Simple Summary
Selenium (Se)-deficient soils result in forages that are deficient in this trace mineral. These forages do not meet the nutritional requirement for Se in grazing beef cattle. To overcome this, producers provide Se as a supplement, typically using an inorganic form (ISe), although organic forms of Se (OSe) are available when cattle graze on forage. This Se is incorporated into a class of proteins (selenoproteins) that function as potent antioxidants, protecting cellular membranes from the toxic effects of exposure to free radicals. We previously reported that heifers supplemented with a 1:1 ratio of organic and inorganic forms of Se (MIX), versus the industry standard of ISe alone, develop longer conceptuses during maternal recognition of pregnancy. This study was designed to investigate the relationship between the form of supplemental Se, the expression of selenoproteins in the intercaruncular (ICAR) endometrium, serum concentrations of glucose, triglycerides, and cholesterol, and the global ICAR transcriptome in heifers during this pivotal period in the establishment of a pregnancy.
Abstract
Selenium (Se)-deficient soils result in Se-deficient forages that do not meet the nutritional requirement for this trace mineral in grazing cattle. Supplementation of Se conventionally occurs through a basal vitamin/mineral mix containing Se in an inorganic form (ISe), although organic Se (OSe) is available when cattle graze on forage. We previously reported that the provision of supplemental Se as a 1:1 mixture of ISe:OSe (MIX), in contrast to the industry standard of ISe alone, increases early luteal phase concentrations of systemic progesterone (P4) and increases the length of the conceptus at day 17 of pregnancy. The objectives herein were to determine the effect of the form of supplemental Se on (1) the abundance of mRNA transcripts encoding the family of selenoproteins in the ICAR endometrium; (2) systemic concentrations of glucose, triglycerides, and cholesterol; and (3) the global transcriptome of the ICAR endometrium. Commercial Angus-cross heifers (N = 20) were subjected to a 45-day period of Se depletion, followed by a 45-day period (Se repletion) where they were supplemented with ISe alone. They were then randomly assigned to receive a vitamin/mineral mix containing 35 ppm Se as either ISe (n = 10) or a 1:1 ratio of ISe and OSe (MIX, n = 10) for at least 90 days prior to synchronization of estrous. Following synchronization of estrous and artificial insemination, heifers were killed on day 17 of presumed pregnancy (maternal recognition of pregnancy, MRP) with their reproductive tracts and conceptuses recovered. When pregnancy was confirmed (n = 6 per treatment), ICAR biopsies were sampled from the uterine horn ipsilateral to the functional corpus luteum (CL) for targeted qPCR and RNA-Seq analyses. Targeted qPCR analysis revealed an effect of treatment (p < 0.05) on the abundance of one selenoprotein transcript (Dio2) during MRP. The quantification of serum parameters (d 0, 7, and 17) revealed treatment effects (p < 0.05) on systemic concentrations of glucose and cholesterol, but not triglycerides. RNASeq analysis revealed significant Se-induced changes in transcripts regulating carbohydrate metabolism. Overall, coinciding with the increase in conceptus length, the form of Se affected serum parameters during the establishment of pregnancy and intercaruncular transcriptomics during MRP.
1. Introduction
It has been well established that selenium (Se) should be provided as a supplement to grazing cattle in regions where the soils and forages are deficient in this trace mineral [1]. Producers conventionally supplement Se in a vitamin/mineral mix formulated with an inorganic form of Se (ISe, sodium selenite or sodium selenate), whereas when cattle naturally consume forage, organic forms (OSe, selenomethionine, and selenocysteine) are available [1]. Functionally, Se is an integral component of selenoproteins, a family of enzymes that include glutathione peroxidases [2], which exert protective antioxidant actions by catalyzing the conversion of cellular hydrogen peroxide (H2O2) into water (H2O [3]).
Studies in our laboratory have been designed to investigate the response to the form of supplemental Se provided as either ISe or a 1:1 mixture of ISe:OSe (MIX) to gain a Se-adequate status, on reproductive function. We initially reported a MIX (versus ISe)-induced increase in concentrations of systemic progesterone (P4) in the early luteal phase (days 6 and 7) of the estrous cycle [4,5]. This increased level of P4 appears to be the result of MIX-induced changes to the uptake of cholesterol by the low-density lipoprotein receptor in the corpus luteum (CL), and to a lesser extent, cholesterol generated from both de novo synthesis and the breakdown of cholesterol esters [4,6].
Increased early luteal phase P4 will affect endometrial development and its ability to support the growth of the post-hatch, pre-implantation conceptus [7,8,9]. Receptivity of the uterine endometrium to the conceptus is dependent on P4 [10], and P4-induced changes in endometrial transcriptomics have been shown to alter the composition of histotroph, which consists of growth factors, glucose, hormones, cytokines, enzymes, ions, adhesion molecules, and transport proteins that are secreted into the uterine lumen and are necessary for survival and growth of the conceptus prior to implantation [11,12].
Subsequently, we directly investigated the effect of the form of Se at the critical time of maternal recognition of pregnancy (MRP) and reported a MIX-induced increase in length of the conceptus at day 17 of pregnancy [13]. Concurrent to this, we observed a MIX-induced increase in the relative abundance of mRNA encoding the pivotal myostatin (MSTN) in the intercaruncular (ICAR) endometrium [13], a protein thought to increase glucose availability in histotroph [14]. Importantly, glucose sequestered from maternal blood and secreted into histotroph is the primary energy source for the post-hatch conceptus [11,12]. It appears that form of Se induced changes to the ICAR, with possible effects on serum metabolites, are affecting development of the early conceptus and its ability to signal MRP. The objectives herein were to determine, in MIX versus ISe supplemented heifers, (1) the relative differences in the abundance of mRNA transcripts encoding selenoproteins in the ICAR endometrium during MRP, (2) systemic concentrations of glucose, triglycerides and cholesterol at the time of insemination, as well as days 7 and 17 (MRP) of pregnancy, and (3) the global transcriptome of the ICAR endometrium at MRP. Understanding the mechanism(s) regulating the form of Se-induced changes in blood parameters and the ICAR endometrium at MRP is necessary to refine supplement recommendations to the producer.
2. Materials and Methods
All procedures were approved by the University of Kentucky’s Institutional Animal Care and Use Committee protocol number 2017-2828, date of approval on 14 December 2020.
2.1. Animals and Experimental Procedure
Angus-cross heifers (N = 20) underwent a 45-day period where they received a vitamin/mineral supplement with no added Se (depletion phase), followed by a 45-day period where they received 35 ppm Se as ISe to re-establish systemic Se to adequate concentrations (repletion phase) [15,16]. Heifers were then randomly assigned to one of two treatments: a vitamin/mineral mix containing 35 ppm Se as either inorganic Se (n = 10, ISe, sodium selenite, Prince Agri Products, Inc., Quincy, IL, USA) or a 1:1 ratio of ISe and OSe (n = 10, MIX, SEL-PLEX; Alltech, Inc., Nicholasville, KY, USA). Heifers were supplemented with their respective dietary treatment for at least 90 days prior to synchronization of estrous and insemination.
Whole blood was collected via jugular venipuncture throughout the duration of this trial (the depletion, repletion, and treatment phases). Whole blood was also collected at estrus, day 7, and day 17 of pregnancy. Concentrations of total blood Se were quantified in whole blood by the University of Kentucky’s Veterinary Diagnostics Laboratory (Lexington, KY, USA) using an Agilent 7900 inductively coupled plasma–mass spectrometer [17]. All animals maintained a Se-adequate status [15,16] throughout the treatment period of the trial; and there tended (p = 0.07) to be a greater concentration of Se in the blood of MIX compared to ISe supplemented heifers [13].
2.2. Experimental Regimen and Tissue Collection
Following at least 90 days of treatment with supplemental Se as either ISe or MIX, heifers were randomly injected with one or two doses of Lutalyse (25 mg dinoprost tromethamine, Zoetis, Parsippany, NJ, USA) to induce regression of the CL and then observed daily for behavioral estrus (d 0), using both visual means and with CowManager technology (Gerverscop 9, The Netherlands). The presence of a preovulatory follicle was confirmed via transrectal ultrasonography using a 5–8 MHz linear transducer (Ibex Pro, E.I. Medical Imaging, Loveland, CO, USA), with artificial insemination performed at 0, 12, and 24 h after the observation of estrus. All heifers were inseminated with commercially available frozen semen from a single bull with a record of high fertility.
