Parental Phasing Study Identified Lineage-Specific Variants Associated with Gene Expression and Epigenetic Modifications in European–Chinese Hybrid Pigs
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
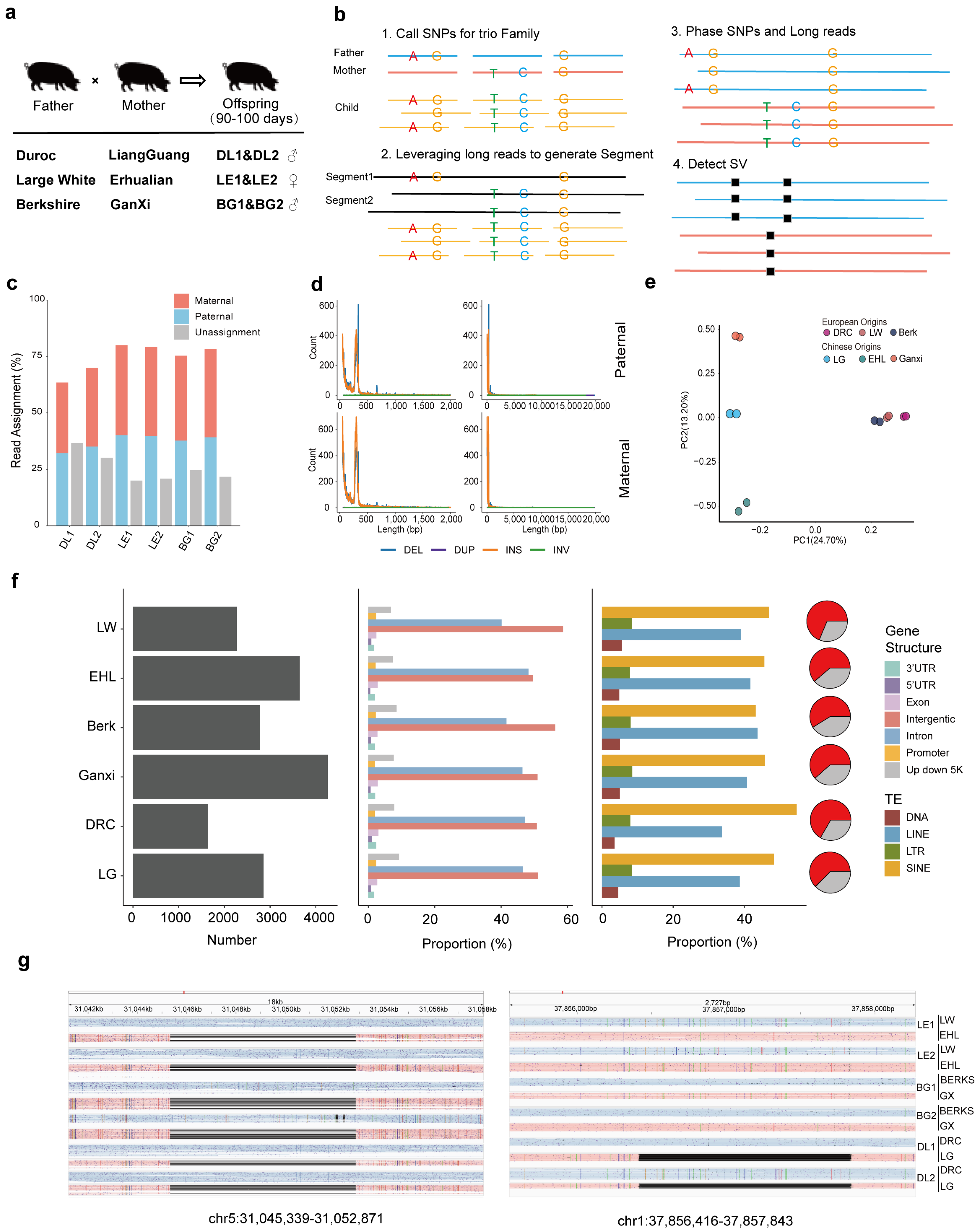
2.1. Biological Sample Collection and Family Identification
2.2. SNP Calling and Phased SNP Calling
2.3. SV Calling and Phased SV Calling
2.4. Genome Transposon Annotation
2.5. Quantification of Phased Gene Expression
2.6. Chromatin State Prediction and Identification of Phased Histone Modifications and CTCF Binding
2.7. Hi-C Data Analysis and Chromatic 3D Structure Identification
2.8. Colocalization of Phased Genetic Variants with Phased Gene Expression and Phased Epigenetic Modifications
2.9. Prediction of Transcription Factor Motifs
2.10. Functional Annotation of Single-Cell Data
3. Results
3.1. Experimental Design and Identification of Genetic Variants Form Different Lineages
3.2. Colocalization of Phased Genetic Variants and Phased Gene Expression
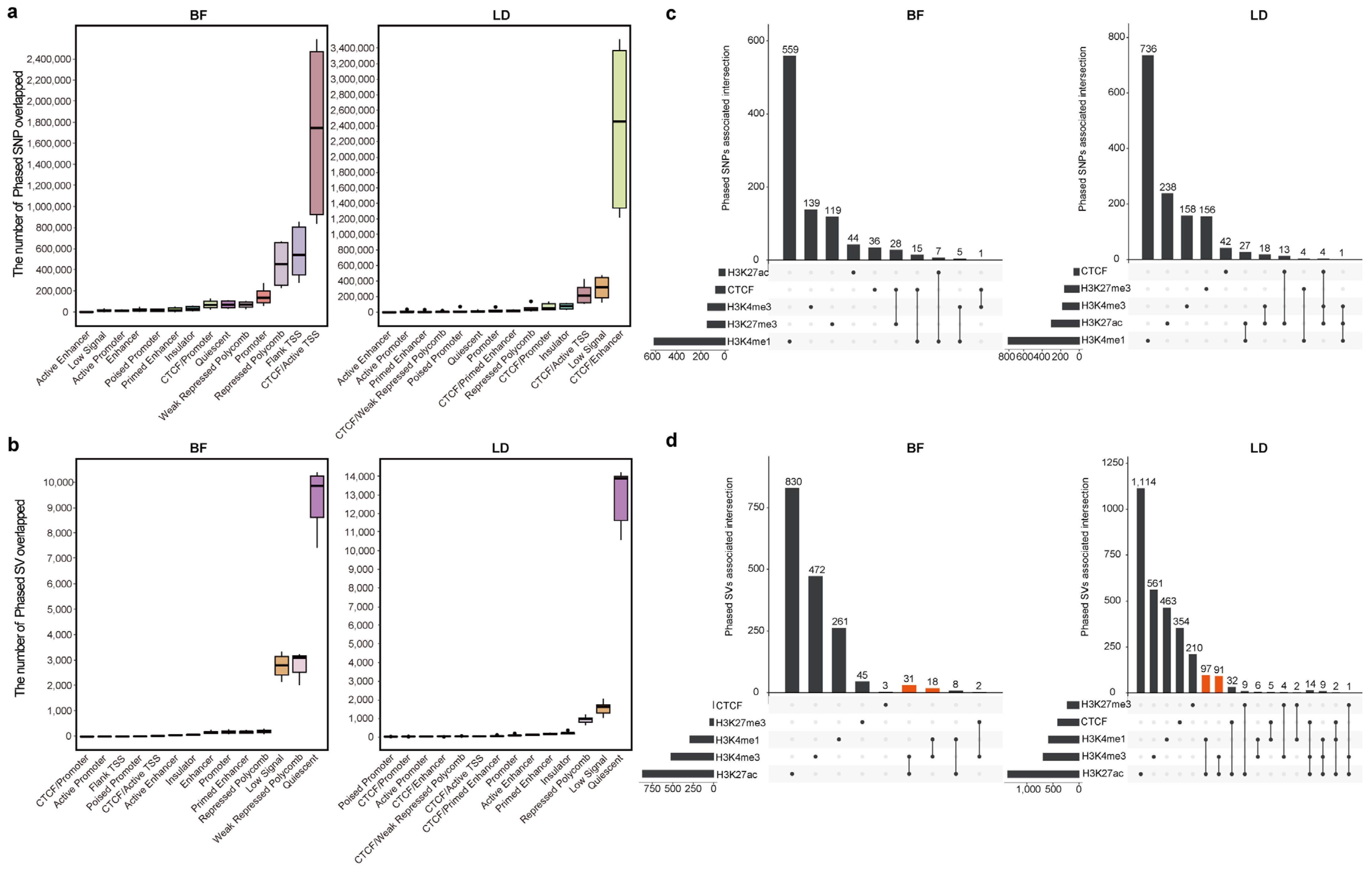
3.3. Associations Between Phased Genetic Variants and Chromatin State
3.4. Lineage-Specific Genetic Variants Modulate Epigenetic Modification to Regulate Lineage-Specific Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pelikan, R.C.; Kelly, J.A.; Fu, Y.; Lareau, C.A.; Tessneer, K.L.; Wiley, G.B.; Wiley, M.M.; Glenn, S.B.; Harley, J.B.; Guthridge, J.M. Enhancer histone-QTLs are enriched on autoimmune risk haplotypes and influence gene expression within chromatin networks. Nat. Commun. 2018, 9, 2905. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Yang, M.; Wang, X.; Cai, G.; Ding, R.; Zhuang, Z.; Zhou, S.; Tan, S.; Ruan, D.; Wu, J. Multi-omic characterization of allele-specific regulatory variation in hybrid pigs. Nat. Commun. 2024, 15, 5587. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Qu, Y.; Jia, Y.; He, S.; Pan, Z.; Wang, L.; Du, X. Assessment of heterosis based on parental genetic distance estimated with SSR and SNP markers in upland cotton (Gossypium hirsutum L.). BMC Genom. 2021, 22, 123. [Google Scholar] [CrossRef] [PubMed]
- Weischenfeldt, J.; Symmons, O.; Spitz, F.; Korbel, J.O. Phenotypic impact of genomic structural variation: Insights from and for human disease. Nat. Rev. Genet. 2013, 14, 125–138. [Google Scholar] [CrossRef]
- Stankiewicz, P.; Lupski, J.R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010, 61, 437–455. [Google Scholar] [CrossRef]
- Sedlazeck, F.J.; Lee, H.; Darby, C.A.; Schatz, M.C. Piercing the dark matter: Bioinformatics of long-range sequencing and mapping. Nat. Rev. Genet. 2018, 19, 329–346. [Google Scholar] [CrossRef]
