Detection of MC1R Genetic Variants and Their Association with Coat Color in Asian Goats
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Sequencing
2.3. Statistical Analysis
2.4. In Silico Protein Structural Analysis
2.5. Sequence Alignment
3. Results
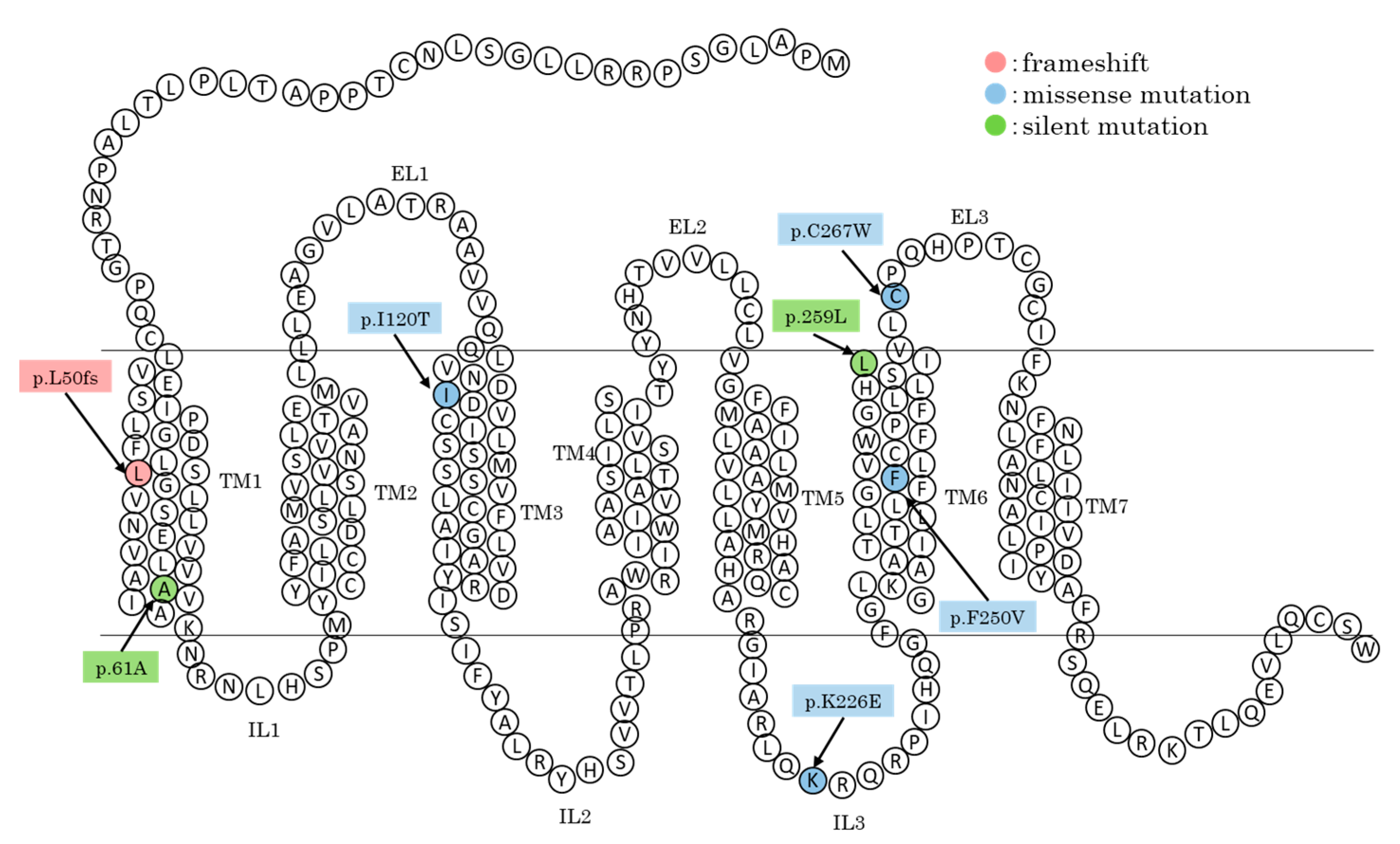
3.1. Genetic Variant Discovery and Characterization
3.2. Associations Between Variants and Coat Colors
3.3. Comparison of MC1R Sequences Among Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MC1R | Melanocortin 1 receptor |
| α-MSH | α-melanocyte-stimulating hormone |
| CDS | Coding sequence |
| RSMD | Root mean square deviation |
| TM | Transmembrane domain |
| EL | Extracellular loop |
| IL | Intracellular loop |
| cAMP | Cyclic adenosine monophosphate |
| pERK | Phosphorylated extracellular signal regulated kinase |
References
- Barsh, G.; Gunn, T.; He, L.; Schlossman, S.; Duke-Cohan, J. Biochemical and genetic studies of pigment-type switching. Pigment. Cell Res. 2000, 8, 48–53. [Google Scholar] [CrossRef]
- Ito, S.; Wakamatsu, K. Human hair melanins: What we have learned and have not learned from mouse coat colour pigmentation. Pigment Cell Melanoma Res. 2010, 24, 63–74. [Google Scholar] [CrossRef]
- Robbins, L.S.; Nadeau, J.H.; Johnson, K.R.; Kelly, M.A.; Roselli-Rehfuss, L.; Baack, E. Pigmentation phenotypes of variant extension locus alleles result from point mutations that alter MSH receptor function. Cell 1993, 72, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Brancalion, L.; Haase, B.; Wade, C.M. Canine coat pigmentation genetics: A review. Anim. Genet. 2021, 53, 3–34. [Google Scholar] [CrossRef]
- Joerg, H.; Fries, H.R.; Meijerink, E.; Stranzinger, G.F. Red coat color in Holstein cattle is associated with a deletion in the MSHR gene. Mamm. Genome 1996, 7, 317–318. [Google Scholar] [CrossRef] [PubMed]
- Klungland, H.; Vage, D.I.; Gomez-Raya, L.; Adalsteinsson, S.; Lien, S. The role of melanocyte-stimulating hormone (MSH) receptor in bovine coat color determination. Mamm. Genome 1995, 6, 636–639. [Google Scholar] [CrossRef] [PubMed]
- Kriegesmann, B.; Dierkes, B.; Leeb, T.; Jansen, S.; Brenig, B. Two breed-specific bovine MC1-R alleles in Brown Swiss and Saler breeds. J. Dairy Sci. 2001, 84, 1768–1771. [Google Scholar] [CrossRef]
- Petersen, J.L.; Kalbfleisch, T.S.; Parris, M.; Tietze, S.M.; Cruickshank, J. MC1R and KIT Haplotypes Associate With Pigmentation Phenotypes of North American Yak (Bos grunniens). J. Hered. 2019, 111, 182–193. [Google Scholar] [CrossRef]