The animals were slaughtered at the USDA-inspected University of Kentucky Meat Laboratory on d 17 of presumed pregnancy, and an intact conceptus was recovered from six heifers per treatment group (ISe, n = 6; MIX, n = 6). Only 6/10 heifers per treatment group were used for the analyses described herein. Endometrial biopsies were collected from the uterine horn ipsilateral to the ovary bearing the CL. This uterine horn was cut longitudinally to expose the lumen, and an 8 mm biopsy punch (Integra LifeSciences Production Corporation, Mansfield, MA, USA) was utilized to collect intercaruncular endometrial samples from each pregnant heifer. Endometrial samples were flash frozen in liquid N2 and then stored at −80 °C until RNA extraction, real-time PCR (qPCR), and RNA sequencing analysis.
2.3. Serum Analyses
On d 0, 7, and 17, approximately 8 mL of blood was collected into additive-free tubes (Vacutainer, Becton, Dickinson and Company, Franklin Lakes, NJ, USA) via jugular venipuncture for quantification of serum glucose, triglycerides, and cholesterol. These analyses were performed at the Cornell University Animal Health Diagnostics Center (Ithaca, NY, USA).
Serum concentrations of glucose were determined using the hexokinase method, which catalyzes the phosphorylation of glucose to form glucose-6-phosphate. This product is then oxidized by glucose-6-phosphate dehydrogenase to form 6-phosphogluconate, with the reaction also resulting in the conversion of NAD+ to NADPH, which is measured at λ = 340 nm; the product measured directly correlates to the concentration of glucose in the sample.
Serum triglycerides were quantified using the GPO-PAP method, an end-point reaction based on the disruption of triglycerides by lipoprotein lipase, resulting in NEFA and glycerol. Briefly, lipoprotein lipase catalyzes the hydrolysis of the triglycerides to yield glycerol and fatty acids. Following this, glycerol kinase catalyzes the phosphorylation of glycerol, and then glycerophosphate oxidase catalyzes the oxidation of glycerol-3-phosphate to hydrogen peroxide (H2O2) and dihydroxyacetone phosphate. The H2O2 product is measured at λ = 500 nm and is directly proportional to the concentration of triglycerides in the sample.
Total cholesterol was quantified using the CHOD-PAP method, an endpoint reaction. First, cholesterol and free fatty acids are released using cholesterol esterase, and then cholesterol is oxidized to cholest-4-en-3-one by cholesterol oxidase. During this second reaction, H2O2 is produced, which oxidizes a product that fluoresces at λex = 500/λem = 550 nm, a product that is proportional to the concentration of cholesterol present in the sample.
2.4. RNA Extraction
TRIzol (Invitrogen Corporation, Carlsbad, CA, USA) was used to extract total RNA from biopsy punches of the ICAR endometrium. The quality/quantity of all samples was determined using a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). All samples were revealed to have 260/280 absorbance ratios of 1.88 or greater.
2.5. RNA Sequencing
RNA-sequencing analysis was conducted on all 12 ICAR samples; library preparation was performed by Zymo Research Corporation (Irvine, CA, USA). Initially, total RNA (500 ng) was used to construct the total RNA-Seq libraries. Ribosomal RNA (rRNA) was removed as described in [18] with some modifications. Libraries were prepared using the Zymo-Seq RiboFree Total RNA Library Prep Kit (Zymo Research Corporation, Irvine, CA, USA). Sequencing of the RNA-Seq libraries was then performed using an Illumina NovaSeq with a sequencing depth of at least 30 million read pairs per sample.
The RNA-Seq pipeline, as used by the Zymo Research Corporation (Irvine, CA, USA), was adapted from the nf-core/rnaseq pipeline v1.4.2 [19] and built using Nextflow Di Tommaso, 2017; Nextflow enables reproducible computational workflows. FastQC v0.11.9 was used to determine the quality of raw reads, with adaptor and low-quality reads trimmed using Trim Galore! v0.6.6. Alignment of the resultant trimmed reads to the reference genome was performed using STAR v2.6.1d [20], with SAMtools v1.9 used for BAM file filtering and indexing [21]. Library quality control was executed using QualiMap v2.2.2-dev [22] and RSeQC v4.0.0 [23]. Duplicated reads were marked using Picard tools v2.23.9 (Broad Institute, Cambridge, MA, USA), and quality control of the duplication rate was analyzed using dupRadar v1.18.0 [24]. Preseq v2.0.3 [25] was used to estimate the complexity of the library; featureCounts v2.0.1 [26] was used to identify reads that overlapped with exons and to apply gene assignments.
Count data was then uploaded to Integrated Differential Expression and Pathway Analysis (iDEP.96, [27]). Initially, count normalization analyses (count per million, CPM) were performed with lowly expressed transcripts removed at <1.0 CPM. Log10-transformed data were subjected to principal component analysis (PCA) and hierarchical clustering of all expressed genes to visualize sample variation. The average mapping percentage of all identified transcripts was 92.39%. Figure 1A reveals the total counts for all samples, and data for each normally distributed sample are shown in Figure 1B. The average correlation among all samples was 0.96 (Figure 2). Differentially expressed genes/transcripts (DEG) expression analysis was performed using DESeq2 v1.28.0 [28], which uses the Wald test for hypothesis testing by calculating the log2 fold change and dividing it by the standard error, resulting in a z-statistic that can be used to ascertain the p-value.
Figure 1.
The (A) ICAR total read counts and (B) ICAR distribution of log2 transformed data derived from RNA-Seq analysis for each sample of ICAR tissue from heifers supplemented with a vitamin/mineral mix containing 35 ppm as ISe (n = 6) or a 1:1 mixture of ISe:OSe (MIX, n = 6).
Figure 2.
The ICAR correlation matrix of data derived from RNA-Seq analysis for each ICAR sample from heifers supplemented with a vitamin/mineral mix containing 35 ppm as ISe (n = 6) or a 1:1 mixture of ISe:Ose (MIX, n = 6). The average correlation among all samples was 0.96.
Functional Analysis
Global effects of the form of Se treatment on the abundance of transcripts in the ICAR during MRP were then assessed with DEGs identified in DESeq2 analysis and analyzed for canonical, functional, and network analyses using QIAGEN’s Ingenuity Pathway Analysis (IPA, QIAGEN, Redwood City, CA, USA). For each sample, the FASTQ file has been deposited into the National Center for Biotechnology Information Sequence Read Archive (accession number: PRJNA1096973).
2.6. Real-Time PCR Analysis
To quantify the relative abundance of mRNA encoding targeted genes from each ICAR sample, real-time PCR (qPCR) was used following a technique routinely reported from our laboratory [4,6,13,29]. Total RNA from each sample (~1 ug) was reverse transcribed into cDNA using SuperScript IV VILO Master Mix with ezDNAse Enzyme (Invitrogen by Thermo Fisher Scientific, Vilnius, Lithuania). A no-reverse transcription control for each sample was included to ensure qPCR results were not a result of contamination with genomic DNA.
The relative abundance of mRNAs encoding members of the family of selenoproteins was determined using a targeted qPCR analysis. We analyzed mRNA transcripts that encode the protein for three iodothyronine deiodinases (DIO1, DIO2, and DIO3), five glutathione peroxidases (GPX1, GPX2, GPX3, GPX4, and GPX6), three thioredoxin reductases (TXNRD1, TXNRD2, and TXNRD3), selenophosphate synthetase (SEPHS2), and other thirteen other identified selenoproteins (SELENOF, SELENOH, SELENOI, SELENOK, SELENOM, SELENON, SELENOO, SELENOP, SELENOR, SELENOS, SELENOT, SELENOV, SELENOW), as well as the selenoprotein P receptors (LRP2, LRP8 and TFRC).
The relative expression of mRNA transcripts identified from RNA-seq data associated with carbohydrate metabolism (Adcy2, Aldob, Apoe, Bmp4, Edn1, Gaa, Gnaq, Manba, Mstn, Neu3, Pygl, Sgsh, Slc1a4, and Spp1) and canonical Wnt signaling (Cpt1a, Ctnnb1, Dkk1, Fzd6, Lrp5, and Lrp6) was quantified by qPCR to corroborate results from the transcriptomic analysis. The NCBI Primer-BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/, accessed on 12 October 2023) was used to design all primers against their respective RefSeq. DNA sequencing of the target product cDNAs was verified by ACGT Inc. (Wheeling, IL, USA), and sequencing results were compared to each respective primer template using the NCBI Nucleotide-BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLAST_SPEC=GeoBlast&PAGE_TYPE=BlastSearch, accessed on 12 October 2023). The GenBank accession numbers, forward and reverse primer sequences, amplicon length of each product and product identify for each transcript of interest are listed in Appendix A, Table A1, Table A2 and Table A3. For qPCR analysis, a total volume of 25 μL containing cDNA (5 uL), a 10 μM stock of each primer (1 uL forward and 1 uL reverse), 2 × SYBR Green PCR Master Mix (12.5 uL, iTaq Universal SYBR Green Supermix, BIO-RAD, Hercules, CA, USA), and nuclease-free water (5.5 uL) was used. Reactions were conducted using a Bio- Rad CFX Maestro thermal cycler (Bio-Rad, Hercules, CA, USA) using a 3-step amplification protocol with optimal annealing temperature determined for each primer pair.