- De Coster, W.; De Rijk, P.; De Roeck, A.; De Pooter, T.; D’Hert, S.; Strazisar, M.; Sleegers, K.; Van Broeckhoven, C. Structural variants identified by Oxford Nanopore PromethION sequencing of the human genome. Genome Res. 2019, 29, 1178–1187. [Google Scholar] [CrossRef]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef]
- Conrad, D.F.; Pinto, D.; Redon, R.; Feuk, L.; Gokcumen, O.; Zhang, Y.; Aerts, J.; Andrews, T.D.; Barnes, C.; Campbell, P. Origins and functional impact of copy number variation in the human genome. Nature 2010, 464, 704–712. [Google Scholar] [CrossRef]
- Li, M.; Tian, S.; Jin, L.; Zhou, G.; Li, Y.; Zhang, Y.; Wang, T.; Yeung, C.K.; Chen, L.; Ma, J. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 2013, 45, 1431–1438. [Google Scholar] [CrossRef]
- Kojima, M.; Nakajima, I.; Arakawa, A.; Mikawa, S.; Matsumoto, T.; Uenishi, H.; Nakamura, Y.; Taniguchi, M. Differences in gene expression profiles for subcutaneous adipose, liver, and skeletal muscle tissues between Meishan and Landrace pigs with different backfat thicknesses. PLoS ONE 2018, 13, e0204135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Meng, S.; Wang, H.; Zhang, C.; Sun, Z.; Huang, L.; Miao, Z. Comparison of growth performance, carcass properties, fatty acid Profile, and genes involved in fat metabolism in Nanyang and Landrace Pigs. Genes 2024, 15, 186. [Google Scholar] [CrossRef] [PubMed]
- Groenen, M.A.; Archibald, A.L.; Uenishi, H.; Tuggle, C.K.; Takeuchi, Y.; Rothschild, M.F.; Rogel-Gaillard, C.; Park, C.; Milan, D.; Megens, H.-J. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 2012, 491, 393–398. [Google Scholar] [CrossRef]
- Ouyang, W.; Zhang, X.; Guo, M.; Wang, J.; Wang, X.; Gao, R.; Ma, M.; Xiang, X.; Luan, S.; Xing, F. Haplotype mapping of H3K27me3-associated chromatin interactions defines topological regulation of gene silencing in rice. Cell Rep. 2023, 42, 112350. [Google Scholar] [CrossRef]
- Lin, Y.; Li, J.; Gu, Y.; Jin, L.; Bai, J.; Zhang, J.; Wang, Y.; Liu, P.; Long, K.; He, M. Haplotype-resolved 3D chromatin architecture of the hybrid pig. Genome Res. 2024, 34, 310–325. [Google Scholar] [CrossRef]
- Li, J.; Lin, Y.; Li, D.; He, M.; Kui, H.; Bai, J.; Chen, Z.; Gou, Y.; Zhang, J.; Wang, T. Building Haplotype-Resolved 3D Genome Maps of Chicken Skeletal Muscle. Adv. Sci. 2024, 11, 2305706. [Google Scholar] [CrossRef]
