Effects of Gallic Acid on In Vitro Ruminal Fermentation, Methane Emission, Microbial Composition, and Metabolic Functions
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design, Animals, and Diet
2.2. In Vitro Incubation
2.3. Sample Collection and Analysis
2.4. DNA Extraction and Metagenome Sequencing
2.5. Statistical Analysis
3. Results
3.1. Nutrient Degradability and Gas Production
3.2. In Vitro Fermentation Parameters
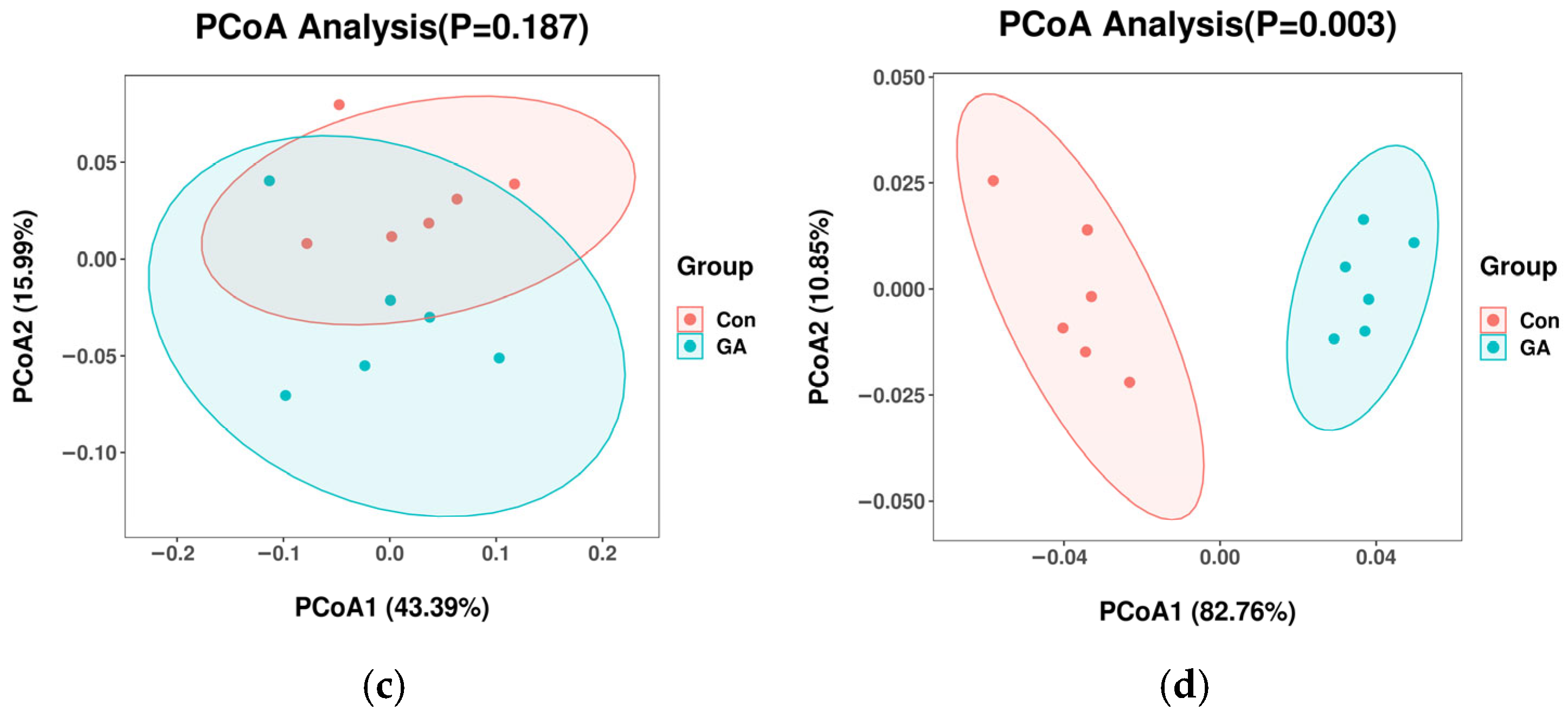
3.3. Alpha Diversity Indices and Principal Coordinate Analysis of Microbial Community
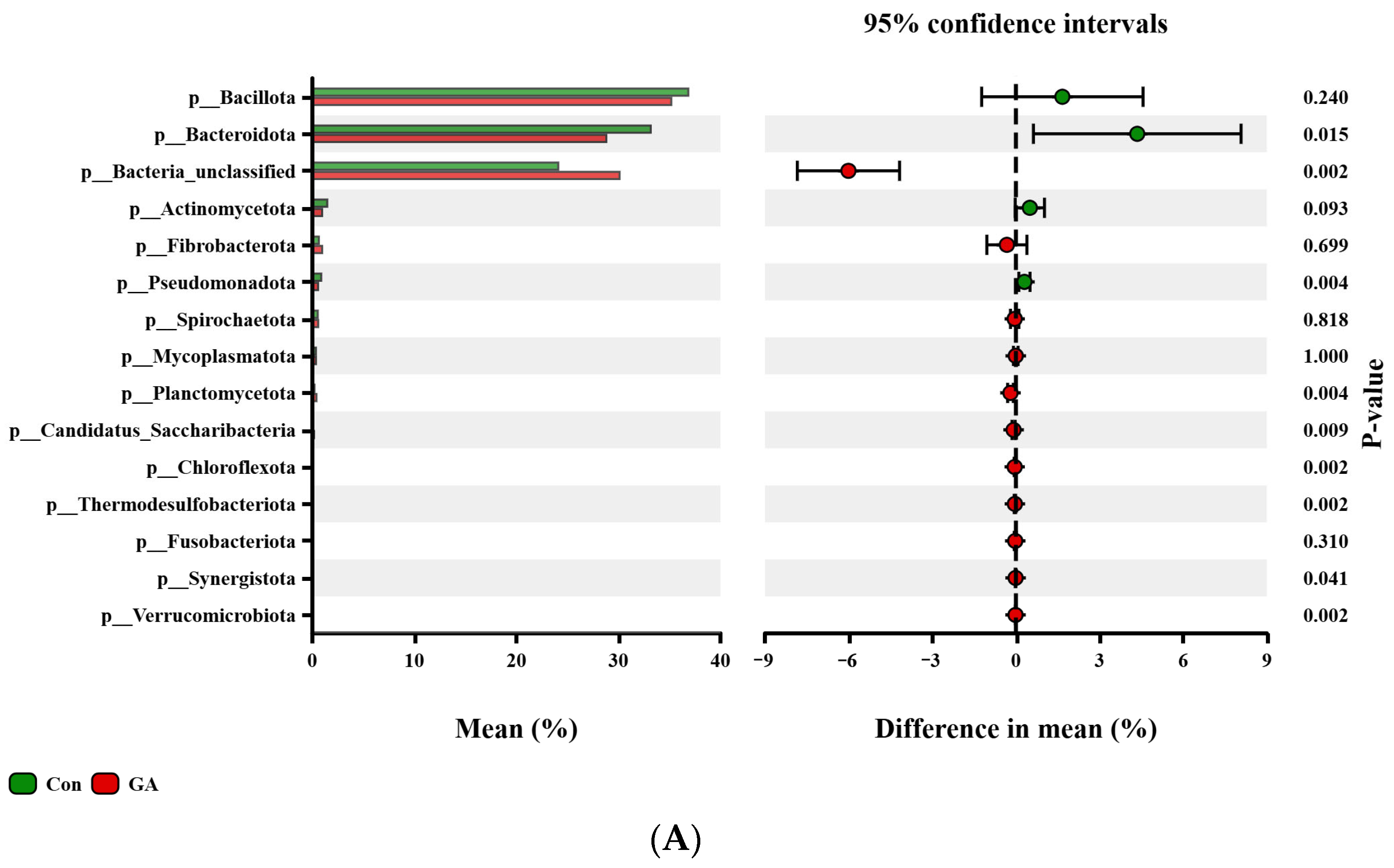
3.4. Differences in Bacterial Community
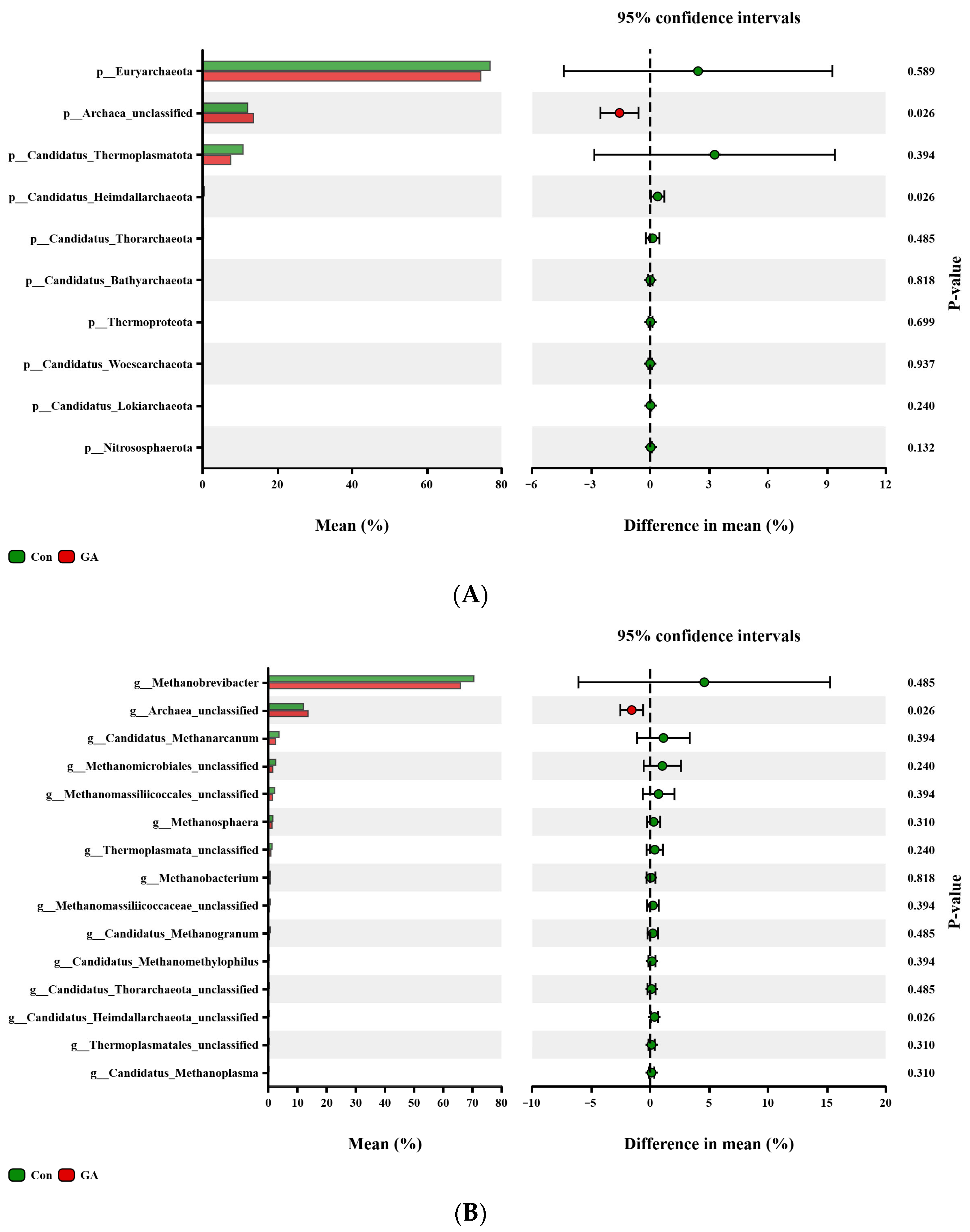
3.5. Differences in Archaeal Community
3.6. Differences in KEGG Functions of Microbial Community
4. Discussion
4.1. Nutrient Digestibility and Gas Production
4.2. Fermentation Parameters
4.3. Microbial Diversity
4.4. Microbial Community
4.5. Microbial Functions in KEGG
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pitta, D.W.; Indugu, N.; Melgar, A.; Hristov, A.; Challa, K.; Vecchiarelli, B.; Hennessy, M.; Narayan, K.; Duval, S.; Kindermann, M.; et al. The effect of 3-nitrooxypropanol, a potent methane inhibitor, on ruminal microbial gene expression profiles in dairy cows. Microbiome 2022, 10, 146. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, K.; Nan, X.; Yang, L.; Wang, Y.; Zhang, F.; Cai, M.; Zhao, Y.; Xiong, B. Effects of combined addition of 3-nitrooxypropanol and vitamin B12 on methane and propionate production in dairy cows by in vitro-simulated fermentation. J. Dairy Sci. 2023, 106, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Bach, A.; Elcoso, G.; Escartín, M.; Spengler, K.; Jouve, A. Modulation of milking performance, methane emissions, and rumen microbiome on dairy cows by dietary supplementation of a blend of essential oils. Animal 2023, 17, 100825. [Google Scholar] [CrossRef]
- Johnson, K.A.; Johnson, D.E. Methane emissions from cattle. J. Anim. Sci. 1995, 73, 2483–2492. [Google Scholar] [CrossRef]
- Martin, C.; Morgavi, D.; Doreau, M. Methane mitigation in ruminants: From microbe to the farm scale. Animal 2010, 4, 351–365. [Google Scholar] [CrossRef]
