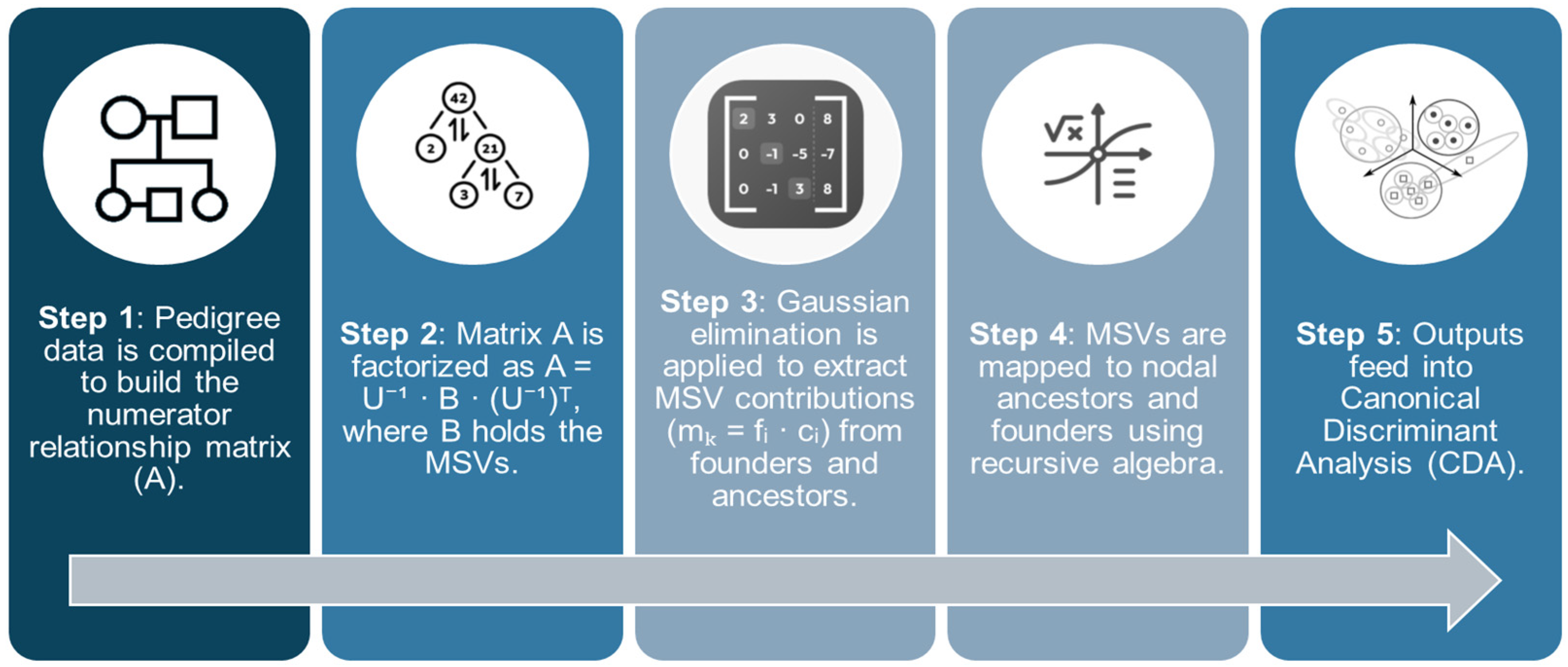
1. Introduction
Genetic diversity refers to the total variety of genetic information within a population. It is a measure of how much variation there is in the genetic makeup of individuals within that population. While populations that enjoy high levels of genetic diversity also enjoy an enhanced ability to adapt to changing environments or challenges like diseases, low genetic diversity frameworks can make a population more vulnerable to such challenges [
1].
On the other hand, genetic drift refers to the random fluctuation in allele frequencies in a population over time due to chance events, especially in small populations. It is a process that leads to changes in the genetic composition of a population independently of natural selection [
2]. Genetic drift can reduce genetic diversity because, over time, some alleles may be lost or become fixed in a population purely by chance, not because they provide any advantage or are selected for.
The genetic diversity of populations, which is closely related to the extent of genetic drift they undergo, has been widely monitored using parameters derived from genealogical data [
3]. The most commonly referenced metrics include the average inbreeding coefficient (
, [
4]), which quantifies the probability of an individual inheriting two alleles that are identical by descent; the average coancestry coefficient (
) [
5], which measures the genetic relatedness between individuals in a population; and the effective population size, which is directly related to the inbreeding rate per unit of time [
6].
Although and remain fundamental metrics in population genetics, neither of these parameters explicitly tracks the specific contributions of genes from each ancestor. Therefore, they provide limited insight into how and when specific ancestors shaped the current genetic structure. Hence, they summarize genetic similarity but do not distinguish the underlying sources of homozygosity or identify the historical contributors to genetic drift and, by extension, do not account for the detailed genetic contributions of individual ancestors.
For these reasons, other parameters and methods have been proposed to support the assessment of genetic diversity more comprehensively, including the founder genome equivalent [
7], the effective number of ancestors [
8], and the effective number of non-founders [
9], and the decomposition of (
) and (
) into the contributions of nodal common ancestors (NCAs) and Mendelian sampling variances (MSVs).