- Valverde, P.; Healy, E.; Jackson, I.; Rees, J.L.; Thody, A.J. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat. Genet. 1995, 11, 328–330. [Google Scholar] [CrossRef]
- Cone, R.D.; Lu, D.; Koppula, S.; Vage, D.I.; Klungland, H.; Boston, B.; Chen, W.; Orth, D.N.; Pouton, C.; Kesterson, R.A. The melanocortin receptors: Agonists, antagonists, and the hormonal control of pigmentation. Recent Prog. Horm. Res. 1996, 51, 287–317. [Google Scholar]
- Kijas, J.M.H.; Wales, R.; Törnsten, A.; Chardon, P.; Moller, M.; Andersson, L. Melanocortin receptor 1 (MC1R) mutations and coat color in pigs. Genetics 1998, 150, 1177–1185. [Google Scholar] [CrossRef] [PubMed]
- Marklund, L.; Moller, M.J.; Sandberg, K.; Andersson, L. A missense mutation in the gene for melanocyte-stimulating hormone receptor (MC1R) is associated with the chestnut coat color in horses. Mamm. Genome 1996, 7, 895–899. [Google Scholar] [CrossRef]
- Våge, D.I.; Klungland, H.; Lu, D.; Cone, R.D. Molecular and pharmacological characterization of dominant black coat color in sheep. Mamm. Genome 1999, 10, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Newton, J.M.; Wilkie, A.L.; He, L.; Jordan, S.A.; Metallinos, D.L.; Holmes, N.G.; Jackson, I.J.; Barsh, G.S. Melanocortin 1 receptor variation in the domestic dog. Mamm. Genome 2000, 11, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Everts, R.E.; Rothuizen, J.; van Oost, B.A. Identification of a premature stop codon in the melanocyte-stimulating hormone receptor gene (MC1R) in Labrador and Golden retrievers with yellow coat colour. Anim. Genet. 2000, 31, 194–199. [Google Scholar] [CrossRef]
- Ritland, K.; Newton, C.; Marshall, H.D. Inheritance and population structure of the white-phased “Kermode” black bear. Curr. Biol. 2001, 11, 1468–1472. [Google Scholar] [CrossRef]
- Eizirik, E.; Yuhki, N.; Johnson, W.E.; Menotti-Raymond, M.; Hannah, S.S.; O’Brien, S.J. Molecular genetics and evolution of melanism in the cat family. Curr. Biol. 2003, 13, 448–453. [Google Scholar] [CrossRef]
- Fontanesi, L.; Tazzoli, M.; Beretti, F.; Russo, V. Mutations in the melanocortin 1 receptor (MC1R) gene are associated with coat colours in the domestic rabbit (Oryctolagus cuniculus). Anim. Genet. 2006, 37, 489–493. [Google Scholar] [CrossRef]
- Nachman, M.W.; Hoekstra, H.E.; D’Agostino, S.L. The genetic basis of adaptive melanism in pocket mice. Proc. Natl. Acad. Sci. USA 2003, 100, 5268–5273. [Google Scholar] [CrossRef]