- Benchaar, C.; McAllister, T.; Chouinard, P. Digestion, ruminal fermentation, ciliate protozoal populations, and milk production from dairy cows fed cinnamaldehyde, quebracho condensed tannin, or yucca schidigera saponin extracts. J. Dairy Sci. 2008, 91, 4765–4777. [Google Scholar] [CrossRef]
- Zhou, R.; Wu, J.; Lang, X.; Liu, L.; Casper, D.P.; Wang, C.; Zhang, L.; Wei, S. Effects of oregano essential oil on in vitro ruminal fermentation, methane production, and ruminal microbial community. J. Dairy Sci. 2020, 103, 2303–2314. [Google Scholar] [CrossRef]
- Romero, P.; Huang, R.; Jiménez, E.; Palma-Hidalgo, J.; Ungerfeld, E.; Popova, M.; Morgavi, D.; Belanche, A.; Yáñez-Ruiz, D. Evaluating the effect of phenolic compounds as hydrogen acceptors when ruminal methanogenesis is inhibited in vitro—Part 2. Dairy goats. Animal 2023, 17, 100789. [Google Scholar] [CrossRef]
- Krumholz, L.R.; Bryant, M.P. Eubacterium oxidoreducens sp. nov. requiring H2 or formate to degrade gallate, pyrogallol, phloroglucinol and quercetin. Arch. Microbiol. 1986, 144, 8–14. [Google Scholar] [CrossRef]
- Huang, R.; Romero, P.; Belanche, A.; Ungerfeld, E.; Yanez-Ruiz, D.; Morgavi, D.; Popova, M. Evaluating the effect of phenolic compounds as hydrogen acceptors when ruminal methanogenesis is inhibited in vitro—Part 1. Dairy cows. Animal 2023, 17, 100788. [Google Scholar] [CrossRef]
- Goel, G.; Makkar, H.P.S. Methane mitigation from ruminants using tannins and saponins. Trop. Anim. Health Prod. 2012, 44, 729–739. [Google Scholar] [CrossRef]
- Rira, M.; Morgavi, D.; Popova, M.; Maxin, G.; Doreau, M. Microbial colonisation of tannin-rich tropical plants: Interplay between digestibility, methane production and tannin disappearance in the rumen. Animal 2022, 16, 100589. [Google Scholar] [CrossRef]
- Wei, C.; Guyader, J.; Collazos, L.; Beauchemin, K.A.; Zhao, G.Y. Effects of gallic acid on in vitro rumen fermentation and methane production using rumen simulation (Rusitec) and batch-culture techniques. Anim. Prod. Sci. 2019, 59, 277–287. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, Q.; Wang, L.; Zhang, C.; Li, Y.; Zhang, Y. Growth performance, digestibility, blood metabolites, ruminal fermentation, and bacterial communities in response to the inclusion of gallic acid in the starter feed of preweaning dairy calves. J. Dairy Sci. 2022, 105, 3078–3089. [Google Scholar] [CrossRef]