These approaches allow us to trace the flow of genes across generations, quantify the impact of influential ancestors, and better understand the temporal dynamics of inbreeding—key elements for managing genetic resources in endangered breeds. Particularly,
and
partition can be helpful in understanding who contributed the most to drift and when. Decomposing these values into ancestral components can provide valuable insights into how genetic diversity has evolved over time, identifying which individuals contributed the most to genetic drift and at what point in history [
9,
10].
Understanding the distribution of genetic diversity across generations is essential for monitoring conservation status, particularly in endangered breeds where historical events, breeding strategies, and demographic shifts have significantly shaped current genetic variability. Probabilities of gene origin derived from pedigree data are valuable tools for monitoring genetic diversity. An analytical relationship has been established between the genetic contributions of nodal common ancestors (NCAs)—those forming inbreeding loops [
4], and the contributions of Mendelian sampling variances (MSVs) from ancestors. This relationship demonstrates that the contributions of MSVs to individual inbreeding coefficients, mean inbreeding, or average coancestry can be efficiently converted into the genetic contributions of NCAs and founders, even in large and complex populations.
Wright’s [
4] path-counting method provides a detailed breakdown of the genetic contributions of NCAs to individual inbreeding coefficients. However, this method presents a limitation when an NCA is a direct descendant of another NCA, as it only provides a marginal contribution for the older ancestor. To address this, Lacy [
11] proposed a decomposition method that attributes inbreeding components to each founder, allowing for a more refined understanding of genetic contributions. Similarly, Caballero and Toro [
9] introduced a general framework for partitioning coancestry into contributions from both founders and non-founder ancestors, enabling the estimation of genetic drift components more accurately. The proposed method provides a powerful approach for identifying the most influential sources of homozygosity within extensive and intricately structured pedigrees.
In this framework, Caballero and Toro [
9] proposed a general method for partitioning
into components due to founders and non-founder ancestors. Later, Sargolzaei and Colleau [
12], using the aforementioned as a reference, would describe a refined model for decomposing
F,
and
into ancestral components through the study of MSVs. This revealed the analytical link between Wright’s path-counting method [
6] and the method by Caballero and Toro [
9] that enhanced the understanding of historical genetic contributions and drift effects.
The contributions of MSVs of ancestors play a crucial role in assessing genetic diversity and inbreeding within animal populations [
12]. MSV refers to the genetic variability that arises due to the random process of gene transmission between generations, which generates inherent variance in the offspring. In pedigree analysis, it is essential to break down an individual’s inbreeding or coancestry level based on the genetic contributions of its ancestors, as these variances provide an accurate measure of how genetic material inherited from previous generations influences the current genetic structure. This approach allows for more detailed identification of the effects of NCAs on the formation of homozygosity and the preservation of genetic diversity, especially in breeds with limited pedigrees or endangered populations. By quantifying MSV contributions, a deeper understanding of how breeding practices and the genetic history of a population impact its sustainability and genetic health can be achieved.
In this framework, canonical discriminant analysis (CDA) arises as an exceptional statistical tool to track NCAs and identify inbreeding loops within pedigrees. By projecting individuals into a reduced multivariate space based on genetic similarity, this technique facilitates the detection of ancestral nodes that disproportionately contribute to inbreeding coefficients and coancestry patterns. Building upon these approaches, this study introduces a novel methodology that decomposes both inbreeding (F) and coancestry (f) into distinct ancestral components. Using discriminant analysis, it becomes possible to assign genetic variance to specific historical lineages, offering deeper insights into the origins and propagation of inbreeding within a population.
This statistical framework is applied to the Andalusian Black cattle, a native Spanish breed from the Autonomous Community of Andalusia. This breed, historically used for draft work in agriculture, has undergone significant demographic and functional shifts due to modernization. With the advent of mechanized farming, its primary role transitioned from labor to meat production, leading to a reduction in population size and a geographic concentration in mountainous grazing systems [
1]. The breed is currently classified as endangered, with an officially declared population of 1797 individuals distributed across 22 farms as of 2020. Despite ongoing conservation efforts—including the implementation of a pedigree book, controlled breeding programs, and performance monitoring—the Andalusian Black cattle remain at risk of further genetic erosion. Analyzing the genetic structure of this population is therefore essential to ensure its long-term sustainability and to develop informed conservation strategies [
1].