- Fontanesi, L.; Beretti, F.; Riggio, V.; Dall’Olio, F.; González, E.Z.; Finocchiaro, R.; Davoli, R.; Russo, V.; Portolano, B. Missense and nonsense mutations in melanocortin 1 receptor (MC1R) gene of different goat breeds: Association with red and black coat colour phenotypes but with unexpected evidences. BMC Genet. 2009, 10, 47. [Google Scholar] [CrossRef]
- Guan, D.; Martínez, A.; Luigi-Sierra, M.G.; Delgado, J.V.; Landi, V.; Castelló, A.; Álvarez, J.F.; Such, X.; Jordana, J.; Amills, M. Detecting the footprint of selection on the genomes of Murciano-Granadina goats. Anim. Genet. 2021, 52, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.L.; Li, X.L.; Liu, Y.Q.; Gong, Y.F.; Liu, Z.Z.; Wang, X.J.; Xin, T.R.; Ji, Q. The red head and neck of Boer goats may be controlled by the recessive allele of the MC1R gene. Anim. Res. 2006, 55, 313–322. [Google Scholar] [CrossRef]
- Badaoui, B.; Manunza, A.; Castelló, A.; D’Andrea, M.; Pilla, F.; Capote, J.; Jordana, J.; Ferrando, A.; Martínez, A.; Cabrera, B.; et al. Advantages and limitations of authenticating Palmera goat dairy products by pyrosequencing the melanocortin 1 receptor (MC1R) gene. J. Dairy Sci. 2014, 97, 7293–7297. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Chai, J.; Chen, M.; Tao, Y.X. Identification and pharmacological analyses of eight naturally occurring caprine melanocortin-1 receptor mutations in three different goat breeds. Gen. Comp. Endocrinol. 2016, 235, 1–10. [Google Scholar] [CrossRef]
- Venkatesh, K.M.; Mishra, C.; Pradhan, S.K.; Behera, K.; Mishra, S.R.; Nayak, G. A novel heterozygote allele in caprine melanocortin 1 receptor (MC1R) gene: An association with heat stress traits. Trop. Anim. Health Prod. 2023, 55, 68. [Google Scholar] [CrossRef]
- Mannen, H.; Iso, K.; Kawaguchi, F.; Sasazaki, S.; Yonezawa, T.; Dagong, M.I.A.; Bugiwati, S.R.A. Indonesian native goats (Capra hircus) reveal highest genetic frequency of mitochondrial DNA haplogroup B in the world. Anim. Sci. J. 2020, 91, e13485. [Google Scholar] [CrossRef]
- VarGoats, C.; Nijman, I.J.; Rosen, B.D.; Bardou, P.; Faraut, T.; Cumer, T.; Daly, K.G.; Zheng, Z.; Cai, Y.; Asadol-lahpour, H.; et al. Geographical contrasts of Y-chromosomal haplogroups from wild and domestic goats reveal ancient migrations and recent introgressions. Mol. Ecol. 2022, 16, 4364–4380. [Google Scholar] [CrossRef]
- Hauser, M.; Signer-Hasler, H.; Küttel, L.; Capitan, A.; Guldbrandtsen, B.; Hinrichs, D.; Flury, C.; Seefried, F.R.; Drögemüller, C. Identification of two new recessive MC1R alleles in red-coloured Evolèner cattle and other breeds. Anim. Genet. 2022, 53, 427–435. [Google Scholar] [CrossRef]
