U.S. Horse Population. In Proceedings of the Annual Convention of the AAEP, San Antonio, TX, USA, 26–29 November 2000. [Google Scholar]
- Keegan, K.G. Evidence-based lameness detection and quantification. Vet. Clin. N. Am. Equine Pract. 2007, 23, 403–423. [Google Scholar] [CrossRef] [PubMed]
- Merridale-Punter, M.S.; Wiethoelter, A.K.; El-Hage, C.M.; Hitchens, P.L. Prevalence and Factors Associated with Working Equid Lameness in Low- and Middle-Income Countries: A Systematic Review and Meta-Analysis. Animals 2022, 12, 3100. [Google Scholar] [CrossRef]
- Ross, M.W.; Dyson, S.J. Diagnosis and Management of Lameness in the Horse; Elsevier Health Sciences: St. Louis, MO, USA, 2010. [Google Scholar]
- Baxter, G.M.; Stashak, T.S.; Keegan, K.G. Examination for Lameness. In Adams and Stashak’s Lameness in Horses; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2020; pp. 67–188. [Google Scholar]
- Pfau, T.; Fiske-Jackson, A.; Rhodin, M. Quantitative assessment of gait parameters in horses: Useful for aiding clinical decision making? Equine Vet. Educ. 2015, 28, 209–215. [Google Scholar] [CrossRef]
- Serra Braganca, F.M.; Rhodin, M.; van Weeren, P.R. On the brink of daily clinical application of objective gait analysis: What evidence do we have so far from studies using an induced lameness model? Vet. J. 2018, 234, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Starke, S.D.; Oosterlinck, M. Reliability of equine visual lameness classification as a function of expertise, lameness severity and rater confidence. Vet. Rec. 2019, 184, 63. [Google Scholar] [CrossRef]
- Hardeman, A.M.; Egenvall, A.; Serra Braganca, F.M.; Swagemakers, J.H.; Koene, M.H.W.; Roepstorff, L.; van Weeren, R.; Bystrom, A. Visual lameness assessment in comparison to quantitative gait analysis data in horses. Equine Vet. J. 2022, 54, 1076–1085. [Google Scholar] [CrossRef]
- EquiGait: Sensor Based Gait Analysis. Available online: www.equigait.co.uk (accessed on 10 January 2024).
- EquiMoves: An Advanced System for Quanitative Gait Analysis. Available online: https://equimoves.nl (accessed on 10 January 2024).
- Equinosis. The Equinosis with Lamness Locator. Available online: https://equinosis.com (accessed on 10 January 2024).
- Qualysis: Animal Motion Capture. Available online: https://www.qualysis.com/life-science/animal-motion-capture (accessed on 10 January 2024).
- Hardeman, A.M.; Serra Braganca, F.M.; Swagemakers, J.H.; van Weeren, P.R.; Roepstorff, L. Variation in gait parameters used for objective lameness assessment in sound horses at the trot on the straight line and the lunge. Equine Vet. J. 2019, 51, 831–839. [Google Scholar] [CrossRef]
- Hardeman, A.M.; Van Weeren, P.R.; Serra Bragança, F.M.; Warmerdam, H.; Bok, H.G.J. A first exploration of perceived pros and cons of quantitative gait analysis in equine clinical practice. Equine Vet. Educ. 2021, 34, E438–E444. [Google Scholar] [CrossRef]
- Pfau, T.; Landsbergen, K.; Davis, B.L.; Kenny, O.; Kernot, N.; Rochard, N.; Porte-Proust, M.; Sparks, H.; Takahashi, Y.; Toth, K.; et al. Comparing Inertial Measurement Units to Markerless Video Analysis for Movement Symmetry in Quarter Horses. Sensors 2023, 23, 8414. [Google Scholar] [CrossRef]