The 2−ΔΔCT method [30] was used to determine the relative abundance of each transcript using three constitutively expressed and normally distributed housekeeping genes that were not affected by Se-treatment: Actb, Gapdh, and Hprt1. Six heifers per treatment, in triplicate, were used for each transcript. Data were normalized to the relative expression level of the ISe-supplemented treatment group.
2.7. Statistical Analysis
The individual heifer was the experimental unit, and data are presented as least square means (±SEM). Data were analyzed for normal distribution and homogeneity. When appropriate, qPCR data were transformed for normalization, and each transformation is indicated in the results below the tables accompanying each data set. To determine the effect of the form of Se on the abundance of each mRNA transcript, data were analyzed using Student’s t-test (n = 6 per treatment) with the PROC TTest procedure of SAS statistical software package (version 9.4; SAS Institute, Inc., Cary, NC, USA). The relative expression for each transcript identified in RNA-sequencing results was subjected to the Wald test using DESeq2 as described above. For all data, results were considered statistically significant at p ≤ 0.05 or a tendency to differ at 0.05 < p ≤ 0.10.
3. Results
3.1. qPCR of Selenoproteins and Selenoprotein P Receptors in ICAR
Twenty-five selenoproteins and three selenoprotein P receptor transcripts were analyzed via targeted qPCR. We observed a decrease (p < 0.05) in the abundance of mRNA for only iodothyronine deiodinase 2 (Dio2) in ICAR from MIX- compared to ISe- supplemented heifers (Table 1). In addition, the relative expression of mRNA encoding the LDL receptor-related protein (Lrp2) tended (p < 0.1) to be decreased in MIX-Se supplemented heifers, and mRNA for iodothyronine deiodinase 1 (Dio1) was unable to be detected.

Table 1.
The relative abundance of mRNA transcripts encoding selenoproteins and selenoprotein P receptors in the intercaruncular (ICAR) tissue of heifers supplemented with ISe (n = 6) or MIX (n = 6) 1.
3.2. Serum Glucose, Triglycerides, and Cholesterol
Concentrations of glucose, triglycerides, and cholesterol were quantified in the serum of all heifers on d 0 (estrus), and d 7 and 17 of pregnancy. Serum glucose was affected by treatment (Figure 3A). The dietary form of Se affected serum concentrations of glucose (p = 0.03), but time did not (p > 0.05), nor was there an interaction between the dietary form of Se and time (p > 0.05, Figure 3A). The dietary form of Se did not affect serum concentration of triglycerides (p > 0.05); however, there was an effect of time (p = 0.03), with no significant interaction between the two (p > 0.05, Figure 3B). Total circulating cholesterol was lower in the MIX vs. ISe treatment group with the main effects of treatment (p = 0.01), and an effect of day of gestation (p < 0.01), but no treatment x day interaction (p > 0.05, Figure 3C). In addition, a tendency for MIX to have a lower concentration of serum cholesterol on d 0 (170.83 ± 15.88 v 134.33 ± 12.22 mg/dL, p < 0.1) was observed, but no difference was observed on d 7 or d 17 of early pregnancy.
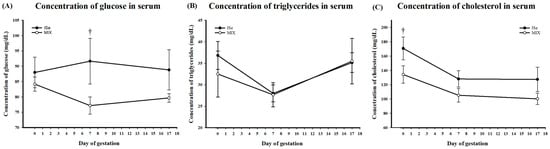
Figure 3.
Concentration of (A) glucose, (B) triglycerides, and (C) cholesterol in serum from heifers at estrus (d 0) and gestational d 7 and 17 in heifers supplemented with either ISe (n = 6) or MIX (n = 6). (A) The dietary form of Se (p = 0.03), but not time (p > 0.05), affected serum concentrations of glucose. There was no interaction between the dietary form of Se and time (p > 0.05). (B) The dietary form of Se did not affect serum concentrations of triglycerides; however, an effect of time (p < 0.05) was observed. There was no interaction between treatment and time of systemic triglycerides (p > 0.05). (C) The dietary form of Se (p = 0.01) and day of gestation (p = 0.02) affected serum concentrations of cholesterol; there was no treatment x day interaction (p > 0.05) observed. Data were analyzed as an ANOVA with repeated measures. A † indicates a tendency to differ at 0.05 < p ≤ 0.01. Comparisons represented are between ISe and MIX at each indicated time point.
3.3. RNA-Sequencing in ICAR
3.3.1. Cluster Analyses
To evaluate the relative relationships and variation among individual heifers, principal component analysis (PCA) of RNA-Seq data was performed. The score plot (Figure 4A) reveals that principal component 1 (PC#1, x-axis) explained 31% of variance among the samples and principal component 2 (PC#2, y-axis) explained 16% of variance. Results indicate that the ISe-supplemented heifers are less closely clustered than the MIX. Hierarchical clustering analysis of the DEGs (Figure 4B) revealed a considerable separation between treatments (ISe versus MIX), but there may be some overlap in transcriptomic profiles of the ICAR, consistent with PCA results.
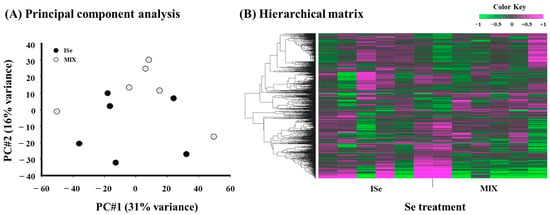
Figure 4.
(A) Score plot and (B) hierarchical matrix of data derived from RNA-Seq analysis for each intercaruncular (ICAR) sample from heifers supplemented with either ISe (n = 6) or MIX (n = 6) in their vitamin/mineral mixes. (A) Principal components 1 and 2 (PC#1 and PC#2) accounted for 31% and 16% of the variance among samples, respectively. (B) Hierarchical cluster analysis comprised the top 1000 differentially expressed transcripts using iDEP.96.
3.3.2. Differentially Expressed Genes
The Wald test of DESeq2 was used to determine changes in the abundance of ICAR transcripts between the ISe- and MIX-supplemented heifers. A total of 838 DEGs, with a total of 427 transcripts upregulated and 411 transcripts downregulated, were identified in MIX vs. ISe-supplemented heifers at p < 0.05. The differentiated genes that are most up-regulated and down-regulated in MIX compared to ISe are provided in Table 2.

Table 2.
Top 10 most highly up- and downregulated DEGs in the ICAR of MIX (n = 6) versus ISe (n = 6)-treated heifers 1.
3.3.3. Pathway and Gene Network Analysis
To determine the effect of the form of Se on global changes in DEGs, bioinformatic analysis was performed using QIAGEN’s Ingenuity Pathway Analysis (IPA, QIAGEN, Redwood City, CA, USA). The canonical pathway analysis revealed the top five pathways (Table 3) based on p-value that were affected by the form of Se are mitochondrial dysfunction (p < 0.0001), cardiac-adrenergic signaling (p < 0.0001), endocannabinoid neuronal synapse pathway (p < 0.0001), G beta gamma signaling (p < 0.0001), and synaptogenesis signaling pathway (p < 0.0001). Further, Figure 5 shows the top canonical pathways in ICAR ranked by z-score. Interestingly, the most positively affected pathways are oxidative phosphorylation (z-score = 3.00), ras homolog family member A (RHOA) signaling (z-score = 2.449), Gα12/13 signaling (z-score = 2.236), and 14-3-3-mediated signaling (z-score = 2.000). The most negatively affected pathways based on z-score are protein kinase A signaling (z-score = −2.324), xenobiotic metabolism PXR signaling pathway (z-score = −2.121), cardiac β-adrenergic signaling (z-score = −1.667), and corticotropin-releasing hormone signaling (z-score = −1.667).

Table 3.
Top 5 canonical pathways identified by IPA of genes differentially expressed from ICAR of heifers supplemented with either ISe (n = 6) or MIX (n = 6) in their vitamin/mineral mixes 1.