- Frändberg, P.A.; Doufexis, M.; Kapas, S.; Chhajlani, V. Amino acid residues in third intracellular loop of melanocortin 1 receptor are involved in G-protein coupling. Biochem. Mol. Biol. Int. 1998, 46, 913–922. [Google Scholar] [CrossRef]
- Huang, H.; Tao, Y.X. Pleiotropic functions of the transmembrane domain 6 of human melanocortin-4 receptor. J. Mol. Endocrinol. 2012, 49, 237–248. [Google Scholar] [CrossRef]
- Wolf Horrell, E.M.; Boulanger, M.C.; D’Orazio, J.A. Melanocortin 1 Receptor: Structure, Function, and Regulation. Front. Genet. 2016, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Remmers, E.F.; Ogunwole, C.B.; Kastner, D.L.; Gregersen, P.K.; Li, W. Effective Sample Size: Quick Estimation of the Effect of Related Samples in Genetic Case-Control Association Analyses. Comput. Biol. Chem. 2011, 35, 40–49. [Google Scholar] [CrossRef] [PubMed]

| Position * | Variant | Codon | Amino Acid Substitution | SNP ID | References |
|---|---|---|---|---|---|
| 16105011 | c.26 G > T | CGG/CTG | p. R9L | - | [22] |
| 16105130 | c.145 G > T | GGG/TGG | p. G49W | - | [22] |
| 16105132 | c.147 delG | GGG/GG | p. L50fs | - | present study |
| 16105168 | c.183 C > T | GCC/GCT | p.61A | - | present study, [20,24,25] |
| 16105227 | c.242 C > T | GCC/GTC | p. A81V | rs646332477 | [20] |
| 16105317 | c.332 C > G | GCC/GGC | p. A111G | - | [25] |
| 16105344 | c.359 T > C | ATT/ACT | p. I120T | - | present study |
| 16105658 | c.673 C > T | CAG/TAG | p. Q225X | - | [20,23] |
| 16105661 | c.676 A > G | AAG/GAG | p. K226E | rs664274065 | present study, [22,23,24] |
| 16105686 | c.701 G > A | GGC/GAC | p. G234D | rs644488799 | [22] |
| 16105733 | c.748 T > G | TTC/GTC | p. F250V | rs657434682 | present study, [20,24,25] |
| 16105749 | c.764 G > A | GGC/GAC | p. G255D | - | [23,24] |
| 16105760 | c.775 C > T | CTG/TTG | p.259L | - | present study |
| 16105778 | c.793 G > A | GTC/ATC | p. V265I | - | [24] |
| 16105786 | c.801 C > G | TGC/TGG | p. C267W | rs669694251 | present study, [20,21,24,25] |
| Variants | Nepal (n = 30) | Philippines (n = 30) | Cambodia (n = 30) | Kazakhstan (n = 30) | Merged (n = 120) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Freq. | n | Freq. | n | Freq. | n | Freq. | n | Freq. | n | ||
| c.147 | GG | 0.83 | 25 | 1.00 | 30 | 1.00 | 30 | 1.00 | 30 | 0.96 | 115 |
| (delG) | G- | 0.13 | 4 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.03 | 4 |