- Lawin, F.J.; Bystrom, A.; Roepstorff, C.; Rhodin, M.; Almlof, M.; Silva, M.; Andersen, P.H.; Kjellstrom, H.; Hernlund, E. Is Markerless More or Less? Comparing a Smartphone Computer Vision Method for Equine Lameness Assessment to Multi-Camera Motion Capture. Animals 2023, 13, 390. [Google Scholar] [CrossRef]
- Crecan, C.M.; Pestean, C.P. Inertial Sensor Technologies-Their Role in Equine Gait Analysis, a Review. Sensors 2023, 23, 6301. [Google Scholar] [CrossRef]
- Braganca, F.M.S.; Brommer, H.; van den Belt, A.J.M.; Maree, J.T.M.; van Weeren, P.R.; van Oldruitenborgh-Oosterbaan, M.M.S. Subjective and objective evaluations of horses for fit-to-compete or unfit-to-compete judgement. Vet. J. 2020, 257, 105454. [Google Scholar] [CrossRef]
- Wade, L.; Needham, L.; McGuigan, P.; Bilzon, J. Applications and limitations of current markerless motion capture methods for clinical gait biomechanics. PeerJ 2022, 10, e12995. [Google Scholar] [CrossRef]
- Clayton, H.M.; Schamhardt, H.C. Measurment Techniques for Gait Analysis. In Equine Locomotion; Back, W., Clayton, H.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Serra Braganca, F.M.; Rhodin, M.; Wiestner, T.; Hernlund, E.; Pfau, T.; van Weeren, P.R.; Weishaupt, M.A. Quantification of the effect of instrumentation error in objective gait assessment in the horse on hindlimb symmetry parameters. Equine Vet. J. 2018, 50, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Feuser, A.K.; Gesell-May, S.; Muller, T.; May, A. Artificial Intelligence for Lameness Detection in Horses-A Preliminary Study. Animals 2022, 12, 2804. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V. Equine Gait Analysis, Body Part Tracking using DeepLabCut and Mask R-CNN and Biomechanical Parameter Extraction. Master’s Thesis, Auburn University, Auburn, AL, USA, 2021. [Google Scholar]
- Kallerud, A.S.; Marques-Smith, P.; Bendiksen, H.K.; Fjordbakk, C.T. Objective movement asymmetry in horses is comparable between markerless technology and sensor-based systems. Equine Vet. J. 2025, 57, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Niknejad, N.; Caro, J.L.; Bidese-Puhl, R.; Bao, Y.; Staiger, E.A. Equine Kinematic Gait Analysis Using Stereo Videography and Deep Learning: Stride Length and Stance Duration Estimation. J. ASABE 2023, 66, 865–877. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J.; Zhang, Y.; Sinnott, R.O. Identifying lameness in horses through deep learning. In Proceedings of the 36th Annual ACM Symposium on Applied Computing (SAC’21), Virtual Event, Republic of Korea, 22–26 March 2021; Association for Computing Machinery: New York, NY, USA, 2021; pp. 976–985. [Google Scholar] [CrossRef]
- Li, C.; Ghorbani, N.; Broomé, S.; Rashid, M.; Black, M.J.; Hernlund, E.; Kjellström, H.; Zuffi, S. hSMAL: Detailed Horse Shape and Pose Reconstruction for Motion Pattern Recognition. arXiv 2021, arXiv:2106.10102. Available online: https://arxiv.org/abs/2106.10102 (accessed on 1 January 2024).
- Li, C.; Mellbin, Y.; Krogager, J.; Polikovsky, S.; Holmberg, M.; Ghorbani, N.; Black, M.J.; Kjellström, H.; Zuffi, S.; Hernlund, E. The Poses for Equine Research Dataset (PFERD). Sci. Data 2024, 11, 497. [Google Scholar] [CrossRef]
- Li, C.; Yang, Y.; Weng, Z.; Hernlund, E.; Zuffi, S.; Kjellström, H. Dessie: Disentanglement for Articulated 3D Horse Shape and Pose Estimation from Images. arXiv 2024, arXiv:2410.03438. Available online: https://arxiv.org/abs/2410.03438 (accessed on 1 January 2024).