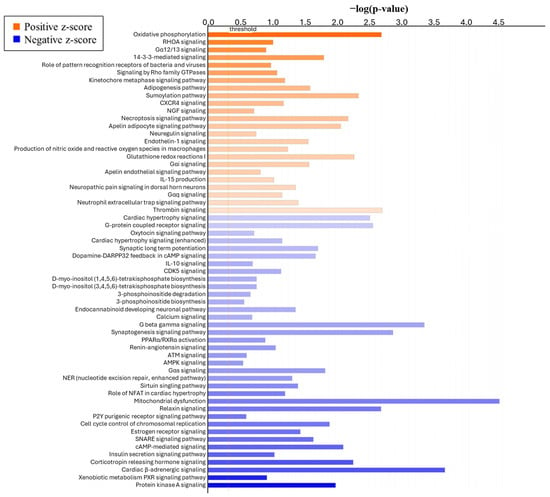
Figure 5.
Top canonical pathways of genes differentially expressed from ICAR of heifers supplemented with either ISe (n = 6) or MIX (n = 6) in their vitamin/mineral mixes, as identified by IPA. Pathways are ranked by z-score, with orange (positive z-score) indicating a pathway that is predicted to be upregulated and blue (negative z-score) indicating a pathway that is predicted to be downregulated.
The top upstream regulators identified using IPA were beta-estradiol, tumor protein p53 (TP53), GLI family zinc finger 1 (GLI1), histidine-rich glycoprotein (HRG), and Wnt family member 3a (WNT3a). The top 5 molecular and cellular functions with specific actions identified by IPA are indicated in Table 4. Only functions with a z-score of greater than or equal to the absolute value of 0.5 are reported. Additionally, key transcripts associated with carbohydrate metabolism and canonical Wnt signaling are reported in Table 5.

Table 4.
Top 5 molecular and cellular functions in ICAR of heifers supplemented with either ISe (n = 6) or MIX (n = 6) in their vitamin/mineral mixes, as identified by IPA 1.

Table 5.
Real-time PCR of selected mRNAs identified by RNA sequencing from ICAR samples during MRP of heifers supplemented with either ISe (n = 6) or MIX (n = 6) in their respective vitamin/mineral mixes 1.
3.3.4. RNA Sequencing Corroboration Using qPCR Analysis
To corroborate the findings of the RNA-seq analysis, qPCR was conducted on select transcripts associated with carbohydrate metabolism and canonical Wnt signaling during MRP (Table 5). A full list of gene transcripts associated with carbohydrate metabolism that were affected is listed in Appendix B, Table A4.
4. Discussion
We previously reported that the length of the preimplantation conceptus during MRP was increased in heifers supplemented with a 1:1 mixture of ISe:OSe (MIX) versus those that received the industry standard of ISe alone [13]. Concurrent to this, we reported a significant increase in the relative abundance of mRNA for Mstn [13]. MSTN protein is thought to increase the availability of glucose [14], the primary energy source for the post-hatch preimplantation conceptus [11], which plausibly provides a relationship between this transcript in ICAR and the observed MIX-induced advancement in conceptus development.
To expand upon these findings, we aimed to define the effects of form of supplemental Se (treatment) on (1) the abundance of mRNA transcripts encoding the functional selenoproteins in the ICAR endometrium during MRP, (2) circulating concentrations of glucose, triglycerides and cholesterol at the establishment of pregnancy, and (3) the global transcriptome of the ICAR endometrium.
4.1. Selenoproteins and Selenoprotein P Receptors in ICAR
We determined the effect of the form of Se required to yield a Se-adequate status on the abundance of mRNAs encoding the 25 identified mammalian selenoproteins and 3 selenoprotein P receptors [3,31] in the ICAR of heifers at d 17 of gestation (MRP). We hypothesized that multiple selenoprotein transcripts with known antioxidant capabilities, such as the glutathione peroxidases, would be significantly more abundant in MIX- versus ISe-supplemented heifers. We only observed a significant effect on the expression of mRNA encoding the intracellular iodothyronine deiodinase 2 (Dio2), which was decreased (p < 0.05) in MIX- versus ISe-treated heifers. This was an unexpected finding, especially given that this same experimental paradigm resulted in the differential expression of multiple selenoprotein transcripts in the CL [4]. It is, however, consistent with the form of Se effects on the expression of selenoprotein mRNAs in the caruncular endometrium, in which MIX-induced decreases in the abundance of only Dio2 and Dio3 were observed [29]. Iodothyronine deiodinases (DIOs) regulate the activity of thyroid hormones by activating and deactivating specific circulating and intracellular thyroid hormones. The regulatory actions of DIOs affect constitutive processes including thermogenesis, cell differentiation and proliferation, energy metabolism, and growth, thus affecting the regulation of metabolism of carbohydrates, proteins, and lipids [32,33,34]. It appears that a subtle form of Se effects on thyroid metabolism in the preimplantation uterine environment exists, noting that a high level of placental DIO3 expression is believed to protect the growing fetus from the activity of maternal thyroid hormones [35]. Of note, low levels of serum T4 can result in an increase in DIO2 activity [36], which may suggest lower systemic concentration of T4 in ISe compared to MIX-treated heifers. Unfortunately, we were unable to quantify circulating thyroid hormone concentrations due to limited sample availability in the present study.
4.2. Serum Glucose, Triglycerides, and Cholesterol
We investigated the effects of treatment (dietary form of Se) on systemic concentrations of glucose, triglycerides, and total cholesterol on d 0 (estrus) and on d 7 and d 17 of gestation. We hypothesized that the concentration of systemic glucose would differ with the form of Se treatment. As the primary source of energy for the post-hatched conceptus prior to implantation and formation of the placenta, glucose is sequestered from the maternal blood and secreted into histotroph [11]. Therefore, changes in glucose availability can have a tremendous impact on conceptus elongation and implantation. Serum concentrations of glucose were higher in ISe compared to MIX-treated heifers. Notably, Moraes et al. [37] observed significantly less plasma glucose on d 17 of gestation in high fertility compared to infertile heifers, with heifers classified as sub-fertile having an intermediate concentration of plasma glucose. The previously observed increase in mRNA abundance for Mstn in MIX-Se form heifers could be a tissue-specific metabolic change to account for the lower amount of this substrate in the blood. Myostatin has been shown to regulate glucose metabolism by promoting glucose uptake, supporting glycolysis, and decreasing the storage of glucose as glycogen [38], with increased MSTN also associated with an increase in glucose in histotroph [14].
We did not observe the expected form of Se-induced difference in the circulating concentration of triglycerides. With a backbone of glycerol and three fatty acids, triglycerides function as a storage depot for excess lipids [39]. In cattle, triglycerides are measured to determine metabolic status, whereas a buildup of triglycerides in the liver decreases liver function and leads to fatty liver disease [39]. Interestingly, we did observe an effect of day on the circulating concentration of triglycerides, with more variation in quantified concentrations observed under a high estrogenic (d 0) versus progestogenic background (d 7 and d 17). The relationship among estradiol, progesterone, and triglycerides has been routinely investigated. Estradiol implants in mature cows resulted in an elevated concentration of triglycerides on day 14 compared to the controls [40]. This relationship is consistent with previous reports at parturition, particularly in dairy cattle that experience a negative energy balance and mobilize NEFA, BHBA, triglycerides, and very low-density lipoproteins during early lactation [41]. Under the high progestogenic conditions before parturition, triglycerides remain relatively low. However, once progesterone drops dramatically in the periparturient period and estrogen undergoes a steep rise in systemic concentrations, triglycerides have been shown to increase as much as 3-fold by the time of calving compared to before parturition [42,43].
Cholesterol is another substrate closely related to glucose and triglycerides in cattle. Cholesterol is the critical precursor for the production of sex steroids, is vital as a cell membrane constituent, and is a modifier of neuronal signaling molecules [44]. Further, cholesterol is the foundation for the synthesis of bile salts to be excreted into the small intestine to solubilize and convert lipids and fats [45]. Relatedly, cholesterol is packaged in lipoproteins and circulated throughout the body, and binding of low-density lipoprotein to its receptor (LDLR) is the primary way that cholesterol is delivered to steroidogenic luteal cells to serve as substrate for the production of P4 [46]. Herein, total systemic cholesterol was higher in ISe- versus MIX-treated heifers. This is consistent with previous findings of early luteal and gestational concentrations of P4 being higher in MIX versus ISe-treated animals [4,6,47], suggesting the form of Se is increasing the abundance of the LDLR, with increased uptake accounting for lower circulating concentrations of cholesterol.
4.3. Global Transcriptomics in ICAR Endometrium
With no panoptic perspective of mRNA transcripts in the ICAR in response to form of Se, and to follow up on previous results from targeted mRNA analyses, we utilized next generation sequencing analysis to perform untargeted transcriptomics and investigate the effects of form of Se on canonical pathways that may be regulating endometrial function to support the elongating conceptus. Results here demonstrate significant effects of the form of Se on carbohydrate metabolism in the ICAR tissue of heifers.