| -- | 0.03 | 1 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.01 | 1 | |
| G | 0.72 | 1.00 | 1.00 | 1.00 | 0.98 | ||||||
| - | 0.28 | 0.00 | 0.00 | 0.00 | 0.02 | ||||||
| c.183 | TT | 0.47 | 14 | 0.67 | 20 | 0.20 | 6 | 0.07 | 2 | 0.35 | 42 |
| (T/C) | TC | 0.40 | 12 | 0.17 | 5 | 0.53 | 16 | 0.53 | 16 | 0.41 | 49 |
| CC | 0.13 | 4 | 0.17 | 5 | 0.27 | 8 | 0.40 | 12 | 0.24 | 29 | |
| T | 0.66 | 0.75 | 0.44 | 0.33 | 0.55 | ||||||
| C | 0.33 | 0.25 | 0.56 | 0.67 | 0.45 | ||||||
| c.359 | TT | 1.00 | 30 | 0.97 | 29 | 1.00 | 30 | 1.00 | 30 | 0.99 | 119 |
| (T/C) | TC | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 |
| CC | 0.00 | 0 | 0.03 | 1 | 0.00 | 0 | 0.00 | 0 | 0.01 | 1 | |
| T | 1.00 | 0.97 | 1.00 | 1.00 | 0.99 | ||||||
| C | 0 | 0.03 | 1.00 | 1.00 | 0.01 | ||||||
| c.676 | AA | 0.53 | 16 | 0.90 | 27 | 0.43 | 13 | 0.33 | 10 | 0.55 | 66 |
| (A/G) | AG | 0.37 | 11 | 0.03 | 1 | 0.37 | 11 | 0.50 | 15 | 0.32 | 38 |
| GG | 0.10 | 3 | 0.07 | 2 | 0.20 | 6 | 0.17 | 5 | 0.13 | 16 | |
| A | 0.72 | 0.92 | 0.66 | 0.58 | 0.71 | ||||||
| G | 0.28 | 0.08 | 0.34 | 0.42 | 0.29 | ||||||
| c.748 | TT | 0.20 | 6 | 0.17 | 5 | 0.27 | 8 | 0.40 | 12 | 0.26 | 31 |
| (T/G) | TG | 0.40 | 12 | 0.17 | 5 | 0.53 | 16 | 0.53 | 16 | 0.41 | 49 |
| GG | 0.40 | 12 | 0.67 | 20 | 0.20 | 6 | 0.07 | 2 | 0.33 | 40 | |
| T | 0.40 | 0.25 | 0.56 | 0.67 | 0.46 | ||||||
| G | 0.60 | 0.75 | 0.44 | 0.33 | 0.54 | ||||||
| c.775 | CC | 1.00 | 30 | 0.97 | 29 | 1.00 | 30 | 1.00 | 30 | 0.99 | 119 |
| (C/T) | CT | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 |
| TT | 0.00 | 0 | 0.03 | 1 | 0.00 | 0 | 0.00 | 0 | 0.01 | 1 | |
| C | 1.00 | 0.97 | 1.00 | 1.00 | 0.99 | ||||||
| T | 0.00 | 0.03 | 0.00 | 0.00 | 0.01 | ||||||
| c.801 | CC | 0.80 | 24 | 0.97 | 29 | 0.77 | 23 | 0.87 | 26 | 0.85 | 102 |
| (C/G) | CG | 0.20 | 6 | 0.03 | 1 | 0.23 | 7 | 0.10 | 3 | 0.14 | 17 |
| GG | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.03 | 1 | 0.01 | 1 | |
| C | 0.90 | 0.98 | 0.91 | 0.92 | 0.92 | ||||||
| G | 0.10 | 0.02 | 0.09 | 0.08 | 0.08 | ||||||
| Nepal (n = 122) | Philippines (n = 110) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| c.147 (delG) | c.359 (T/C) | c.775 (C/T) | |||||||
| Genotype frequency | GG | 0.88 | 107 | TT | 0.95 | 105 | CC | 0.95 | 105 |
| G- | 0.10 | 12 | TC | 0.04 | 4 | CT | 0.04 | 4 | |
| -- | 0.02 | 3 | CC | 0.01 | 1 | TT | 0.01 | 1 | |
| Allele frequency | G | 0.93 | T | 0.97 | C | 0.97 | |||
| - | 0.07 | C | 0.03 | T | 0.03 | ||||