- AOAC International. Official Methods of Analysis, 17th ed.; AOAC International: Arlington, VA, USA, 2000. [Google Scholar]
- Broderick, G.A.; Kang, J.H. Automated simultaneous determination of ammonia and amino acids in ruminal fluids and in vitro media. J. Dairy Sci. 1980, 80, 2964–2971. [Google Scholar] [CrossRef]
- Hu, W.L.; Liu, J.X.; Ye, J.A.; Wu, Y.M.; Guo, Y.Q. Effect of tea saponin on rumen fermentation in vitro. Anim. Feed Sci. Tech. 2005, 120, 333–339. [Google Scholar] [CrossRef]
- Yu, Z.; Morrison, M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 2004, 36, 808–812. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, R.; Yu, C.; Li, Y.; Lam, T.-W.; Yiu, S.-M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (refseq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005, 33, D61–D65. [Google Scholar] [CrossRef]
- Bhatta, R.; Uyeno, Y.; Tajima, K.; Takenaka, A.; Yabumoto, Y.; Nonaka, I.; Enishi, O.; Kurihara, M. Difference in the nature of tannins on in vitro ruminal methane and volatile fatty acid production and on methanogenic archaea and protozoal populations. J. Dairy Sci. 2009, 92, 5512–5522. [Google Scholar] [CrossRef]
- Animut, G.; Puchala, R.; Goetsch, A.; Patra, A.; Sahlu, T.; Varel, V.; Wells, J. Methane emission by goats consuming diets with different levels of condensed tannins from lespedeza. Anim. Feed Sci. Tech. 2008, 144, 212–227. [Google Scholar] [CrossRef]
- Waghorn, G. Beneficial and detrimental effects of dietary condensed tannins for sustainable sheep and goat production-progress and challenges. Anim. Feed Sci. Tech. 2008, 147, 116–139. [Google Scholar] [CrossRef]
- Jayanegara, A.; Goel, G.; Makkar, H.P.; Becker, K. Divergence between purified hydrolysable and condensed tannin effects on methane emission, rumen fermentation and microbial population in vitro. Anim. Feed. Sci. Technol. 2015, 209, 60–68. [Google Scholar] [CrossRef]