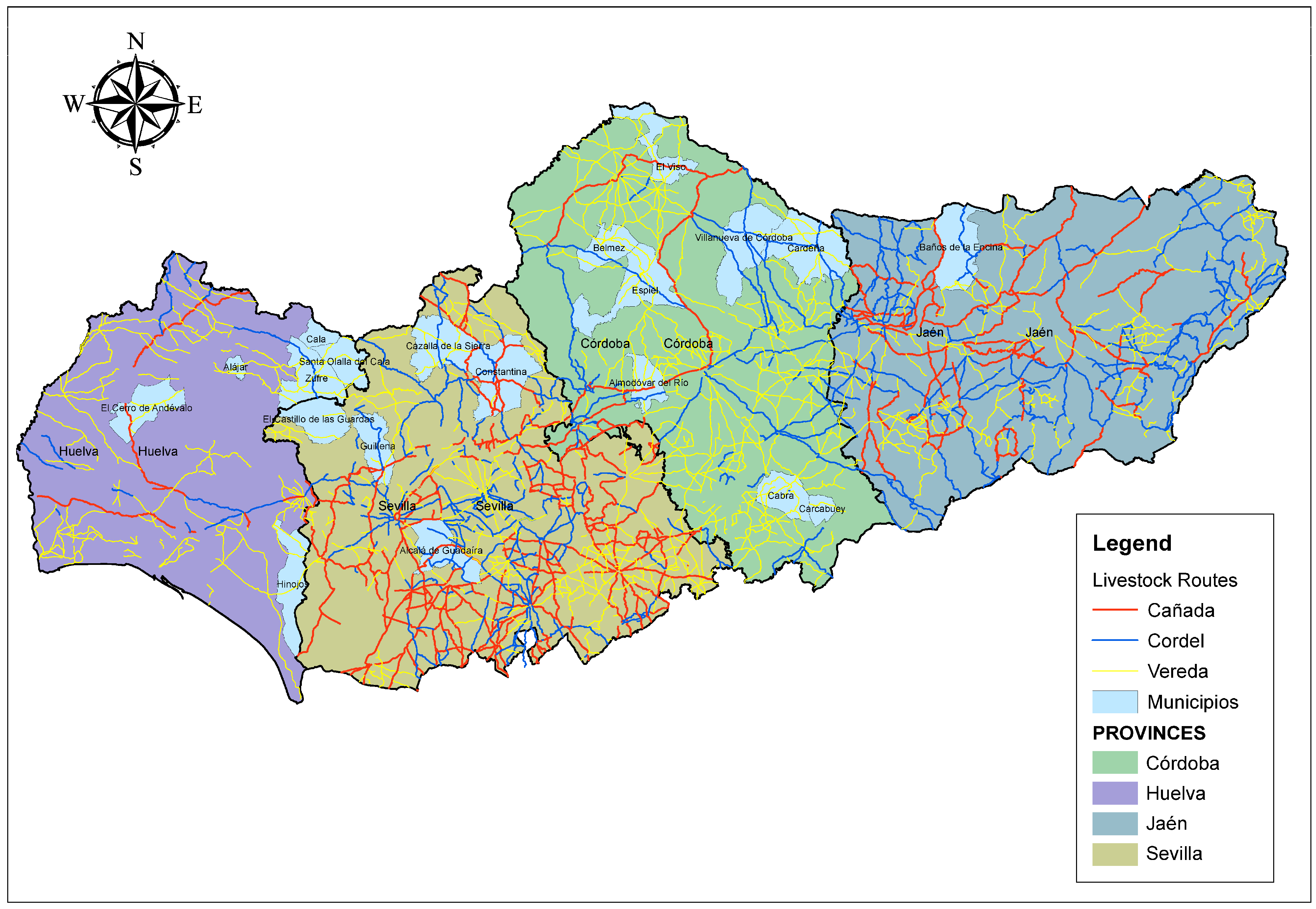
One of the key aspects influencing the genetic diversity of the Andalusian Black cattle is its historical connection to transhumance routes, specifically the Cañadas Reales. In parallel, since transhumance is a prehistoric practice for cattle, goats, and sheep, it is tied to native breeds that provide environmental benefits, landscape conservation, and the production of traditional local products with protected designation of origin. This activity is also linked to summer pastures in public mountain areas and winter pastures that are usually privately owned dehesas (Directorate for Sustainable Rural Development, 2012 [
13]).
These ancient pathways, used for seasonal livestock migration, played a crucial role in the exchange of genetic material among Iberian breeds. The movement of cattle along these routes not only contributed to the breed’s adaptability to diverse environmental conditions but also influenced the structure of its genetic diversity. By integrating genealogical data with a discriminant analysis approach, this study aims to uncover the historical pathways that shaped the genetic makeup of the breed. This approach enables a more detailed understanding of past genetic bottlenecks, drift events, and the specific ancestors that played a significant role in shaping the current population structure.
The primary objective of this study is to utilize the decomposition of inbreeding and coancestry into ancestral components to identify the key centers of creation, distribution, and propagation of genetic diversity within the Andalusian Black cattle breed. By analyzing genetic variability in relation to the population structure, the study aims to quantify the risk of genetic diversity loss and pinpoint the most critical lineages for conservation. This approach will provide a comprehensive understanding of the genetic flux within the breed, highlighting key genetic reservoirs that should be prioritized in conservation programs. Additionally, the findings will inform more effective conservation strategies, ensuring the maintenance of genetic variability and the long-term sustainability of the breed.
4. Discussion
Our results contribute to tracing the historical evidence surrounding the origin and evolution of the Andalusian Black cattle breed. Rooted in ancient bovine lineages, the Andalusian Black cattle reflect a blend of Near Eastern, Italian, and North African genetic influences. While this background provides essential context, its key implication lies in the breed’s resilience and adaptation across diverse Iberian terrains, traits that are mirrored in its retained diversity despite modern demographic pressures. These early domesticates gradually spread across Europe, where regional selection pressures and breeding practices led to the development of the diverse cattle breeds observed today [
23].
In the Iberian Peninsula, the so-called “Iberian trunk” is derived from local aurochs populations. This becomes particularly evident in breeds that are typically characterized by dark coat colors, including black, grey (
cárdena), and black-and-white or red-and-white (
berrenda) varieties [
24]. These breeds adapted to the granitic–siliceous substrates and calcium-poor pastures typical of central and western Iberia, with a historical presence notably concentrated in regions such as the Sistema Central and La Rioja (
Figure 8).