Results from IPA revealed a MIX-induced decrease in transcripts regulating carbohydrate metabolism, and an upregulation of those involved in oxidative phosphorylation. Taken together, it is possible that ICAR may be responding to changes in carbohydrate availability to maintain adequate production of energy. In this case, the greater abundance of mRNA encoding proteins across electron transport and ATP synthase may be accounting for a potential decrease in glucose and other carbohydrate inputs to drive oxidative phosphorylation and the production of ATP.
Interestingly, we observed more abundant expressions of mRNAs encoding the key proteins APOE, MSTN, and SLC1A4 in MIX compared to ISe ICAR samples, and these proteins affect the availability and usability of carbohydrates, fats, and lipid intermediaries [38,48,49]. APOE is involved in the catabolism of triglyceride-rich lipoprotein constituents and is primarily responsible for packaging and transporting cholesterol from the peripheral tissues to be metabolized in the liver and for regulating the availability of glucose to the cell [50]. MSTN has been shown to regulate glucose metabolism in muscle by promoting glucose uptake, supporting glycolysis, and decreasing the storage of glucose as glycogen [38], with increased MSTN also associated with an increase in glucose in the histotroph [14].
The observed greater abundance of mRNA encoding Slc1a4 in MIX compared to ISe is curious, as this protein (SLC1A4) is a sodium-dependent neutral amino acid transporter that transports alanine, serine, cysteine, proline, and threonine, among others. Interestingly, it also transports glutamine and has been speculated to be involved in the transport of selenoamino acids [51]. Glutamine is the most abundant free amino acid in the muscle tissue of cattle [49], and this is attributed to a potential increase in storage of nitrogen and energy for utilization, particularly in the instance of high metabolic demand [52]. Furthermore, glutamine also acts as a precursor for glutathione, which is necessary for the reduction in harmful ROS [52]. Both of the aforementioned provide a plausible link among the greater abundance of mRNA encoding Slc1a4 in the ICAR, changes in carbohydrate metabolism, and longer conceptuses of MIX versus ISe heifers.
The observed MIX-induced decrease in abundance of mRNA encoding the proteins ALDOB, PYGL, and MANBA is also relevant. The reversible conversion of fructose-1,6-bisphosphate to glyceraldehyde 3-phosphate and dihydroxyacetone phosphate, affecting glycolysis, is catalyzed by ALDOB [53]. PYGL catalyzes the cleavage of alpha-1,4-glycosidic bonds to release glucose-1-phosphate and is vital for glucose homeostasis [54]. MANBA is a glycosidase involved in catalyzing the removal of mannose monosaccharides from glycans, which collectively may slow the mobilization of glucose from glycogen stores [55].
It appears that the changes to carbohydrate metabolism may allow more substrate, particularly glucose, to be available to histotroph and thus the developing conceptus. Additionally, in response to changes in carbohydrate metabolism, the ICAR may be up-regulating oxidative phosphorylation as indicated by the RNA-sequencing results to account for the lesser available inputs to maintain needed levels of ATP. Here, MIX-Se supplementation may allow the endometrium to overcome an increased production of ROS due to upregulation of the electron transport chain. Overall, there are changes in carbohydrate metabolism that positively influence the growth of conceptus from MIX-supplemented heifers when compared to ISe-supplementation alone.
5. Conclusions
In investigating the effects of the form of supplemental Se provided to achieve a Se-adequate status on selenoprotein transcripts in the ICAR, key blood metabolites, and the transcriptomic profile of ICAR, we identified marked relationships between previous reports of endometrial changes and longer conceptuses during MRP. Targeted qPCR analysis revealed an effect of treatment on the abundance of one selenoprotein transcript (Dio2) during MRP, a finding consistent with the form of Se-induced changes in selenoprotein mRNAs in the caruncular endometrium, but in stark contrast to the multiple mRNAs affected by the form of Se in the CL. Quantifying serum parameters revealed treatment effects (p < 0.05) on systemic concentrations of glucose and cholesterol, but not triglycerides. Effects on systemic glucose can be tied to treatment-induced differences in energy availability for conceptus development, while effects on cholesterol appear related to increased uptake for steroidogenesis through the LDLR. Finally, RNASeq analysis confirmed significant Se-induced changes in carbohydrate metabolism and oxidative phosphorylation, again appearing to alter glucose availability in the ICAR and affect the production of ATP and the cell’s ability to overcome oxidative stress. Overall, the form of Se affected serum parameters during the establishment of pregnancy and ICAR transcriptomics during MRP, which appear to support the early development of the conceptus.
Author Contributions
Conceptualization and methodology, P.J.B. and S.N.C.; funding acquisition P.J.B. and S.N.C.; project administration, P.J.B.; investigation, P.J.B., S.N.C., B.R.C. and K.S.; writing—original draft preparation, S.N.C.; writing—review and editing, P.J.B. and B.R.C. All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by Agriculture and Food Research Initiative Competitive grant no. 2018-67015-27613 from the USDA National Institute of Food and Agriculture (P.J.B.) and by Agricultural and Food Research Initiative Postdoctoral Fellowship grant no. 2023-67012-39788 from the USDA National Institute of Food and Agriculture (S.N.C.).
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Review Board of the University of Kentucky (IACUC #2017-2828, date of approval: 14 December 2020).
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data (FASTQ files) are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA, accession number PRJNA1096973).
Acknowledgments
The authors would like to thank the faculty and staff at the University of Kentucky Research and Education Center (Princeton, KY, USA) and the C. Oran Little Animal Research Center (Versailles, KY, USA) for managing the animals used in this experiment.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A

Table A1.
Primer sets and product identities of reference transcripts for qPCR analysis.
Table A1.
Primer sets and product identities of reference transcripts for qPCR analysis.
| Gene | Gene Name | Accession Number 1 | Oligonucleotide Primer Design (5′ to 3′) Direction | Amplicon Length (bp) | Product Identity (%) 2 |
|---|---|---|---|---|---|
| Reference transcripts | |||||
| Actb | Actin beta | NM_173979.3 | F: GAGCGGGAAATCGTCCGTGAC R: GTGTTGGCGTAGAGGTCCTTGC | 278 | 99 |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | NM_001034034.2 | F: ACATCAAGTGGGGTGATGCT R: GGCATTGCTGACAATCTTGA | 289 | 99 |
| Hprt1 | Hypoxanthine phosphoribosyltransferase 1 | NM_001034035.2 | F: GCCAGCCGGCTACGTTAT R: ATCCAACAGGTCGGCAAAGA | 256 | 100 |
1 These contents are associated with each gene symbol and are the accession numbers of the sequences retrieved from the NCBI RefSeq database for designing primers and probes. 2 All qPCR products were validated by sequencing. The identity values (%) presented are the base pair ratios between the total amplicon length and the number of identical base pairs.

Table A2.
Primer sets and product identities for qPCR analysis of selenoprotein and selenoprotein P transcripts.
Table A2.
Primer sets and product identities for qPCR analysis of selenoprotein and selenoprotein P transcripts.