- Patra, A.K.; Saxena, J. Dietary phytochemicals as rumen modifiers: A review of the effects on microbial populations. Anton. Leeuw. Int. J. G. 2009, 96, 363–375. [Google Scholar] [CrossRef]
- Goel, G.; Puniya, A.K.; Aguilar, C.N.; Singh, K. Interaction of gut microflora with tannins in feeds. Naturwissenschaften 2005, 92, 497–503. [Google Scholar] [CrossRef]
- Rira, M.; Morgavi, D.P.; Genestoux, L.; Djibiri, S.; Sekhri, I.; Doreau, M. Methanogenic potential of tropical feeds rich in hydrolyzable tannins. J. Anim. Sci. 2019, 97, 2700–2710. [Google Scholar] [CrossRef]
- Hassanat, F.; Benchaar, C. Assessment of the effect of condensed (acacia and quebracho) and hydrolysable (chestnut and valonea) tannins on rumen fermentation and methane production in vitro. J. Sci. Food Agr. 2013, 93, 332–339. [Google Scholar] [CrossRef]
- Conradt, D.; Hermann, B.; Gerhardt, S.; Einsle, O.; Müller, M. Biocatalytic properties and structural analysis of phloroglucinol reductases. Angew. Chem. Int. Ed. 2016, 55, 15531–15534. [Google Scholar] [CrossRef]
- Lotfi, R. A commentary on methodological aspects of hydrolysable tannins metabolism in ruminant: A perspective view. Lett. Appl. Microbiol. 2020, 71, 466–478. [Google Scholar] [CrossRef]
- Getachew, G.; Pittroff, W.; Putnam, D.; Dandekar, A.; Goyal, S.; DePeters, E. The influence of addition of gallic acid, tannic acid, or quebracho tannins to alfalfa hay on in vitro rumen fermentation and microbial protein synthesis. Anim. Feed Sci. Tech. 2009, 140, 444–461. [Google Scholar] [CrossRef]
- McSweeney, C.S.; Palmer, B.; McNeill, D.M.; Krause, D.O. Microbial interactions with tannins: Nutritional consequences for ruminants. Anim. Feed Sci. Tech. 2001, 91, 83–93. [Google Scholar] [CrossRef]
- Hackmann, T.J.; Firkins, J.L. Electron transport phosphorylation in rumen butyrivibrios: Unprecedented ATP yield for glucose fermentation to butyrate. Front. Microbiol. 2015, 6, 622. [Google Scholar] [CrossRef]
- Geerkens, C.H.; Schweiggert, R.M.; Steingass, H.; Boguhn, J.; Rodehutscord, M.; Carle, R. Influence of apple and citrus pectins, processed mango peels, a phenolic mango peel extract, and gallic acid as potential feed supplements on in vitro total gas production and rumen methanogenesis. J. Agric. Food Chem. 2013, 61, 5727–5737. [Google Scholar] [CrossRef]