- Marshall, T.; Slate, J.; Kruuk, L.; Pemberton, J. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 1998, 7, 639–655. [Google Scholar] [CrossRef]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Patterson, M.; Marschall, T.; Pisanti, N.; Van Iersel, L.; Stougie, L.; Klau, G.W.; Schönhuth, A. WhatsHap: Weighted haplotype assembly for future-generation sequencing reads. J. Comput. Biol. 2015, 22, 498–509. [Google Scholar] [CrossRef]
- Smolka, M.; Paulin, L.F.; Grochowski, C.M.; Horner, D.W.; Mahmoud, M.; Behera, S.; Kalef-Ezra, E.; Gandhi, M.; Hong, K.; Pehlivan, D. Detection of mosaic and population-level structural variants with Sniffles2. Nat. Biotechnol. 2024, 42, 1571–1580. [Google Scholar] [CrossRef]
- Bao, W.; Kojima, K.K.; Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. Dna 2015, 6, 11. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Feng, J.; Liu, T.; Qin, B.; Zhang, Y.; Liu, X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012, 7, 1728–1740. [Google Scholar] [CrossRef]
- Ross-Innes, C.S.; Stark, R.; Teschendorff, A.E.; Holmes, K.A.; Ali, H.R.; Dunning, M.J.; Brown, G.D.; Gojis, O.; Ellis, I.O.; Green, A.R. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 2012, 481, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Kellis, M. ChromHMM: Automating chromatin-state discovery and characterization. Nat. Methods 2012, 9, 215–216. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef]
- Durand, N.C.; Shamim, M.S.; Machol, I.; Rao, S.S.; Huntley, M.H.; Lander, E.S.; Aiden, E.L. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016, 3, 95–98. [Google Scholar] [CrossRef]
- Wolff, J.; Bhardwaj, V.; Nothjunge, S.; Richard, G.; Renschler, G.; Gilsbach, R.; Manke, T.; Backofen, R.; Ramírez, F.; Grüning, B.A. Galaxy HiCExplorer: A web server for reproducible Hi-C data analysis, quality control and visualization. Nucleic Acids Res. 2018, 46, W11–W16. [Google Scholar] [CrossRef]
- Roayaei Ardakany, A.; Gezer, H.T.; Lonardi, S.; Ay, F. Mustache: Multi-scale detection of chromatin loops from Hi-C and Micro-C maps using scale-space representation. Genome Biol. 2020, 21, 256. [Google Scholar] [CrossRef]
- Shabalin, A.A. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics 2012, 28, 1353–1358. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.-A.; Kwok, I.W.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2019, 37, 38–44. [Google Scholar] [CrossRef]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Miyake, S.; Oka, M.; Kanai, A.; Kawai, Y.; Nagasawa, S.; Shiraishi, Y.; Tokunaga, K.; Kohno, T.; Seki, M. Phasing analysis of lung cancer genomes using a long read sequencer. Nat. Commun. 2022, 13, 3464. [Google Scholar] [CrossRef]