- Li, C.; Hernlund, E.; Kjellstrom, H.; Zuffi, S. CLHOP: Combined Audio-Video Learning for Horse 3D Pose and Shape Estimation; Computer Vision and Pattern Recognition (cs.CV). arXiv 2024, arXiv:2407.01244. [Google Scholar]
- Zuffi, S.; Mellbin, Y.; Li, C.; Hoeschle, M.; Kjellström, H.; Polikovsky, S.; Hernlund, E.; Black, M.J. VAREN: Very Accurate and Realistic Equine Network. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 16–20 June 2024. [Google Scholar]
- Frazer, L.; Templin, T.; Eliason, T.D.; Butler, C.; Hando, B.; Nicolella, D. Identifying special operative trainees at-risk for musculoskeletal injury using full body kinematics. Front. Bioeng. Biotechnol. 2023, 11, 1293923. [Google Scholar] [CrossRef] [PubMed]
- Eliason, T.D.; Nicolella, D. Development of a Deep Learning Based Markerless Motion Capture System. In Proceedings of the World Congress of Biomechanics Conference, Dublin, Ireland, 8–12 July 2018. [Google Scholar]
- Templin, T.; Eliason, T.D.; Medjaouri, O.; Chambers, D.; Saylor, K.; Nicolella, D. The Effect of Synthetic Training Data on the Performance of a Deep Learning Based Markerless Biomechanics System; Southwest Research Institute: San Antonio, TX, USA, 2020. [Google Scholar]
- Bonilla Lemos Pizzi, G.L.; Holz, K.; Kowalski, E.A.; Fonseca Ribeiro, P.; Blake, R.; Ferreira Martins, C. 2D Kinematic Analysis of the Esbarrada and Volta Sobre Patas Manoeuvres of Criollo Breed Horses Competing in Freio de Ouro. Animals 2024, 14, 2410. [Google Scholar] [CrossRef] [PubMed]
- Rohlf, C.M.; Garcia, T.C.; Marsh, L.J.; Acutt, E.V.; le Jeune, S.S.; Stover, S.M. Effects of Jumping Phase, Leading Limb, and Arena Surface Type on Forelimb Hoof Movement. Animals 2023, 13, 2122. [Google Scholar] [CrossRef] [PubMed]
- Symons, J.E.; Garcia, T.C.; Stover, S.M. Distal hindlimb kinematics of galloping Thoroughbred racehorses on dirt and synthetic racetrack surfaces. Equine Vet. J. 2014, 46, 227–232. [Google Scholar] [CrossRef]
- Banik, P.; Li, L.; Xishuang, D. A Novel Dataset for Keypoint Detection of Quadruped Animals from Images. arXiv. 2021, arXiv:2108.13958. [Google Scholar]
- Yu, H.; Xu, Y.; Zhang, J.; Zhao, W.; Guan, Z.; Tao, D. AP-10K: A Benchmark for Animal Pose Estimation in the Wild. arXiv 2021, arXiv:2108.12617. Available online: https://arxiv.org/abs/2108.12617 (accessed on 1 January 2024).
- Kaiming, H.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Newell, A.; Yang, K.; Deng, J. Stacked Hourglass Networks for Human Pose Estimation. arXiv 2016, arXiv:1603.06937. [Google Scholar]
- Sun, K.; Xiao, B.; Dong, L.; Wang, J. Deep High-Resolution Representation Learning for Human Pose Estimation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019. [Google Scholar]
- Fischler, M.A.; Bolles, R.C. Random Sample Consensus: A Paradigm for Model Fitting with Applications to Image Analysis and Automated Cartography. Commun. ACM 1981, 24, 381–395. [Google Scholar] [CrossRef]
- Torralba, A.; Isola, P.; Freeman, W.T. Foundations of Computer Vision, Section 39: Camera Modeling and Calibration; Adaptive Computation and Machine Learning Series; MIT Press: Cambridge, MA, USA, 2024; Available online: https://mitpress.mit.edu/9780262048972/foundations-of-computer-vision/ (accessed on 1 January 2024).