- Soder, K.J.; Roca-Fernandez, A.; Dillard, S.L. Ruminal fermentation and enteric methane production of legumes containing condensed tannins fed in continuous culture. J. Anim. Sci. 2019, 97, 297–298. [Google Scholar] [CrossRef]
- Lan, W.; Yang, C. Ruminal methane production: Associated microorganisms and the potential of applying hydrogen-utilizing bacteria for mitigation. Sci. Total Environ. 2019, 654, 1270–1283. [Google Scholar] [CrossRef]
- Korsa, G.; Konwarh, R.; Masi, C.; Ayele, A.; Haile, S. Microbial cellulase production and its potential application for textile industries. Ann. Microbiol. 2023, 73, 13. [Google Scholar] [CrossRef]
- Zhang, X.; Ke, W.; Ding, Z.; Xu, D.; Wang, M.; Chen, M.; Guo, X. Microbial mechanisms of using feruloyl esterase-producing Lactobacillus plantarum A1 and grape pomace to improve fermentation quality and mitigate ruminal methane emission of ensiled alfalfa for cleaner animal production. J. Environ. Manag. 2022, 308, 114637. [Google Scholar] [CrossRef]
- Ransom-Jones, E.; Jones, D.L.; McCarthy, A.J.; McDonald, J.E. The fibrobacteres: An important phylum of cellulose-degrading bacteria. Microb. Ecol. 2012, 63, 267–281. [Google Scholar] [CrossRef]
- Gharechahi, J.; Sarikhan, S.; Han, J.-L.; Ding, X.-Z.; Salekdeh, G.H. Functional and phylogenetic analyses of camel rumen microbiota associated with different lignocellulosic substrates. NPJ Biofilms Microbi. 2022, 8, 46. [Google Scholar] [CrossRef]
- Xie, Y.; Sun, H.; Xue, M.; Liu, J. Metagenomics reveals differences in microbial composition and metabolic functions in the rumen of dairy cows with different residual feed intake. Anim. Microbiome 2022, 4, 19. [Google Scholar] [CrossRef]
- Melgar, A.; Harper, M.; Oh, J.; Giallongo, F.; Young, M.; Ott, T.; Duval, S.; Hristov, A. Effects of 3-nitrooxypropanol on rumen fermentation, lactational performance, and resumption of ovarian cyclicity in dairy cows. J. Dairy Sci. 2020, 103, 410–432. [Google Scholar] [CrossRef]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The genus alistipes: Gut bacteria with emerging implications to inflammation, cancer, and mental health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef]