Breeds from the Iberian trunk, such as Serrana de Teruel, Serrana de Soria, Avileña, Preta Portuguesa, Morucha, Sayaguesa, Caldelá, Cárdena Andaluza, and Andalusian Black (Negra Andaluza) have played an essential role in the development of regional livestock systems. These breeds are traditionally found in the mountainous areas of La Rioja, Soria, Teruel, and the Central System, regions characterized by significant territorial mobility that extended into southern territories such as Extremadura and the Alcudia Valley. Historically, these cattle were used for hauling timber from the pine forests of the Central System and other Iberian mountain ranges—a role that earned them the designation
pinariegas cattle—and contributed to their dispersal across Andalusia [
1].
The use of these breeds as draft animals by the Royal Guild of Carters (Real Cabaña de Carreteros) further facilitated their expansion southward, particularly into the Tagus Valley (
Figure 8 and
Figure 9). Although populations of this lineage likely existed since ancient times in the Portuguese foothills of the Central System (Serra da Estrela), their expansion southward and westward was undoubtedly intensified by their role as draught animals in agricultural contexts. With the mechanization of agriculture, these functions have become obsolete, and today, these breeds are reared extensively for their meat production potential [
26,
27].
Among their former uses, dragging and hauling timber to sawmills and transporting wood to markets have been widely documented. This historical use played a key role in the emergence of the Andalusian Black cattle breed, particularly in regions such as Córdoba and Seville, where gene flow resulting from transhumant movements contributed to shaping the breed’s genetic architecture. Selection for traits such as size, strength, and docility was essential to meet the demands of draft work. The prominent role of these cattle within the Royal Guild of Carters further reinforced their geographical spread and consolidation [
1].
One hypothesis suggests that the formation of the Andalusian Black breed in the 19th century was a response to increasing demand for draft animals during the transition from livestock-based to agricultural land use. This transition created a pressing need for
yuntas (draft pairs) for agricultural labor. To meet this demand, animals from provinces such as Ávila and Salamanca were introduced, leading to crossbreeding processes that gave rise to distinct lines, such as the
Negra de las Campiñas and the
Cárdena Andaluza. This period of agricultural expansion was instrumental in consolidating the genetic basis of the Andalusian Black breed. Furthermore, the practice of transhumance—particularly along livestock routes (
cañadas,
veredas, and
cordeles)—further influenced both the genetic diversity and geographical distribution of the breed [
1,
28,
29,
30,
31].
Transhumance, a prehistoric practice associated with the seasonal movement of bovine, caprine, and ovine species, represents a cornerstone of the Andalusian Black cattle’s genetic heritage. These movements, connected to public-use mountain areas and privately managed
dehesas, have played a fundamental role in shaping both the spatial distribution and genetic diversity of the breed. The main transhumance areas in Andalusia include the Sierra Cordobesa, Sierra de Aracena, and the Guadalquivir Valley—regions that today remain crucial reservoirs of genetic diversity for this breed [
1,
32].
The core distribution area of the Andalusian Black cattle breed extends from the Sierra of Córdoba to the Sierra de Aracena, also encompassing zones such as the northern Sierra of Seville, the Valle de Los Pedroches and the Valle del Guadiato. According to Columela [
33], traditional Andalusian cattle included three main types:
Morena,
Roja, and
Rubia. The
Morena, described as eumetric and brevilinear, was considered representative of the fine
casta cattle and was frequently used in crossbreeding to improve temperament and enhance functional traits in working animals. Its compact body—marked by a voluminous trunk, shorter and finer limbs and head, and a lighter skeleton—also made it advantageous for meat production, reinforcing its agricultural and economic value [
32].
From the crossing of the
Morena,
Roja, and
Rubia types, several local varieties emerged, often quite similar in phenotype. However, breeder selection for straight, black-coated, and compact sires led to the emergence of a relatively uniform type: the
Negra Andaluza. This process was further supported by the incorporation of
Avileña Negra Ibérica bulls as improvers. Historically known as
Negra Campiñesa or
Negra de las Campiñas, the breed was thus distinguished from the
Avileña Negra Ibérica, more common in the
dehesas of Extremadura and Castile [
33].
The resulting population, characterized by black coats and functional conformation, established itself mainly in the southern provinces of Córdoba and Seville, where the breed’s genetic base consolidated.