| Gene | Gene Name | Accession Number 1 | Oligonucleotide Primer Design (5′ to 3′) Direction | Amplicon Length (bp) | Product Identity (%) 2 |
|---|---|---|---|---|---|
| Enzymatic transcripts | |||||
| Dio1 | Iodothyronine deiodinase 1 | NM_001122593.2 | F: TCCTGTAGTCCGCCTGTCA R: TCCGGTGATTCTTGATGTCCA | 242 | 99 |
| Dio2 | Iodothyronine deiodinase 2 | NM_001010992.4 | F: GATGGGCATCCTCAGCGTAG R: TTCTCCTGGGCACCATTTCC | 315 | 100 |
| Dio3 | Iodothyronine deiodinase 3 | NM_001010993.3 | F: AAGTGGAGCTCAACAGCGAT R: AGTCGAGGATGTGCTGGTTC | 213 | 100 |
| Glutathione peroxidases | |||||
| Gpx1 | Glutathione peroxidase 1 | NM_174076.3 | F: GCAACCAGTTTGGGCATCAG R: TAGGGTCGGTCATGAGAGCA | 210 | 100 |
| Gpx2 | Glutathione peroxidase 2 | NM_001163139.2 | F: AACAGCCTCAAGTACGTCCG R: TCGGTCATGAGGGAAAACGG | 158 | 100 |
| Gpx3 | Glutathione peroxidase 3 | NM_174077.5 | F: GCACCATCTATGAGTACGGGG R: CCCCATTCACATCGCCTTTC | 315 | 100 |
| Gpx4 | Glutathione peroxidase 4 | NM_174770.3 | F: GATCAAAGAGTTCGCCGCTG R: CCATACCGCTTCACCACACA | 198 | 100 |
| Gpx6 | Glutathione peroxidase 6 | NM_001163142.1 | F: CACTGTTCCTGGTCGGCTTA R: CCCAGCACAACTACACCGAA | 259 | 100 |
| Thioredoxin reductases | |||||
| Txnrd1 | Thioredoxin reductase 1 | NM_174625.5 | F: AAGGCCGCGTTATTTGGGTA R: CCTGGTGTCCCTGCTTCAAT | 306 | 100 |
| Txnrd2 | Thioredoxin reductase 2 | NM_174626.2 | F: CAAATGGCTTCGCTGGTCAC R: TTCGTATGCACACCAGCCTT | 230 | 100 |
| Txnrd3 | Thioredoxin reductase 3 | XM_015468824.1 | F: CGGCGTATGACTACGACCTC R: GACTGTACTCCCAGCCGAAC | 249 | 100 |
| Other selenoproteins | |||||
| Selenof | Selenoprotein F | NM_001034759.2 | F: GCAGCTCCTGTGATTTGCTT R: TTTAGCACAGGGTCTGAACCG | 241 | 100 |
| Selenoh | Selenoprotein H | NM_001321327.1 | F: CACGAGCTGACGAGTCTACG R: CTTCTTCAGCTCCTCCAGCA | 235 | 100 |
| Selenoi | Selenoprotein I | NM_001075257.2 | F: TCTGGCTTTCTGCTGGTTGT R: TGGTCAAAAAGCTCCCCCAG | 212 | 100 |
| Selenok | Selenoprotein K | NM_001037489.3 | F: CCGTTTTGTCGATTCACGGC R: CAGATGAGCTTCCGTAGCCT | 278 | 100 |
| Selenom | Selenoprotein M | NM_001163171.2 | F: CCCACTCTACCACAACCTGG R: ACCTAAAGGTCTGCGTGGTC | 249 | 100 |
| Selenon | Selenoprotein N | NM_001114976.2 | F: GTGGCCATGTACCCCTTCAA R: GGGATGGGTTCTCCTGGTTG | 265 | 100 |
| Selenoo | Selenoprotein O | NM_001163193.2 | F: TGGACAGGTATGACCCCGAT R: ATCTTCTGCAGGTAGTGCCG | 202 | 100 |
| Selenop | Selenoprotein P | NM_174459.3 | F: TCAGGTCTTCATCACCACCA R: GTGGCAACAGCAGCTACTCA | 201 | 100 |
| Selenor | Selenoprotein R, Methionine sulfoxide reductase B1 (MSRB1) | NM_001034810.2 | F: GAACCACTTTGAGCCGGGTA R: GGCCATCGTTCAGGAACTCA | 221 | 100 |
| Selenos | Selenoprotein S | NM_001046114.3 | F: CCCACCCTCGAGACCGA R: GCCCAGGACTGTCTTCTTCC | 394 | 100 |
| Selenot | Selenoprotein T | NM_001103103.2 | F: TGGTCACCTTCCATCCATGC R: AAGAGGTACAACGAGCCTGC | 240 | 100 |
| Selenov | Selenoprotein V | NM_001163244.2 | F: ACTCCATTGGCCACCGATTT R: AGGCCACAGTAAACCACTCG | 224 | 100 |
| Selenow | Selenoprotein W | NM_001163225.1 | F: AGTGTTCGTAGCGGGAAAGC R: CGCGAGAACATCAGGGAAGG | 233 | 98 |
| Selenophosphate synthetase | |||||
| Sephs2 | Selenophosphate synthetase 2 | NM_001114732.2 | F: GATCCCTACATGATGGGGCG R: GTTTACCACCGTTTGCCCAC | 219 | 100 |
| Selenoprotein P receptors | |||||
| Lrp2 | LDL receptor related protein 2 | XM_024983502.1 | F: GTGGTTTGGGTTACCGTTGC R: GGCACCCTGTTAGCTGTGAT | 304 | 99 |
| Lrp8 | LDL receptor related protein 8 | NM_001097565.1 | F: AGCCACCCTTTTGGGATAGC R: AAGGCACAGGTACTCACAGC | 231 | 100 |
| Tfrc | Transferrin receptor | NM_001206577.1 | F: CCAGGTTTAGTCTGGCTCGG R: GGTCTGCCCAGAATATGCGA | 339 | 99 |
1 These contents are associated with each gene symbol and are the accession numbers of the sequences retrieved from the NCBI RefSeq database for designing primers and probes. 2 All qPCR products were validated by sequencing. The identity values (%) presented are the base pair ratios between the total amplicon length and the number of identical base pairs.

Table A3.
Primer sets and product identities for qPCR analysis of transcripts used to corroborate RNA-sequencing results.
Table A3.
Primer sets and product identities for qPCR analysis of transcripts used to corroborate RNA-sequencing results.
| Gene | Gene Name | Accession Number 1 | Oligonucleotide Primer Design (5′ to 3′) Direction | Amplicon Length (bp) | Product Identity (%) 2 |
|---|---|---|---|---|---|
| Carbohydrate Metabolism | |||||
| Adcy2 | PREDICTED: Adenylate cyclase 2 | XM_024981538.1 | F: ATCAGCACCACGGATGTACC R: GAAGATCAGGCAAGCGCAAG | 267 | 99 |
| Aldob | Aldolase, fructose-bisphosphate B | NM_001034485.2 | F: CAGTTCCGCGAACTCCTCTT R: AGCGTTCAGAAAGGCCATCA | 232 | 100 |
| Apoe | Apolipoprotein E | NM_173991.2 | F: ACGCTGACGACCTGAAGAAG R: CCTCTAGCTGCTGGCGTATC | 261 | 100 |
| Bmp4 | Bone morphogenetic protein 4 | NM_001045877.1 | F: ACTTCGAGGCCACACTTCTG R: AGAGTTTTCGCTGGTCCCTG | 245 | 100 |
| Edn1 | Endothelin 1 | NM_181010.2 | F: CTTCTAGGTCCAAGCGCTCC R: CTGATGGCCTCCAACCTTCTT | 259 | 99 |
| Gaa | Alpha glucosidase | NM_173913.2 | F: CATGACTTGGAGGTGGTCCC R: GTGGTGGTCAGGTTCTCCAG | 308 | 100 |
| Gnaq | Guanine nucleotide binding protein (G protein), q polypeptide | NM_001110002.1 | F: CGAGCACAATAAGGCTCATGC R: TTGTTGCGTAGGCAGGTAGG | 223 | 100 |
| Manba | Mannosidase beta | NM_174387.2 | F: CAGTCGCAGCGAGATAGTGA R: TCTGAGTGAAGGTGTGACGC | 22 | 99 |
| Mstn * | Myostatin | NM_001001525.3 | F: TGCCCACGGAGTCTGATCTT R: TGCCTGGGTTCATGTCAAGT | 237 | 100 |
| Neu3 | Neuraminidase 3 | NM_174122.3 | F: ATGGAGGAGCCGGGGTT R: CAGGGTTCCTGCCTGACATA | 400 | 100 |
| Pygl | Glycogen phosphorylase L | NM_001075203.2 | F: GAAGTGCCCCAAGAGGGTTT R: ATGGAATCCAGGAAGCAGGC | 213 | 100 |
| Sgsh | N-sulfoglucosamine sulfohydrolase | NM_001102189.2 | F: ACAACAGCGCCATCTCTACC R: AGGAATTTCCGGACCAGCAG | 367 | 99 |
| Slc1a4 | Solute carrier family 1 member 4 | NM_001081577.1 | F: AGTGACCTACAACACGAGCG R: ACATGATGCCCACAGGTACG | 233 | 100 |
| Spp1 | Secreted phosphoprotein 1 | NM_174187.2 | F: GCCTGACCCATCTCAGAAGC R: TCTGAACGTTAGATCGGCGG | 386 | 99 |
| Canonical Wnt Signaling | |||||
| Cpt1a | Carnitine palmitoyltransferase 1A | NM_001304989.2 | F: GGGTCTACGATTCCGCTCTG R: GGCATCCAGAGACTGCTTGT | 339 | 100 |
| Ctnnb1 | Catenin beta 1 | NM_001076141.1 | F: CAGCAGTTTGTGGAGGGAGT R: GAACTGGTCAGCTCAACCGA | 371 | 100 |
| Dkk1 * | Dickkopf WNT signaling pathway inhibitor 1 | NM_001205544.1 | F: GGCAGCAAGTACCAGACCAT R: AGAAGGCATGCATATCCCGTT | 207 | 100 |
| Fzd6 | PREDICTED: Frizzled class receptor 6, transcript variant X1 | XM_010812158.3 | F: TGCACAGAATGGGCTGGATT R: GTACCATGATTTGCCGTCGC | 212 | 100 |
| Lrp5 | PREDICTED: LDL receptor related protein 5, transcript variant X1 | XM_024987583.1 | F: CTGGAGGAGTTCTCAGCCCAC R: TTCAGAGAGGCGTCGCAGTC | 386 | 100 |
| Lrp6 | PREDICTED: LDL receptor related protein 6, transcript variant X1 | XM_005207028.4 | F: GTGCCCTGGAACATGTGGTA R: TTCACGGTTGAGTCCAAGCA | 398 | 100 |
1 These contents are associated with each gene symbol and are the accession numbers of the sequences retrieved from the NCBI RefSeq database for designing primers and probes. 2 All qPCR products were validated by sequencing. The identity values (%) presented are the base pair ratios between the total amplicon length and the number of identical base pairs. * Primer pair published in Crites et al. [13]
Appendix B

Table A4.