- Desmarais, Y.; Mottet, D.; Slangen, P.; Montesinos, P. A review of 3D human pose estimation algorithms for markerless motion capture. Comput. Vis. Image Underst. 2021, 212, 103275. [Google Scholar] [CrossRef]
- Matsuura, A.; Ohta, E.; Ueda, K.; Nakatsuji, H.; Kondo, S. Influence of Equine Conformation on Rider Oscillation and Evaluation of Horses for Therapeutic Riding. J. Equine Sci. 2008, 19, 9–18. [Google Scholar] [CrossRef] [PubMed] [PubMed Central][Green Version]
- van Bijlert, P.A.; Geijtenbeek, T.; Smit, I.H.; Schulp, A.S.; Bates, K.T. Muscle-Driven Predictive Physics Simulations of Quadrupedal Locomotion in the Horse. Integr. Comp. Biol. 2024, 64, 694–714. [Google Scholar] [CrossRef]
- He, H.; Palm-Vlasak, L.S.; Chen, C.; Banks, S.A.; Biedrzycki, A.H. Quantification of equine stifle passive kinematics. Am. J. Vet. Res. 2023, 84. [Google Scholar] [CrossRef]
- Setterbo, J.; Garcia, T.; Campbell, I.; Kim, S.; Hubbard, M.; Stover, S. Forelimb Kinematics of Galloping Thoroughbred Racehorses Measured on Dirt, Synthetic, and Turf Track Surfaces (P235). In The Engineering of Sport 7; Springer: Paris, France, 2009; pp. 437–446. [Google Scholar]
- Ripic, Z.; Nienhus, M.; Signorile, J.; Best, T.; Jacobs, K.; Eltoukhy, M. A comparison of three-dimensional kinematics between markerless and marker-based motion capture in overground gait. J. Biomech. 2023, 159, 111793. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Vallat, R. Pingouin: Statistics in Python. J. Open Source Softw. 2018, 3, 1026. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef]
- Uchida, T.K.; Seth, A. Conclusion or Illusion: Quantifying Uncertainty in Inverse Analyses From Marker-Based Motion Capture due to Errors in Marker Registration and Model Scaling. Front. Bioeng. Biotechnol. 2022, 10, 874725. [Google Scholar] [CrossRef] [PubMed]
- Chateau, H.; Degueurce, C.; Denoix, J.M. Effects of 6 degree elevation of the heels on 3D kinematics of the distal portion of the forelimb in the walking horse. Equine Vet. J. 2004, 36, 649–654. [Google Scholar] [CrossRef]
- Chateau, H.; Degueurce, C.; Denoix, J.M. Three-dimensional kinematics of the equine distal forelimb: Effects of a sharp turn at the walk. Equine Vet. J. 2005, 37, 12–18. [Google Scholar] [CrossRef]
- Chateau, H.; Degueurce, C.; Denoix, J.M. Three-dimensional kinematics of the distal forelimb in horses trotting on a treadmill and effects of elevation of heel and toe. Equine Vet. J. 2006, 38, 164–169. [Google Scholar] [CrossRef]
- Clayton, H.M.; Sha, D.; Stick, J.; Elvin, N. 3D kinematics of the equine metacarpophalangeal joint at walk and trot. Vet. Comp. Orthop. Traumatol. 2007, 20, 86–91. [Google Scholar]
- Halley, S.E.; Bey, M.J.; Haladik, J.A.; Lavagnino, M.; Arnoczky, S.P. Three dimensional, radiosteriometric analysis (RSA) of equine stifle kinematics and articular surface contact: A cadaveric study. Equine Vet. J. 2014, 46, 364–369. [Google Scholar] [CrossRef]
- Shaffer, S.K.; Shelly, K.; Garcia, T.C.; Samol, M.A.; Hill, A.E.; Fyhrie, D.P.; Stover, S.M. In vitro motions of the medial and lateral proximal sesamoid bones under mid-stance load conditions are consistent with racehorse fracture configurations. J. Biomech. 2022, 130, 110888. [Google Scholar] [CrossRef] [PubMed]






| Model | PCKh0.1 | PCKh0.25 | PCKh0.5 | PCKh1.0 | Average PCKh |
|---|---|---|---|---|---|
| PFERD_base | 47.4 | 78.6 | 88.2 | 92.1 | 76.5 |
| PFERD-SwRI_Horse | 49.8 | 82.6 | 92.4 | 95.7 | 80.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaffer, S.K.; Medjaouri, O.; Swenson, B.; Eliason, T.; Nicolella, D.P. A Markerless Approach for Full-Body Biomechanics of Horses. Animals 2025, 15, 2281. https://doi.org/10.3390/ani15152281
Shaffer SK, Medjaouri O, Swenson B, Eliason T, Nicolella DP. A Markerless Approach for Full-Body Biomechanics of Horses. Animals. 2025; 15(15):2281. https://doi.org/10.3390/ani15152281
Chicago/Turabian StyleShaffer, Sarah K., Omar Medjaouri, Brian Swenson, Travis Eliason, and Daniel P. Nicolella. 2025. "A Markerless Approach for Full-Body Biomechanics of Horses" Animals 15, no. 15: 2281. https://doi.org/10.3390/ani15152281
APA StyleShaffer, S. K., Medjaouri, O., Swenson, B., Eliason, T., & Nicolella, D. P. (2025). A Markerless Approach for Full-Body Biomechanics of Horses. Animals, 15(15), 2281. https://doi.org/10.3390/ani15152281