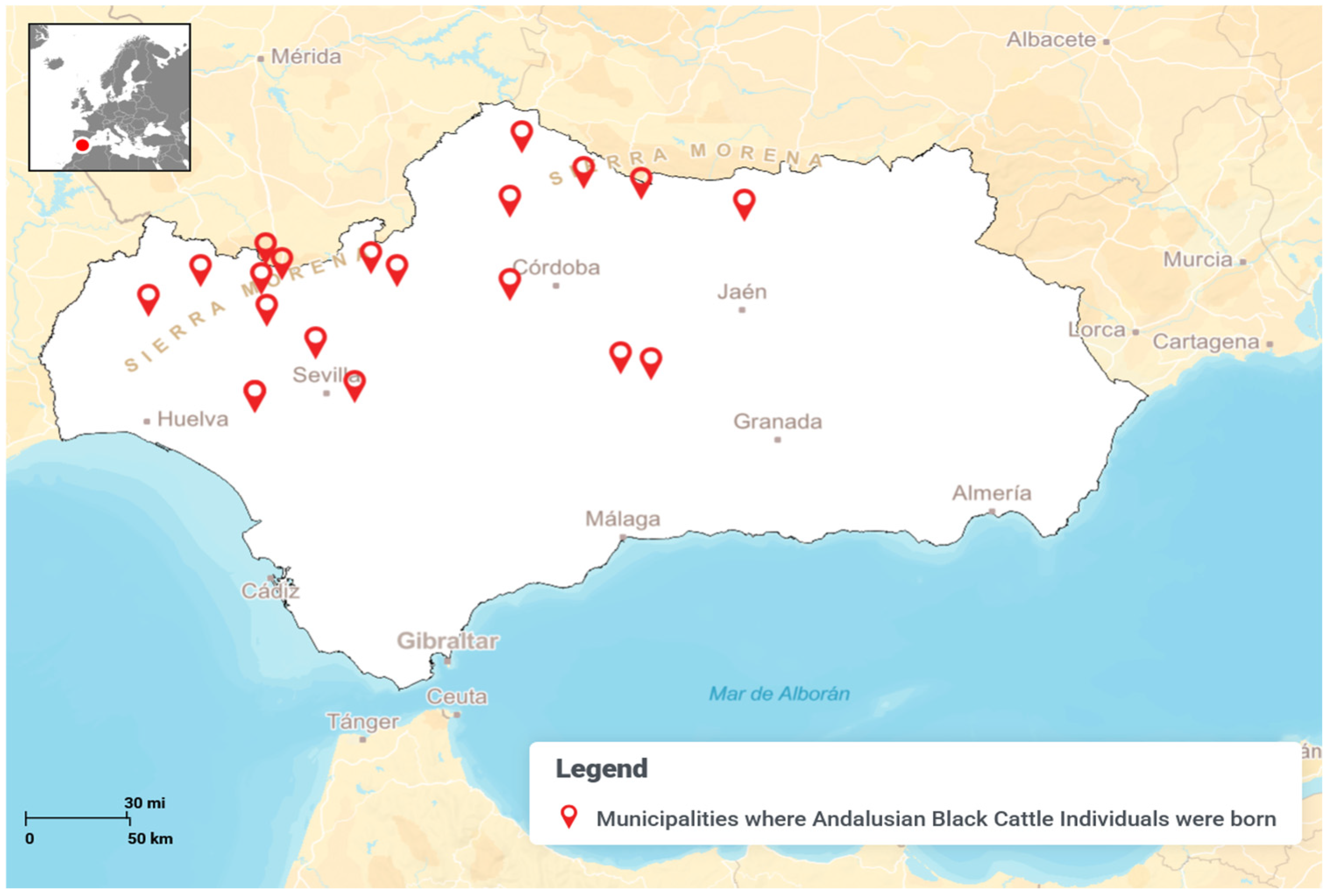
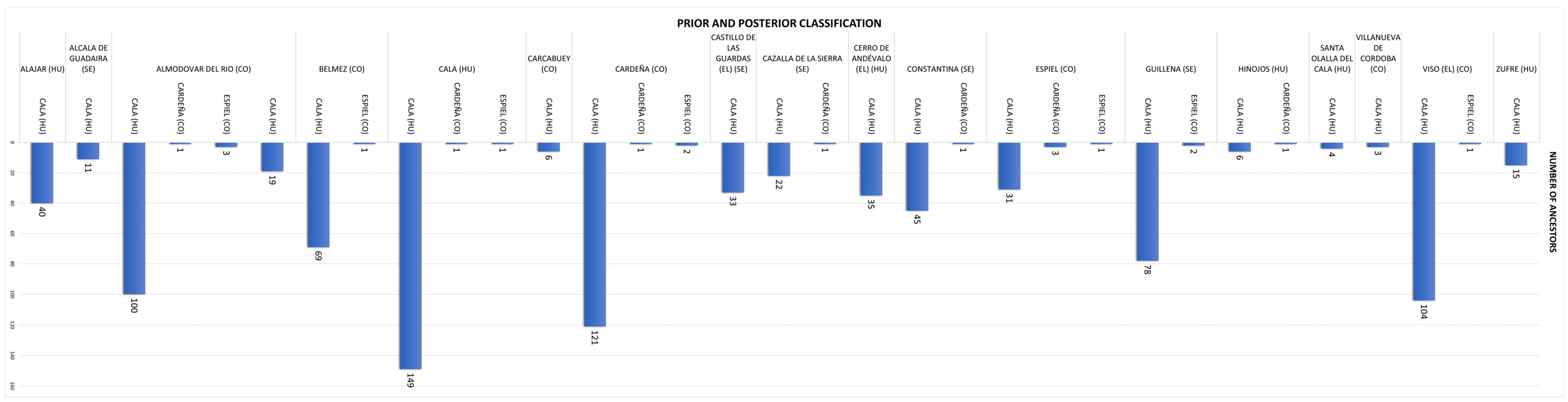
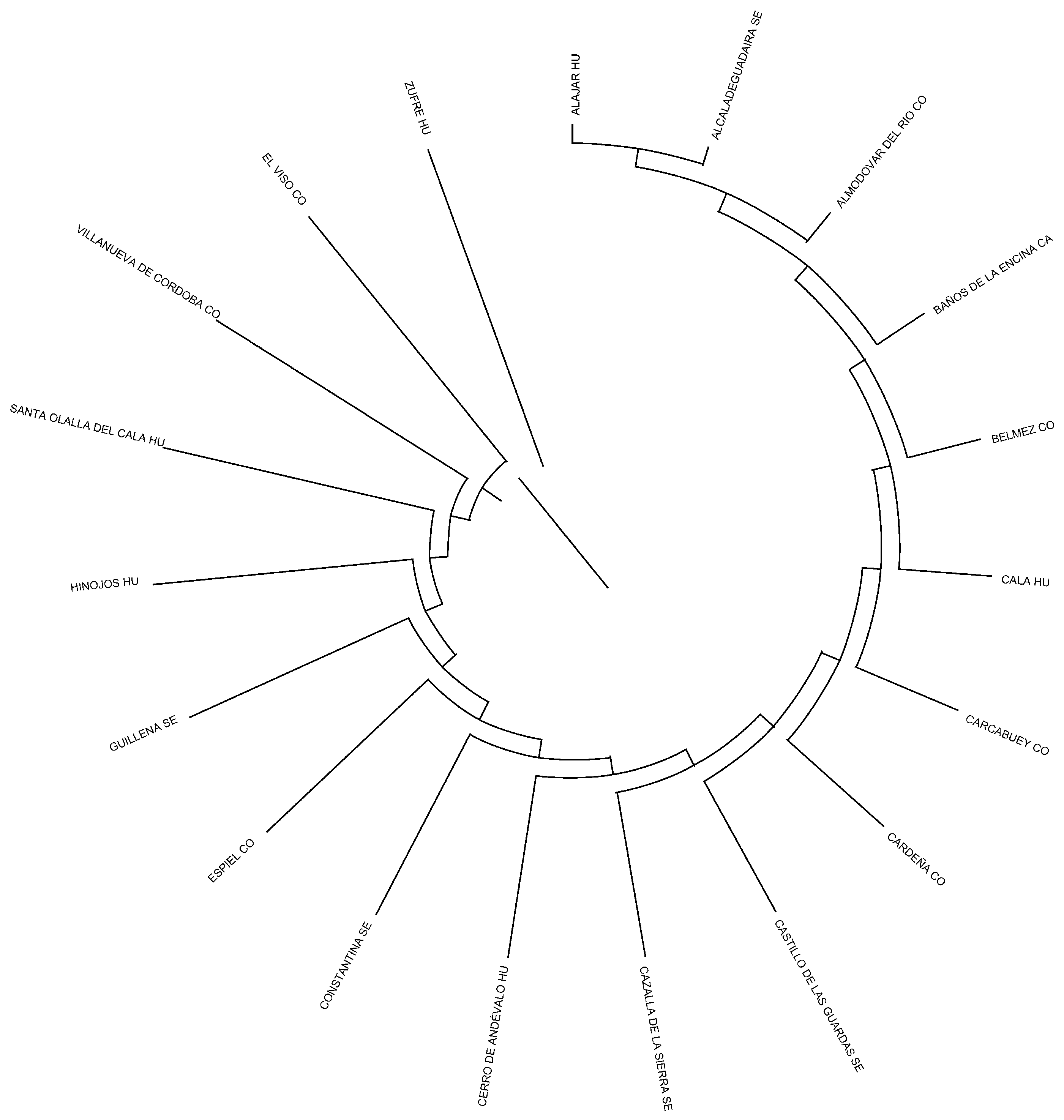
The genetic structure of the Negra Andaluza has been shaped by founder effects, inbreeding loops, and the influence of nodal ancestors. Analyses indicate that Córdoba is the primary genetic nucleus, with a significantly higher number of recorded ancestors (448) compared to Huelva (252) and Seville (193). This concentration of genealogical contributions suggests that Córdoba played a central role in shaping the breed’s genetic architecture.