- Mahoney-Kurpe, S.C.; Palevich, N.; Noel, S.J.; Gagic, D.; Biggs, P.J.; Soni, P.; Reid, P.M.; Koike, S.; Kobayashi, Y.; Janssen, P.H.; et al. Aristaeella hokkaidonensis gen. nov. sp. nov. and Aristaeella lactis sp. nov., two rumen bacterial species of a novel proposed family, Aristaeellaceae fam. nov. Int. J. Syst. Evol. Micr. 2023, 73, 005831. [Google Scholar] [CrossRef]
- Benchaar, C.; Romero-Pérez, G.; Chouinard, P.; Hassanat, F.; Eugene, M.; Petit, H.; Côrtes, C. Supplementation of increasing amounts of linseed oil to dairy cows fed total mixed rations: Effects on digestion, ruminal fermentation characteristics, protozoal populations, and milk fatty acid composition. J. Dairy Sci. 2012, 95, 4578–4590. [Google Scholar] [CrossRef]
- Zhang, X.M.; Wang, M.; Yu, Q.; Ma, Z.Y.; Beauchemin, K.A.; Wang, R.; Wen, J.N.; Lukuyu, B.A.; Tan, Z.L. Liquid hot water treatment of rice straw enhances anaerobic degradation and inhibits methane production during in vitro ruminal fermentation. J. Dairy Sci. 2020, 103, 4252–4261. [Google Scholar] [CrossRef]
- Miller, T.L.; Lin, C. Description of Methanobrevibacter gottschalkii sp nov., Methanobrevibacter thaueri sp nov., Methanobrevibacter woesei sp nov and Methanobrevibacter wolinii sp nov. Int. J. Syst. Evol. Micr. 2002, 52, 819–822. [Google Scholar] [CrossRef]
- Lee, J.H.; Kumar, S.; Lee, G.H.; Chang, D.H.; Rhee, M.S.; Yoon, M.H.; Kim, B.C. Methanobrevibacter boviskoreani sp nov., isolated from the rumen of Korean native cattle. Int. J. Syst. Evol. Micr. 2013, 63, 4196–4201. [Google Scholar] [CrossRef] [PubMed]




| Ingredient 1 | % | Nutrient Level 2 | % |
|---|---|---|---|
| Corn silage | 31.7 | NEL (Mcal/Kg) | 1.8 |
| Alfalfa hay | 17.8 | CP | 18 |
| Soybean meal | 11.1 | NDF | 38.3 |
| Rapeseed meal | 2.3 | ADF | 25.8 |
| Cottonseed meal | 2.2 | Ca | 0.66 |
| Steam flaked corn | 18.7 | P | 0.36 |
| Dried distillers’ grains with solubles | 4.9 | ||
| Wheat bran | 9.6 | ||
| Minerals and vitamins premix | 1.7 | ||