- Gigante, S.; Gouil, Q.; Lucattini, A.; Keniry, A.; Beck, T.; Tinning, M.; Gordon, L.; Woodruff, C.; Speed, T.P.; Blewitt, M.E. Using long-read sequencing to detect imprinted DNA methylation. Nucleic Acids Res. 2019, 47, e46. [Google Scholar] [CrossRef]
- Yang, L.; Yin, H.; Bai, L.; Yao, W.; Tao, T.; Zhao, Q.; Gao, Y.; Teng, J.; Xu, Z.; Lin, Q. Mapping and functional characterization of structural variation in 1060 pig genomes. Genome Biol. 2024, 25, 116. [Google Scholar] [CrossRef]
- Logsdon, G.A.; Ebert, P.; Audano, P.A.; Loftus, M.; Porubsky, D.; Ebler, J.; Yilmaz, F.; Hallast, P.; Prodanov, T.; Yoo, D. Complex genetic variation in nearly complete human genomes. bioRxiv 2024, preprint. [Google Scholar]
- Hénault, M.; Marsit, S.; Charron, G.; Landry, C.R. The genomic landscape of transposable elements in yeast hybrids is shaped by structural variation and genotype-specific modulation of transposition rate. eLife 2024, 12, RP89277. [Google Scholar] [CrossRef]
- Lazarescu, O.; Ziv-Agam, M.; Haim, Y.; Hekselman, I.; Jubran, J.; Shneyour, A.; Muallem, H.; Zemer, A.; Rosengarten-Levin, M.; Kitsberg, D. Human subcutaneous and visceral adipocyte atlases uncover classical and nonclassical adipocytes and depot-specific patterns. Nat. Genet. 2025, 57, 413–426. [Google Scholar] [CrossRef]
- Li, M.; Ang, K.S.; Teo, B.; Rom, U.; Nguyen, M.N.; Maurer-Stroh, S.; Chen, J. Rediscovering publicly available single-cell data with the DISCO platform. Nucleic Acids Res. 2025, 53, D932–D938. [Google Scholar] [CrossRef]
- Oh, G.S.; Yoon, J.; Kim, G.; Kim, G.H.; Kim, D.S.; Choi, B.; Chang, E.J.; Lee, E.S.; Kim, S.W. Regulation of adipocyte differentiation by clusterin-mediated Krüppel-like factor 5 stabilization. FASEB J. 2020, 34, 16276–16290. [Google Scholar] [CrossRef]
- Tsai, M.; Valent, P.; Galli, S.J. KIT as a master regulator of the mast cell lineage. J. Allergy Clin. Immunol. 2022, 149, 1845–1854. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Ruan, H.-B.; Xian, L.; Chen, W.; Jiang, S.; Song, A.; Wang, Q.; Shi, P.; Gu, X.; Gao, X. The stem cell factor/Kit signalling pathway regulates mitochondrial function and energy expenditure. Nat. Commun. 2014, 5, 4282. [Google Scholar] [CrossRef] [PubMed]
- Rubin, C.-J.; Megens, H.-J.; Barrio, A.M.; Maqbool, K.; Sayyab, S.; Schwochow, D.; Wang, C.; Carlborg, Ö.; Jern, P.; Jørgensen, C.B. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. USA 2012, 109, 19529–19536. [Google Scholar] [CrossRef]
- Moller, M.J.; Chaudhary, R.; Hellmén, E.; Höyheim, B.; Chowdhary, B.; Andersson, L. Pigs with the dominant white coat color phenotype carry a duplication of the KIT gene encoding the mast/stem cell growth factor receptor. Mamm. Genome 1996, 7, 822–830. [Google Scholar] [CrossRef]