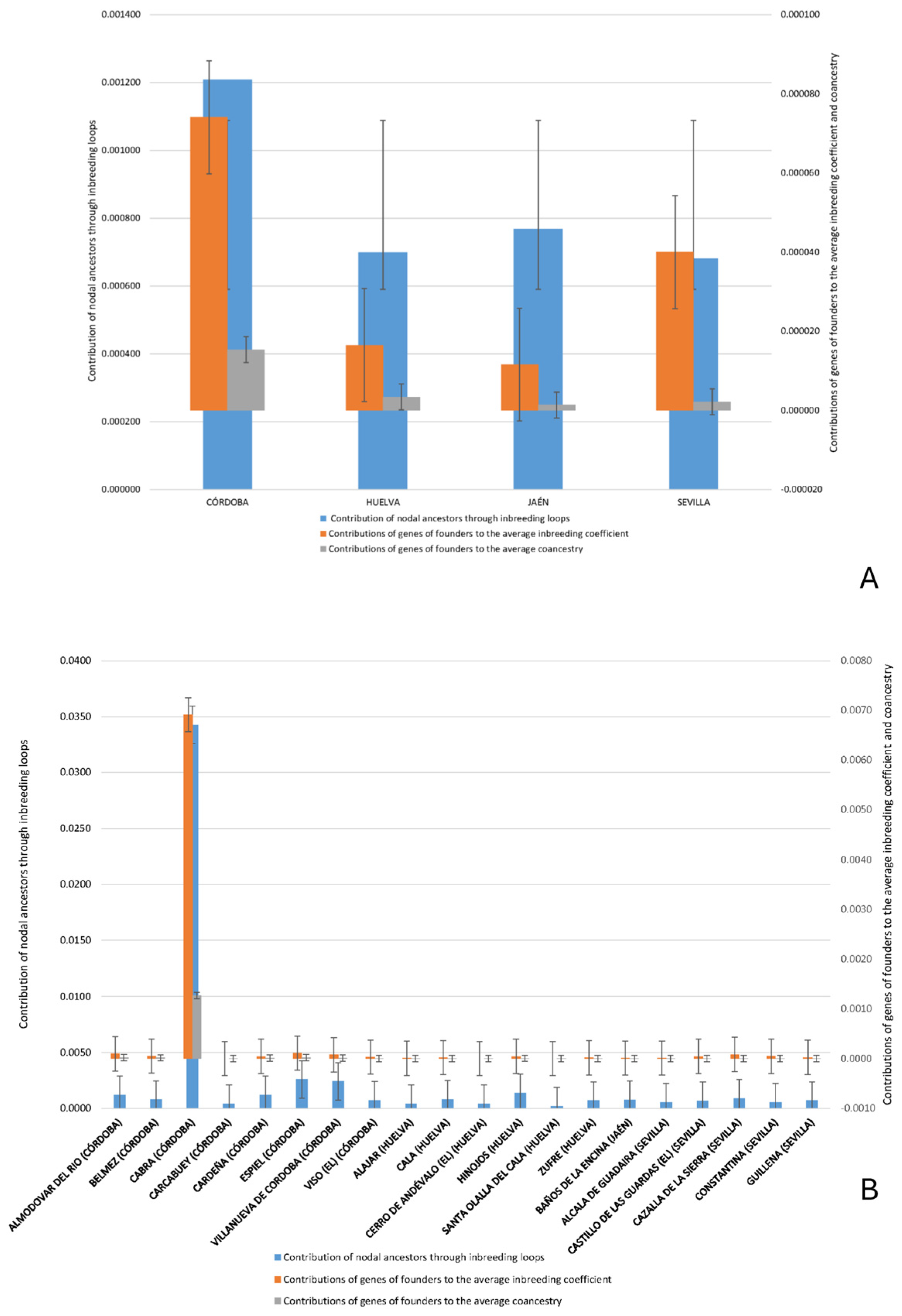
Despite its broad range, two major zones of concentration can be identified: one in the Sierra Morena and its foothills (Córdoba and Huelva) and another in the lowlands of Seville, Cádiz, and Huelva. As shown in
Figure 4, the highest nodal contributions through inbreeding loops are concentrated in Córdoba. Similarly, the greatest contributions of founder genes to both average inbreeding and coancestry coefficients have been observed in Córdoba and Seville, as reported by Cartuche Macas et al. [
1] using Wright’s parameters. These findings support the hypothesis that the
Cañada Real Soriana Oriental was a major axis for the genetic establishment of the breed in the region, a process reinforced by Andalusia’s livestock trails (
cordeles,
veredas, and
coladas; see
Figure 9).
Within Córdoba, the municipality of Cabra stands out for its exceptionally high contribution of nodal ancestors and founder genes, both to inbreeding and coancestry (
Figure 5). This prominence may be due to its relative geographical isolation in the southern part of the province, with limited connectivity to other nuclei, which would have restricted gene flow and led to reduced genetic diversity. In contrast, areas better connected through transhumance routes (
cañadas and
cordeles) exhibit greater genetic exchange and heterogeneity. Similarly, the Hinojos nucleus in Huelva, due to its geographical isolation, exhibited a greater contribution to both inbreeding and coancestry, underscoring the critical role of isolation in shaping the genetic makeup of Andalusian Black Cattle [
32]. The influence of nodal ancestors is key to understanding regional differentiation, especially in Córdoba, Seville, and Huelva, where historic livestock routes (cañadas, veredas, cordeles, and coladas) facilitated gene flow across the region [
1].
Notably, the Cañada Real Soriana Oriental linked Córdoba and Seville, promoting the spread of founder genes that significantly shaped the breed’s current genetic structure. Conversely, Huelva’s connection through the Cañada Real Soriana Occidental and the Cañada Real de la Plata—linked to the Leonesa Occidental and Oriental and Segoviana routes—allowed for a more limited but still meaningful genetic exchange. The relatively low genetic input from Jaén is likely due to its weak connection via the Real Conquense and Soriana Oriental routes [
1,
28,
30,
31].
Regarding the number of identified ancestors, Jaén recorded the fewest (19), while Córdoba had the most (448), followed by Huelva (252) and Seville (193). This underscores Córdoba’s genetic prominence, with more than double the number of ancestral contributions compared to Huelva and Seville. Wright’s parameters for coancestry and inbreeding further confirm Córdoba as the primary center of genetic consolidation, reinforcing its pivotal role in the development of the breed’s distinct genetic traits.
The origin and structure of Andalusian Black Cattle can be traced through inbreeding loops, MSVs, and contributions from nodal ancestors. CDA was employed to evaluate how these variables—essential components of genetic architecture—contribute to population differentiation across Andalusia [
1].