| Total | 100 |
| Item | GA Levels (mg/g of DM) | SEM | p-Value 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Con | 5 | 10 | 20 | 40 | L | Q | C | ||
| Nutrient digestibility 1 | |||||||||
| DMD, % | 56.1 ab | 53.7 b | 52.2 b | 59.8 a | 60.2 a | 1.75 | 0.22 | 0.006 | 0.29 |
| CPD, % | 56.6 ab | 53.4 bc | 50.8 c | 55.5 ab | 59.5 a | 1.73 | 0.46 | 0.02 | 0.39 |
| NDFD, % | 29.8 | 29.4 | 30.3 | 30.3 | 29.7 | 0.92 | 0.57 | 0.79 | 0.61 |
| ADFD, % | 20.1 | 19.2 | 19.8 | 22.0 | 21.5 | 1.25 | 0.36 | 0.21 | 0.50 |
| Gas production | |||||||||
| Total gas, mL | 230.4 b | 230.1 b | 215.3 c | 229.6 b | 243.3 a | 3.69 | 0.30 | 0.05 | 0.01 |
| CH4, mL | 54.8 c | 49.4 d | 45.7 e | 58.9 b | 64.0 a | 1.09 | 0.09 | <0.0001 | 0.003 |
| CH4/total gas% | 23.8 b | 21.5 c | 21.2 c | 25.7 a | 26.3 a | 0.33 | 0.0006 | <0.0001 | 0.09 |
| CO2, mL | 149.2 b | 147.9 b | 135.4 c | 155.4 b | 166.8 a | 3.12 | 0.66 | 0.001 | 0.003 |
| CO2/total gas% | 64.7 b | 64.3 b | 62.9 b | 67.8 a | 68.6 a | 0.92 | 0.06 | 0.006 | 0.09 |
| Item | GA Levels (mg/g of DM) | SEM | p-Value 1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Con | 5 | 10 | 20 | 40 | L | Q | C | ||
| pH | 6.74 | 6.73 | 6.69 | 6.69 | 6.69 | 0.021 | 0.06 | 0.99 | 0.57 |
| NH3-N (mg/dL) | 32.61 a | 29.39 b | 29.30 b | 28.36 b | 27.79 b | 1.066 | 0.01 | 0.29 | 0.41 |
| MCP (mg/mL) | 29.43 b | 30.61 b | 39.77 a | 39.48 a | 38.64 a | 2.165 | 0.0003 | 0.74 | 0.08 |
| Total VFA (mM) | 79.49 b | 75.29 b | 74.14 b | 85.16 a | 87.02 a | 1.961 | 0.08 | 0.0004 | 0.31 |
| Concentration (mol/100 mol) | |||||||||
| Acetate | 56.54 b | 56.44 b | 55.82 b | 58.24 a | 58.85 a | 0.540 | 0.07 | 0.03 | 0.15 |
| Propionate | 24.14 a | 23.26 ab | 23.43 ab | 22.91 b | 23.19 ab | 0.341 | 0.03 | 0.61 | 0.26 |
| Butyrate | 12.51 bc | 13.69 b | 14.96 a | 12.93 bc | 11.68 c | 0.427 | 0.20 | 0.0006 | 0.08 |
| Isobutyrate | 2.68 | 2.79 | 2.14 | 2.20 | 2.31 | 0.266 | 0.09 | 0.93 | 0.23 |
| Valerate | 2.29 ab | 2.00 bc | 1.85 c | 2.13 abc | 2.45 a | 0.125 | 0.27 | 0.03 | 0.58 |
| Isovalerate | 1.84 a | 1.83 a | 1.80 a | 1.59 ab | 1.52 b | 0.086 | 0.06 | 0.27 | 0.70 |