- Tong, X.; Chen, D.; Hu, J.; Lin, S.; Ling, Z.; Ai, H.; Zhang, Z.; Huang, L. Accurate haplotype construction and detection of selection signatures enabled by high quality pig genome sequences. Nat. Commun. 2023, 14, 5126. [Google Scholar] [CrossRef]
- Yang, R.; Guo, X.; Zhu, D.; Tan, C.; Bian, C.; Ren, J.; Huang, Z.; Zhao, Y.; Cai, G.; Liu, D. Accelerated deciphering of the genetic architecture of agricultural economic traits in pigs using a low-coverage whole-genome sequencing strategy. Gigascience 2021, 10, giab048. [Google Scholar] [CrossRef]
- Ding, R.; Savegnago, R.; Liu, J.; Long, N.; Tan, C.; Cai, G.; Zhuang, Z.; Wu, J.; Yang, M.; Qiu, Y. The SWine IMputation (SWIM) haplotype reference panel enables nucleotide resolution genetic mapping in pigs. Commun. Biol. 2023, 6, 577. [Google Scholar] [CrossRef]
- Kim, H.; Song, K.D.; Kim, H.J.; Park, W.; Kim, J.; Lee, T.; Shin, D.-H.; Kwak, W.; Kwon, Y.-j.; Sung, S. Exploring the genetic signature of body size in Yucatan miniature pig. PLoS ONE 2015, 10, e0121732. [Google Scholar] [CrossRef]
- Teng, J.; Gao, Y.; Yin, H.; Bai, Z.; Liu, S.; Zeng, H.; Consortium, P.; Bai, L.; Cai, Z.; Zhao, B. A compendium of genetic regulatory effects across pig tissues. Nat. Genet. 2024, 56, 112–123. [Google Scholar] [CrossRef]
- Ebert, P.; Audano, P.A.; Zhu, Q.; Rodriguez-Martin, B.; Porubsky, D.; Bonder, M.J.; Sulovari, A.; Ebler, J.; Zhou, W.; Serra Mari, R. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 2021, 372, eabf7117. [Google Scholar] [CrossRef]
- Schöpflin, R.; Melo, U.S.; Moeinzadeh, H.; Heller, D.; Laupert, V.; Hertzberg, J.; Holtgrewe, M.; Alavi, N.; Klever, M.-K.; Jungnitsch, J. Integration of Hi-C with short and long-read genome sequencing reveals the structure of germline rearranged genomes. Nat. Commun. 2022, 13, 6470. [Google Scholar] [CrossRef]
- Low, W.Y.; Tearle, R.; Liu, R.; Koren, S.; Rhie, A.; Bickhart, D.M.; Rosen, B.D.; Kronenberg, Z.N.; Kingan, S.B.; Tseng, E. Haplotype-resolved genomes provide insights into structural variation and gene content in Angus and Brahman cattle. Nat. Commun. 2020, 11, 2071. [Google Scholar] [CrossRef]
- Di Siena, S.; Gimmelli, R.; Nori, S.L.; Barbagallo, F.; Campolo, F.; Dolci, S.; Rossi, P.; Venneri, M.; Giannetta, E.; Gianfrilli, D. Activated c-Kit receptor in the heart promotes cardiac repair and regeneration after injury. Cell Death Dis. 2016, 7, e2317. [Google Scholar] [CrossRef][Green Version]