| Population | Coat Color | n | c.676A > G | c.748T > G | c.801C > G | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | TT | TG | GG | CC | CG | GG | |||
| Nepal (n = 30) | Black | 17 | 10 | 5 | 2 | 2 | 7 | 8 | 12 | 5 | 0 |
| Brown | 7 | 3 | 4 | 0 | 3 | 3 | 1 | 7 | 0 | 0 | |
| White | 6 | 3 | 2 | 1 | 1 | 2 | 3 | 5 | 1 | 0 | |
| Philippines (n = 30) | Black | 17 | 17 | 0 | 0 | 1 | 3 | 13 | 16 | 1 | 0 |
| Brown | 9 | 8 | 0 | 1 | 2 | 0 | 9 | 9 | 0 | 0 | |
| White | 4 | 2 | 1 | 1 | 2 | 2 | 0 | 4 | 0 | 0 | |
| Cambodia (n = 30) | Black | 21 | 11 | 7 | 3 | 5 | 11 | 5 | 16 | 5 | 0 |
| Brown | 5 | 2 | 2 | 1 | 1 | 3 | 1 | 3 | 2 | 0 | |
| White | 4 | 0 | 2 | 2 | 2 | 2 | 0 | 4 | 0 | 0 | |
| Kazakhstan (n = 30) | Black | 21 | 8 | 9 | 4 | 8 | 11 | 2 | 20 | 0 | 1 |
| Brown | 4 | 2 | 1 | 1 | 1 | 3 | 0 | 2 | 2 | 0 | |
| White | 5 | 0 | 5 | 0 | 3 | 2 | 0 | 5 | 0 | 0 | |
| Merged (n = 120) | Black | 76 | 46 | 21 | 9 | 16 | 32 | 28 | 64 | 11 | 1 |
| (0.61) | (0.28) | (0.11) | (0.21) | (0.42) | (0.37) | (0.84) | (0.14) | (0.01) | |||
| Brown | 25 | 15 | 7 | 3 | 7 | 9 | 11 | 21 | 4 | 0 | |
| (0.60) | (0.28) | (0.12) | (0.28) | (0.36) | (0.44) | (0.84) | (0.16) | (0.00) | |||
| White | 19 | 5 | 10 | 4 | 8 | 8 | 3 | 18 | 1 | 0 | |
| (0.26) | (0.53) | (0.21) | (0.42) | (0.42) | (0.16) | (0.95) | (0.05) | (0.00) | |||
| p-value | Black vs. Brown | 0.097 † | 0.077 † | 0.084 † | |||||||
| Black vs. White | 0.001 * | 0.002 * | 0.005 * | ||||||||
| Brown vs. White | 0.084 † | 0.309 | 0.267 | ||||||||
| Black vs. Others | 0.189 | 0.547 | 0.935 | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawaguchi, F.; Shaku, A.; Shah, M.K.; Masangkay, J.S.; Mannen, H.; Sasazaki, S. Detection of MC1R Genetic Variants and Their Association with Coat Color in Asian Goats. Animals 2025, 15, 2026. https://doi.org/10.3390/ani15142026
Kawaguchi F, Shaku A, Shah MK, Masangkay JS, Mannen H, Sasazaki S. Detection of MC1R Genetic Variants and Their Association with Coat Color in Asian Goats. Animals. 2025; 15(14):2026. https://doi.org/10.3390/ani15142026
Chicago/Turabian StyleKawaguchi, Fuki, Amane Shaku, Manoj Kumar Shah, Joseph S. Masangkay, Hideyuki Mannen, and Shinji Sasazaki. 2025. "Detection of MC1R Genetic Variants and Their Association with Coat Color in Asian Goats" Animals 15, no. 14: 2026. https://doi.org/10.3390/ani15142026
APA StyleKawaguchi, F., Shaku, A., Shah, M. K., Masangkay, J. S., Mannen, H., & Sasazaki, S. (2025). Detection of MC1R Genetic Variants and Their Association with Coat Color in Asian Goats. Animals, 15(14), 2026. https://doi.org/10.3390/ani15142026