Gene transcripts associated with carbohydrate metabolism that were affected by form of selenium (MIX compared to ISe).
Table A4.
Gene transcripts associated with carbohydrate metabolism that were affected by form of selenium (MIX compared to ISe).
| ID | Gene | Expr Log Ratio |
|---|---|---|
| ENSBTAG00000019988 | GNA15 | 1.284 |
| ENSBTAG00000002982 | PITPNM3 | 1.041 |
| ENSBTAG00000011808 | MSTN | 0.988 |
| ENSBTAG00000004855 | PRDX6 | 0.975 |
| ENSBTAG00000018223 | CHI3L1 | 0.935 |
| ENSBTAG00000010123 | APOE | 0.784 |
| ENSBTAG00000019079 | PLCB2 | 0.656 |
| ENSBTAG00000016021 | GAA | 0.650 |
| ENSBTAG00000004150 | NRG1 | 0.612 |
| ENSBTAG00000038062 | FUT4 | 0.598 |
| ENSBTAG00000015267 | SGSH | 0.563 |
| ENSBTAG00000018119 | AOAH | 0.527 |
| ENSBTAG00000008096 | EDN1 | 0.490 |
| ENSBTAG00000046155 | RGN | 0.433 |
| ENSBTAG00000037527 | OAS1 | 0.386 |
| ENSBTAG00000001420 | ABHD12 | 0.354 |
| ENSBTAG00000000134 | MPDU1 | 0.349 |
| ENSBTAG00000030434 | FUCA1 | 0.328 |
| ENSBTAG00000010336 | TALDO1 | 0.326 |
| ENSBTAG00000021504 | TIRAP | 0.322 |
| ENSBTAG00000014534 | EEF1A1 | 0.303 |
| ENSBTAG00000025931 | NEU3 | 0.294 |
| ENSBTAG00000010136 | CMAS | 0.280 |
| ENSBTAG00000004587 | DUSP6 | 0.273 |
| ENSBTAG00000011056 | IDS | 0.258 |
| ENSBTAG00000005371 | DPAGT1 | 0.257 |
| ENSBTAG00000016845 | GALNT7 | 0.234 |
| ENSBTAG00000009789 | GNAQ | −0.185 |
| ENSBTAG00000011709 | IMPA1 | −0.203 |
| ENSBTAG00000034436 | PDPK1 | −0.204 |
| ENSBTAG00000011494 | PYGL | −0.291 |
| ENSBTAG00000005997 | ABCB1 | −0.296 |
| ENSBTAG00000011761 | LRP6 | −0.348 |
| ENSBTAG00000021999 | CPT1A | −0.423 |
| ENSBTAG00000005359 | TGFB2 | −0.426 |
| ENSBTAG00000050602 | HAS3 | −0.540 |
| ENSBTAG00000001879 | PER2 | −0.556 |
| ENSBTAG00000010206 | UAP1 | −0.579 |
| ENSBTAG00000000507 | NR4A1 | −0.591 |
| ENSBTAG00000002699 | KIT | −0.630 |
| ENSBTAG00000023600 | APOD | −0.631 |
| ENSBTAG00000019026 | EXTL2 | −0.634 |
| ENSBTAG00000014674 | CHRM2 | −0.745 |
| ENSBTAG00000001649 | ZFPM2 | −0.779 |
| ENSBTAG00000019761 | MANBA | −0.841 |
| ENSBTAG00000024957 | SNCA | −0.903 |
| ENSBTAG00000019892 | HAS2 | −0.961 |
| ENSBTAG00000019210 | ADCY2 | −1.030 |
| ENSBTAG00000015358 | ALDOB | −1.077 |
| ENSBTAG00000005260 | SPP1 | −1.311 |
References
- Ammerman, C.B.; Miller, S.M. Selenium in ruminant nutrition: A review. J. Dairy Sci. 1975, 58, 1561–1577. [Google Scholar] [CrossRef]
- Flohe, L.; Gunzler, W.A.; Schock, H.H. Glutathione peroxidase: A selenoenzyme. FEBS Lett. 1973, 32, 132–134. [Google Scholar] [CrossRef]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef]
- Carr, S.N.; Crites, B.R.; Pate, J.L.; Hughes, C.H.K.; Matthews, J.C.; Bridges, P.J. Form of supplemental selenium affects the expression of mRNA transcripts encoding selenoproteins, and proteins regulating cholesterol uptake, in the corpus luteum of grazing beef cows. Animals 2022, 12, 313. [Google Scholar] [CrossRef]
- Cerny, K.L.; Anderson, L.; Burris, W.R.; Rhoads, M.; Matthews, J.C.; Bridges, P.J. Form of supplemental selenium fed to cycling cows affects systemic concentrations of progesterone but not those of estradiol. Theriogenology 2016, 85, 800–806. [Google Scholar] [CrossRef]
- Crites, B.R.; Carr, S.N.; Matthews, J.C.; Bridges, P.J. Form of dietary selenium affects mRNA encoding cholesterol biosynthesis and immune response elements in the early luteal phase bovine corpus luteum. J. Anim. Sci. 2022, 100, skac135. [Google Scholar] [CrossRef]
- Spencer, T.E.; Bazer, F.W. Biology of progesterone action during pregnancy recognition and maintenance of pregnancy. Front. Biosci. 2002, 7, 1879–1898. [Google Scholar] [CrossRef]
- Carter, F.; Forde, N.; Duffy, P.; Wade, M.; Fair, T.; Crowe, M.; Evans, A.; Kenny, D.; Roche, J.; Lonergan, P. Effect of increasing progesterone concentration from Day 3 of pregnancy on subsequent embryo survival and development in beef heifers. Reprod. Fertil. Dev. 2008, 20, 368–375. [Google Scholar] [CrossRef]
- Garrett, J.E.; Geisert, R.D.; Zavy, M.T.; Morgan, G.L. Evidence for maternal regulation of early conceptus growth and development in beef cattle. J. Reprod. Fertil. 1988, 84, 437–446. [Google Scholar] [CrossRef]
- Mansouri-Attia, N.; Aubert, J.; Reinaud, P.; Giraud-Delville, C.; Taghouti, G.; Galio, L.; Everts, R.E.; Degrelle, S.; Richard, C.; Hue, I.; et al. Gene expression profiles of bovine caruncular and intercaruncular endometrium at implantation. Physiol. Genom. 2009, 39, 14–27. [Google Scholar] [CrossRef]
- Atkinson, B.A.; King, G.J.; Amoroso, E.C. Development of the caruncular and intercaruncular regions in the bovine endometrium. Biol. Reprod. 1984, 30, 763–774. [Google Scholar] [CrossRef]
- Martal, J.; Chêne Camous, N.; Huynh, L.; Lantier, F.; Hermier, P.; L’haridon, R.; Charpigny, G.; Charlier, M.; Chaouat, G. Recent developments and potentialities for reducing embryo mortality in ruminants: The role of IFN-t and other cytokines in early pregnancy. Reprod. Fertil. Dev. 1997, 9, 355–380. [Google Scholar] [CrossRef]
- Crites, R.B.; Carr, N.S.; Anderson, H.L.; Matthews, C.J.; Bridges, J.P. Form of dietary selenium affects mRNA encoding inter-feron-stimulated and progesterone-induced genes in the bovine endometrium and conceptus length at maternal recognition of pregnancy. J. Anim. Sci. 2022, 100, skac137. [Google Scholar] [CrossRef]
- Forde, N.; Carter, F.; Fair, T.; Crowe, M.A.; Evans, A.C.O.; Spencer, T.E.; Bazer, F.W.; McBride, R.; Boland, M.P.; O’Gaora, P.; et al. Progesterone-regulated changes in endometrial gene expression contribute to advanced conceptus development in cattle. Biol. Reprod. 2009, 81, 784–794. [Google Scholar] [CrossRef]
- Gerloff, B.J. Effect of selenium supplementation on dairy cattle. J. Anim. Sci. 1992, 70, 3934–3940. [Google Scholar] [CrossRef]
- Dargatz, D.A.; Ross, P.F. Blood selenium concentrations in cows and heifers on 253 cow-calf operations in 18 states. J. Anim. Sci. 1996, 74, 2891–2895. [Google Scholar] [CrossRef]
- Wahlen, R.; Evans, L.; Turner, J.; Hearn, R. The use of collision/ reaction cell ICP-MS for the determination of elements in blood and serum samples. Spectroscopy 2005, 20, 84–89. [Google Scholar]