- Marino, F.; Scalise, M.; Cianflone, E.; Mancuso, T.; Aquila, I.; Agosti, V.; Torella, M.; Paolino, D.; Mollace, V.; Nadal-Ginard, B. Role of c-kit in myocardial regeneration and aging. Front. Endocrinol. 2019, 10, 371. [Google Scholar] [CrossRef]
- Sun, L.; Wu, R. Mapping complex traits as a dynamic system. Phys. Life Rev. 2015, 13, 155–185. [Google Scholar] [CrossRef]
- Zhabotynsky, V.; Huang, L.; Little, P.; Hu, Y.-J.; Pardo-Manuel de Villena, F.; Zou, F.; Sun, W. eQTL mapping using allele-specific count data is computationally feasible, powerful, and provides individual-specific estimates of genetic effects. PLoS Genet. 2022, 18, e1010076. [Google Scholar] [CrossRef]
- Turner, R.T.; Wong, C.P.; Iwaniec, U.T. Effect of Reduced c-Kit Signaling on Bone Marrow Adiposity. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 2011, 294, 1126–1134. [Google Scholar] [CrossRef]
- Zhang, Z.; Hong, Y.; Gao, J.; Xiao, S.; Ma, J.; Zhang, W.; Ren, J.; Huang, L. Genome-wide association study reveals constant and specific loci for hematological traits at three time stages in a White Duroc× Erhualian F2 resource population. PLoS ONE 2013, 8, e63665. [Google Scholar] [CrossRef]
- Pan, C.; Yang, C.; Wang, S.; Ma, Y. Identifying key genes and functionally enriched pathways of diverse adipose tissue types in cattle. Front. Genet. 2022, 13, 790690. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, H.; Xu, Q.; Liu, M.; Chao, X.; Chen, J.; Zhou, B. Genome-Wide Analysis Reveals Copy Number Variant Gene TGFBR3 Regulates Pig Back Fat Deposition. Animals 2024, 14, 2657. [Google Scholar] [CrossRef]
- Matsumoto, S.; Kurimoto, T.; Taketo, M.M.; Fujii, S.; Kikuchi, A. The WNT/MYB pathway suppresses KIT expression to control the timing of salivary proacinar differentiation and duct formation. Development 2016, 143, 2311–2324. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Lee, J.; Yang, M.J.; Kim, Y.-C.; Hong, S.P.; Kim, J.M.; Hwang, G.-S.; Koh, G.Y. Endothelial cell-derived stem cell factor promotes lipid accumulation through c-Kit-mediated increase of lipogenic enzymes in brown adipocytes. Nat. Commun. 2023, 14, 2754. [Google Scholar] [CrossRef] [PubMed]
- Fontanesi, L.; D’Alessandro, E.; Scotti, E.; Liotta, L.; Crovetti, A.; Chiofalo, V.; Russo, V. Genetic heterogeneity and selection signature at the KIT gene in pigs showing different coat colours and patterns. Anim. Genet. 2010, 41, 478–492. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Lee, S.; Shrestha, P.; Kim, J.; Park, J.A.; Ko, Y.; Ban, Y.H.; Park, D.-Y.; Ha, S.-J.; Koh, G.Y. AMIGO2, a novel membrane anchor of PDK1, controls cell survival and angiogenesis via Akt activation. J. Cell Biol. 2015, 211, 619–637. [Google Scholar] [CrossRef]