| A/P | 2.34 b | 2.44 ab | 2.40 ab | 2.54 b | 2.54 b | 0.054 | 0.03 | 0.63 | 0.19 |
| Item | Con | GA | SEM | p-Value |
|---|---|---|---|---|
| Bacteria | ||||
| Observed species | 15,793.83 | 16,195.83 | 133.821 | 0.06 |
| Chao1 | 16,196.87 | 16,475.24 | 127.563 | 0.15 |
| Shannon | 7.94 | 7.61 | 0.019 | <0.01 |
| Simpson | 0.956 | 0.922 | 0.0035 | <0.01 |
| Archaea | ||||
| Observed species | 353.83 | 344.83 | 8.968 | 0.49 |
| Chao1 | 370.84 | 370.55 | 5.067 | 0.97 |
| Shannon | 4.238 | 3.900 | 0.2252 | 0.31 |
| Simpson | 0.886 | 0.865 | 0.0156 | 0.37 |
| Protozoa | ||||
| Observed species | 88.33 | 95.50 | 1.755 | 0.02 |
| Chao1 | 94.82 | 101.42 | 3.004 | 0.15 |
| Shannon | 4.29 | 4.33 | 0.048 | 0.54 |
| Simpson | 0.909 | 0.909 | 0.0025 | 0.99 |
| Fungi | ||||
| Observed species | 1230.33 | 1246.33 | 6.548 | 0.11 |
| Chao1 | 1267.89 | 1278.08 | 9.80 | 0.48 |
| Shannon | 2.513 | 2.419 | 0.0207 | 0.009 |
| Simpson | 0.617 | 0.630 | 0.0044 | 0.054 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, W.; Guo, J.; Li, X.; Li, Y.; Song, L.; Li, Y.; Feng, B.; Bao, X.; Li, J.; Gao, Y.; et al. Effects of Gallic Acid on In Vitro Ruminal Fermentation, Methane Emission, Microbial Composition, and Metabolic Functions. Animals 2025, 15, 1959. https://doi.org/10.3390/ani15131959
Zhu W, Guo J, Li X, Li Y, Song L, Li Y, Feng B, Bao X, Li J, Gao Y, et al. Effects of Gallic Acid on In Vitro Ruminal Fermentation, Methane Emission, Microbial Composition, and Metabolic Functions. Animals. 2025; 15(13):1959. https://doi.org/10.3390/ani15131959
Chicago/Turabian StyleZhu, Wei, Jianjun Guo, Xin Li, Yan Li, Lianjie Song, Yunfei Li, Baoshan Feng, Xingnan Bao, Jianguo Li, Yanxia Gao, and et al. 2025. "Effects of Gallic Acid on In Vitro Ruminal Fermentation, Methane Emission, Microbial Composition, and Metabolic Functions" Animals 15, no. 13: 1959. https://doi.org/10.3390/ani15131959
APA StyleZhu, W., Guo, J., Li, X., Li, Y., Song, L., Li, Y., Feng, B., Bao, X., Li, J., Gao, Y., & Xu, H. (2025). Effects of Gallic Acid on In Vitro Ruminal Fermentation, Methane Emission, Microbial Composition, and Metabolic Functions. Animals, 15(13), 1959. https://doi.org/10.3390/ani15131959