- Bogdanova, E.A.; Barsova, E.V.; Shagina, I.A.; Scheglov, A.; Anisimova, V.; Vagner, L.L.; Lukyanov, S.A.; Shagin, D.A. Normalization of full-length-enriched cDNA. In cDNA Libraries: Methods and Applications; Methods in Molecular Biology; Lu, C., Browse, J., Wallis, J.G., Eds.; Humana Press: Totawa, NJ, USA, 2011. [Google Scholar] [CrossRef]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alnebert, J.; Wilm, A.; Garcia, M.U.; Di Tommaso, P.; Nahnsen, S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- García-Alcalde, F.; Okonechnikov, K.; Carbonell, J.; Cruz, L.M.; Götz, S.; Tarazona, S.; Dopazo, J.; Meyer, T.F.; Conesa, A. Qualimap: Evaluating next-generation sequencing alignment data. Bioinformatics 2012, 28, 2678–2679. [Google Scholar] [CrossRef]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef]
- Sayols, S.; Scherzinger, D.; Klein, H. dupRadar: A Bioconductor package for the assessment of PCR artifacts in RNA-Seq data. BMC Bioinform. 2016, 17, 428. [Google Scholar] [CrossRef]
- Daley, T.; Smith, A.D. Predicting the molecular complexity of sequencing libraries. Nat. Methods. 2013, 10, 325–327. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Carr, S.N.; Crites, B.R.; Shinde, H.; Bridges, P.J. Transcriptomic chnges in response to form of selenium on the interferon-tau signaling mechanism in the caruncular tissue of beef heifers at maternal recognition of pregnancy. Int. J. Mol. Sci. 2023, 24, 17327. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, X.D.; Zhao, Z.P.; Zhao, J.C.; Lei, X.G. Evolution, regulation, and function of porcine selenogenome. Free Radic. Biol. Med. 2018, 127, 116–123. [Google Scholar] [CrossRef]
- Bianco, A.C.; Salvatore, D.; Gereben, B.; Berry, M.J.; Larsen, P.R. Biochemistry, cellular and molecular biology, and physiological roles of the iodothyronine selenodeiodinases. Endocr. Rev. 2002, 23, 38–89. [Google Scholar] [CrossRef] [PubMed]
- Mullur, R.; Liu, Y.Y.; Brent, G.A. Thyroid hormone regulation of metabolism. Physiol. Rev. 2014, 94, 355–382. [Google Scholar] [CrossRef]
- Silva, J.F.; Ocarino, N.M.; Serakides, R. Thyroid hormones and female reproduction. Biol. Reprod. 2018, 99, 907–921. [Google Scholar] [CrossRef]
- Forhead, A.J.; Fowden, A.L. Thyroid hormones in fetal growth and prepartum maturation. J. Endocrinol. 2014, 221, R87–R103. [Google Scholar] [CrossRef] [PubMed]
- Gereben, B.; Zeöld, A.; Dentice, M.; Salvatore, D.; Bianco, A.C. Activation and inactivation of thyroid hormone by deiodinases: Local action with general consequences. Cell. Mol. Life Sci. 2008, 65, 570–590. [Google Scholar] [CrossRef] [PubMed]
- Moraes, J.G.N.; Behura, S.K.; Geary, T.W.; Spencer, T.E. Analysis of the uterine lumen in fertility-classified heifers: I. Glucose, prostaglandins, and lipids. Biol. Reprod. 2020, 102, 456–474. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, J.; Cao, L.; Zhang, Y.; Xia, W.; Zhu, D. Myostatin regulates glucose metabolism via the AMP-activated protein kinase pathway in skeletal muscle cells. Int. J. Biochem. Cell Biol. 2010, 42, 2072–2081. [Google Scholar] [CrossRef]
- Bobe, G.; Young, J.W.; Beitz, D.C. Invited review: Pathology, etiology, prevention, and treatment of fatty liver in dairy cows. J. Dairy Sci. 2004, 87, 3105–3124. [Google Scholar] [CrossRef]
- Miles, E.D. Effects of Estradiol Supplementation on Blood Estradiol and Metabolite Levels, and Hepatic Protein Expression, in Growing, Mature, and Senescent Beef Cattle. Ph.D. Thesis, University of Kentucky, Lexington, KY, USA, 2013. [Google Scholar]
- Goff, J.P.; Horst, R.L. Physiological changes at parturition and their relationship to metabolic disorders. J. Dairy Sci. 1997, 80, 1260–1268. [Google Scholar] [CrossRef]
- Bertics, S.J.; Grummer, R.R.; Cadorniga-Valino, C.; Stoddard, E.E. Effect of prepartum dry matter intake on liver triglyceride concentration and early lactation. J. Dairy Sci. 1992, 75, 1914–1922. [Google Scholar] [CrossRef]
- Drackley, J.K.; Veenhuizen, J.J.; Richard, M.J.; Young, J.W. Metabolic changes in blood and liver of dairy cows during either feed restriction or administration of 1,3-butanediol. J. Dairy Sci. 1991, 74, 4254–4264. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, Z.; Shen, W.J.; Azhar, S. Cellular cholesterol delivery, intracellular processing and utilization for biosynthesis of steroid hormones. Nutr. Metab. 2010, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.W. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 2003, 72, 137–174. [Google Scholar] [CrossRef] [PubMed]
- Pate, J.L.; Condon, W.A. Regulation of steroidogenesis and cholesterol synthesis by prostaglandin F-2 alpha and lipoproteins in bovine luteal cells. J. Reprod. Fertil. 1989, 87, 439–446. [Google Scholar] [CrossRef]
- Carr, S.N.; Jia, Y.; Crites, B.R.; Hamilton, C.H.; Burris, W.R.; Edwards, J.L.; Matthews, J.C.; Bridges, P.J. Form of supplemental selenium in vitamin-mineral premixes differentially affects early luteal and gestational concentrations of progesterone, and postpartum concentrations of prolactin in beef cows. Animals 2020, 10, 967. [Google Scholar] [CrossRef]
- Baboota, R.K.; Blüher, M.; Smith, U. Emerging role of bone morphogenetic protein 4 in metabolic disorders. Diabetes 2021, 70, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Meijer, G.A.; Van der Meulen, J.; Bakker, J.G.; Van der Koelen, C.J.; Van Vuuren, A.M. Free amino acids in plasma and muscle of high yielding dairy cows in early lactation. J. Dairy Sci. 1995, 78, 1131–1141. [Google Scholar] [CrossRef]
- Getz, S.G.; Reardon, A.C. Apoprotein E as a lipid transport and signaling protein in the blood, liver, and artery wall. J. Lipid Res. 2009, 50, S156–S161. [Google Scholar] [CrossRef] [PubMed]
- Cousins, R.J.; Liuzzi, J.P. Trace Metal Absorption and Transport. In Physiology of the Gastrointestinal Tract, 6th ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 1485–1498. [Google Scholar]
- Cruzat, V.; Rogero, M.M.; Keane, N.K.; Curi, R.; Newsholme, P. Glutamine: Metabolism and immune function, supplementation and clinical translation. Nutrients 2018, 10, 1564. [Google Scholar] [CrossRef]
- UniProt. Q3T0S5·ALDOB_BOVIN; UniProt: Cambridge, UK, 2025. [Google Scholar]
- National Center for Biotechnology Information (NCBI). Protein [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; 1988—Accession No. Q0VCM4.1, RecName: Full=Glycogen Phosphorylase, liver Form. Available online: https://www.ncbi.nlm.nih.gov/protein/Q0VCM4.1 (accessed on 20 February 2025).
- Chen, H.; Leipprandt, R.J.; Traviss, E.C.; Sopher, L.B.; Jones, Z.M.; Cavanagh, T.K.; Friderici, H.K. Molecular cloning and characterization of bovine β-mannosidase. J. Biol. Chem. 1995, 270, 3841–3848. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).