- Tian, Z.; Zhou, D.; Jiang, R.; Zhou, B. Role of AMIGO2 in cancer progression: Novel insights. Oncol. Lett. 2024, 28, 434. [Google Scholar] [CrossRef]


| SNP Region (Clustered by Window of 1 Mb) | SV Region | Gene | H3K4me3 | H3K27ac | H3K4me1 | H3K27me3 | CTCF | Tissue |
|---|---|---|---|---|---|---|---|---|
| chr5:77483571-78328698 | (INS) chr5:77568931-77569226 | AMIGO2 | - | chr5:77754058-77755070 chr5:77757811-77758325 | - | - | - | BF |
| chr7:135107363-135351353 | - | ENSSSCG00000036983 | - | - | - | - | chr7:135297819-135299247 | BF |
| - | (DUP) chr8:41223208-41783661 | KIT | chr8:41401921-41403769 | chr8:41401817-41403400 | - | - | - | BF |
| (DUP) chr8:41223208-41783661 | KIT | chr8:41401866-41403769 | chr8:41402038-41403489 | - | - | - | LD | |
| chr12:19702952-20479694 | - | PSME3 | - | chr12:19713381-19714949 | - | - | chr12:19713354-19714899 | LD |
| Breeds | Variants | Ref_Allele_Freq (%) | Alt_Allele_Freq (%) |
|---|---|---|---|
| DRC | chr5:77568931-77569226 | 71.08 | 28.92 |
| LW | 20.81 | 79.19 | |
| LaiWu | 13.64 | 86.36 | |
| EHL | 8.02 | 91.98 | |
| DRC | chr5:77754641-77754642 | 90.17 | 9.83 |
| LW | 1.94 | 98.06 | |
| LaiWu | 2.27 | 97.73 | |
| EHL | 21.96 | 78.04 | |
| DRC | chr5:77757817-77757818 | 90.03 | 9.97 |
| LW | 0.00 | 100.00 | |
| LaiWu | 2.27 | 97.73 | |
| EHL | 13.08 | 86.92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Ge, M.; Long, K.; Han, Z.; Li, J.; Li, M.; Zhang, Z. Parental Phasing Study Identified Lineage-Specific Variants Associated with Gene Expression and Epigenetic Modifications in European–Chinese Hybrid Pigs. Animals 2025, 15, 1494. https://doi.org/10.3390/ani15101494
Li C, Ge M, Long K, Han Z, Li J, Li M, Zhang Z. Parental Phasing Study Identified Lineage-Specific Variants Associated with Gene Expression and Epigenetic Modifications in European–Chinese Hybrid Pigs. Animals. 2025; 15(10):1494. https://doi.org/10.3390/ani15101494
Chicago/Turabian StyleLi, Chenyu, Mei Ge, Keren Long, Ziyin Han, Jing Li, Mingzhou Li, and Zhiyan Zhang. 2025. "Parental Phasing Study Identified Lineage-Specific Variants Associated with Gene Expression and Epigenetic Modifications in European–Chinese Hybrid Pigs" Animals 15, no. 10: 1494. https://doi.org/10.3390/ani15101494
APA StyleLi, C., Ge, M., Long, K., Han, Z., Li, J., Li, M., & Zhang, Z. (2025). Parental Phasing Study Identified Lineage-Specific Variants Associated with Gene Expression and Epigenetic Modifications in European–Chinese Hybrid Pigs. Animals, 15(10), 1494. https://doi.org/10.3390/ani15101494