Multicollinearity was observed among variables such as the founder contributions to average inbreeding, average coancestry, and non-random mating (α), along with the contribution of nodal ancestors through inbreeding loops and coancestry (C). These interdependencies, both conceptual and mathematical, reflect overlapping aspects of population structure and mating patterns. To ensure model robustness and interpretability, collinear variables were excluded, retaining only those offering distinct, non-redundant information.
The analysis revealed statistically significant yet modest differences between groups, with the first discriminant function (F1) emerging as the most informative. Inbreeding loops and nodal ancestor contributions were central to explaining group structure.
Previous studies have established that domestic cattle, including Andalusian Black Cattle, descend from the extinct Eurasian aurochs, with the Iberian Peninsula acting as a major center for the development of native breeds. The Andalusian Black is thought to have originated from historical breeding events in the fertile regions of Andalucía, influenced by the introgression of genetic diversity from areas like Ávila and Salamanca [
1,
23,
34,
35].
Inbreeding and coancestry are central to the breed’s genetic architecture, with inbreeding loops playing a significant role in maintaining specific traits. Our analysis showed that MSVs enabled a quantitative decomposition of inbreeding, highlighting the critical contribution of nodal ancestors—particularly in Córdoba—where these loops were more prevalent and impactful [
28,
36,
37,
38,
39].
The regional variation in the contribution to inbreeding was most pronounced in Córdoba, suggesting a potential genetic bottleneck due to limited gene flow. Although the analysis revealed pronounced inbreeding loops, the breed appears to have retained a moderate level of genetic diversity, as indicated by the coancestry mean of 0.000475 and inbreeding loop contribution mean of 0.000949 (
Table 2). This resilience may be attributed to historical transhumance, which enabled genetic exchange across regions despite isolation (see
Figure 9). Previous work by Cartuche Macas et al. [
1] and Navas González et al. [
15] also supports this finding, highlighting the buffering effect of migratory breeding on genetic erosion.
The observed discrepancy between inbreeding and coancestry may be attributed to the complex interplay of genetic drift, selection, and gene flow. While inbreeding loops can lead to the fixation of alleles, connectivity through historical transhumance routes likely prevented excessive genetic homogenization [
35,
37].
The primary driver of inbreeding in Andalusian Black Cattle appears to be the influence of nodal ancestors. CDA revealed their disproportionate impact on the breed’s genetic structure, particularly in Córdoba and Huelva, where their contributions to inbreeding and coancestry exceeded expectations. These findings suggest the persistence of distinct genetic lineages tied to specific founders.
Wright’s parameters further corroborated Córdoba’s strong genetic signature, characterized by a higher proportion of ancestral contributions to inbreeding loops and coancestry. LOOCV confirmed this pattern, with Córdoba achieving high classification accuracy, while Huelva and Seville showed lower discriminant precision. This emphasizes Córdoba’s genetic isolation and its central role in shaping the breed’s diversity [
1,
35,
36].
The geographic concentration of the breed and the clear regional variation in genetic composition reflect distinct selection pressures. Córdoba and Huelva exhibited higher relatedness and fewer external genetic inputs, whereas Seville displayed a more homogenized profile, likely facilitated by greater gene flow through transhumant livestock routes.
The model’s inability to classify individuals from Huelva and Jaén may be due to several factors. First, limited sample sizes (especially in Jaén, with only 19 individuals) reduce classification accuracy. Second, sampling bias may exist if animals from these regions are underrepresented in pedigree completeness or geographic coverage. Additionally, genetic homogeneity—either from bottlenecks or founder effects—could obscure distinctiveness in the discriminant space. These issues suggest a need for enhanced sampling, particularly in these provinces, and potentially incorporating molecular markers to uncover hidden genetic structures.
CDA identified three discriminant functions, with the first function (F1) explaining the vast majority of the total variance (79.72%). This dominance suggests that the primary axis of genetic differentiation among the groups is strongly associated with the variables contributing most to F1. Notably, this function was chiefly influenced by inbreeding loops and the genetic contributions of nodal ancestors, highlighting their central role in shaping the observed genetic structure.
The secondary (F2) and tertiary (F3) functions contributed comparatively less to the overall variance, implying that while they capture additional dimensions of genetic differentiation, their explanatory power is limited. These functions may still reflect biologically meaningful patterns, potentially related to more subtle demographic or historical factors, but they do not overshadow the primary signal associated with inbreeding and key ancestral contributions.
Interestingly, while coancestry was also included in the analysis, its influence appears secondary. This suggests that, in the studied population, patterns of genetic differentiation are driven more by internal lineage structures (e.g., repeated inbreeding and strong founder effects from nodal ancestors) than by broader patterns of relatedness among individuals or subpopulations. In practical terms, this underscores the importance of managing inbreeding and monitoring influential ancestors in conservation or breeding programs, as these factors may have a disproportionate effect on genetic diversity and population structure [
1,
35,
37].
The prominence of nodal ancestors highlights the historical influence of selective breeding, where specific individuals or family lines were favored for traits such as size, temperament, or adaptation to local environments. These past practices have left a lasting genetic legacy in the Andalusian Black Cattle.
In conservation terms, the breed has maintained high genetic diversity despite the influence of inbreeding. Historical gene flow across extensive livestock routes likely played a crucial role in preventing significant genetic erosion. Nonetheless, the continued presence of inbreeding loops signals the need for future breeding programs to manage genetic diversity carefully and mitigate risks associated with further inbreeding.