A Canine c-kit Novel Mutation Isolated from a Gastrointestinal Stromal Tumor (GIST) Retains the Ability to Form Dimers but Lacks Autophosphorylation
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Histological Analysis
2.2. Sample Preparation and Sequencing
2.3. Cells and Cell Culture
2.4. Generation of Hemagglutinin-Tagged KIT Mutants into the Mammalian Expression Vectors
2.5. Transfection and Adding SCF
2.6. Western Blot Analysis
2.7. Cloning of c-kit-Expressing Cells
2.8. Cell Migration Assay with SCF Stimulation
2.9. Immunostaining
2.10. Membrane Protein Extraction
2.11. Halo-Tag Pull-Down Assay
2.12. Structure Prediction of KIT F436S by Modeling
2.13. Statistical Analysis
3. Results
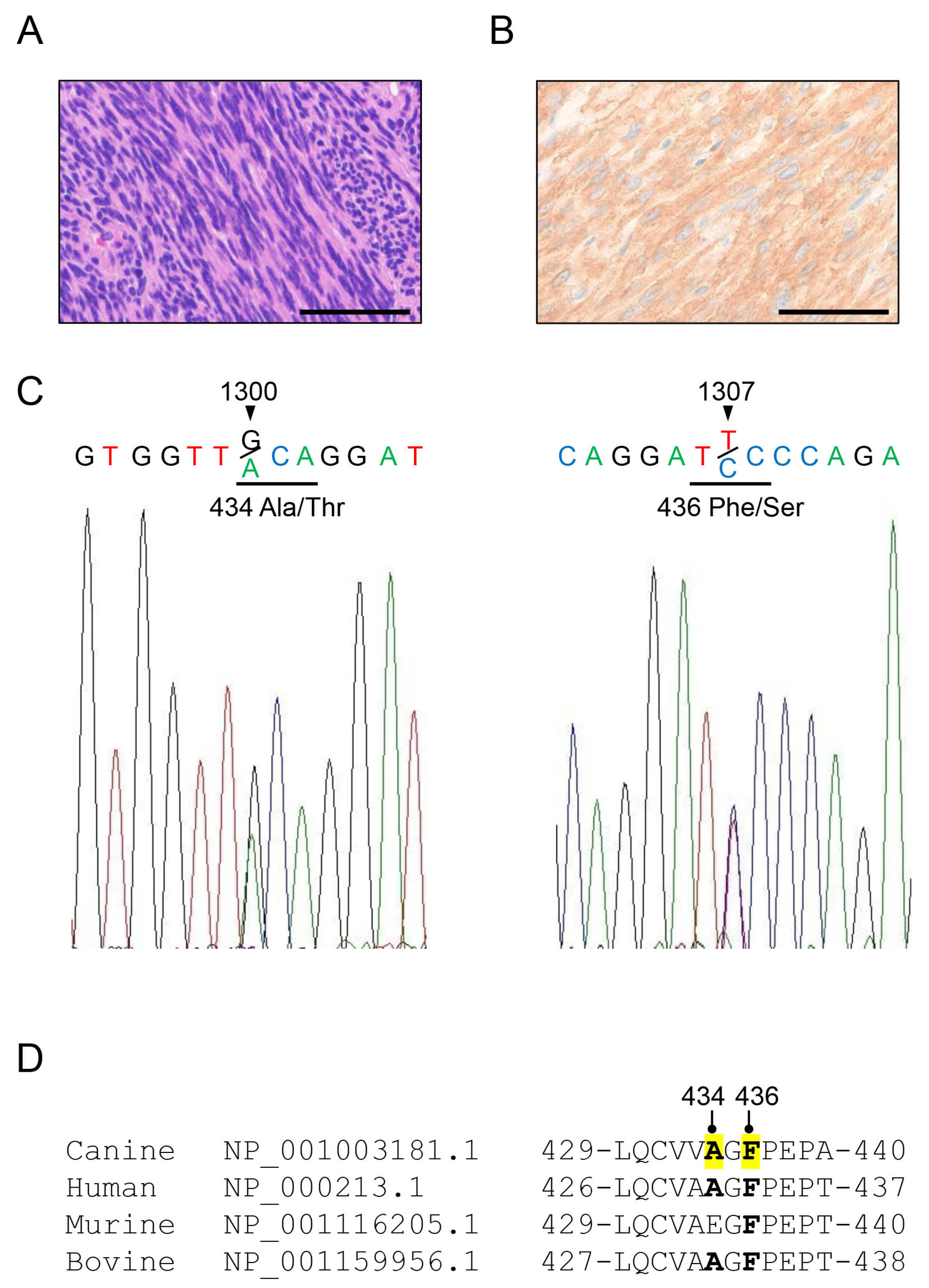
3.1. Novel c-kit Mutations Have Also Been Detected in Canine GIST Tissues
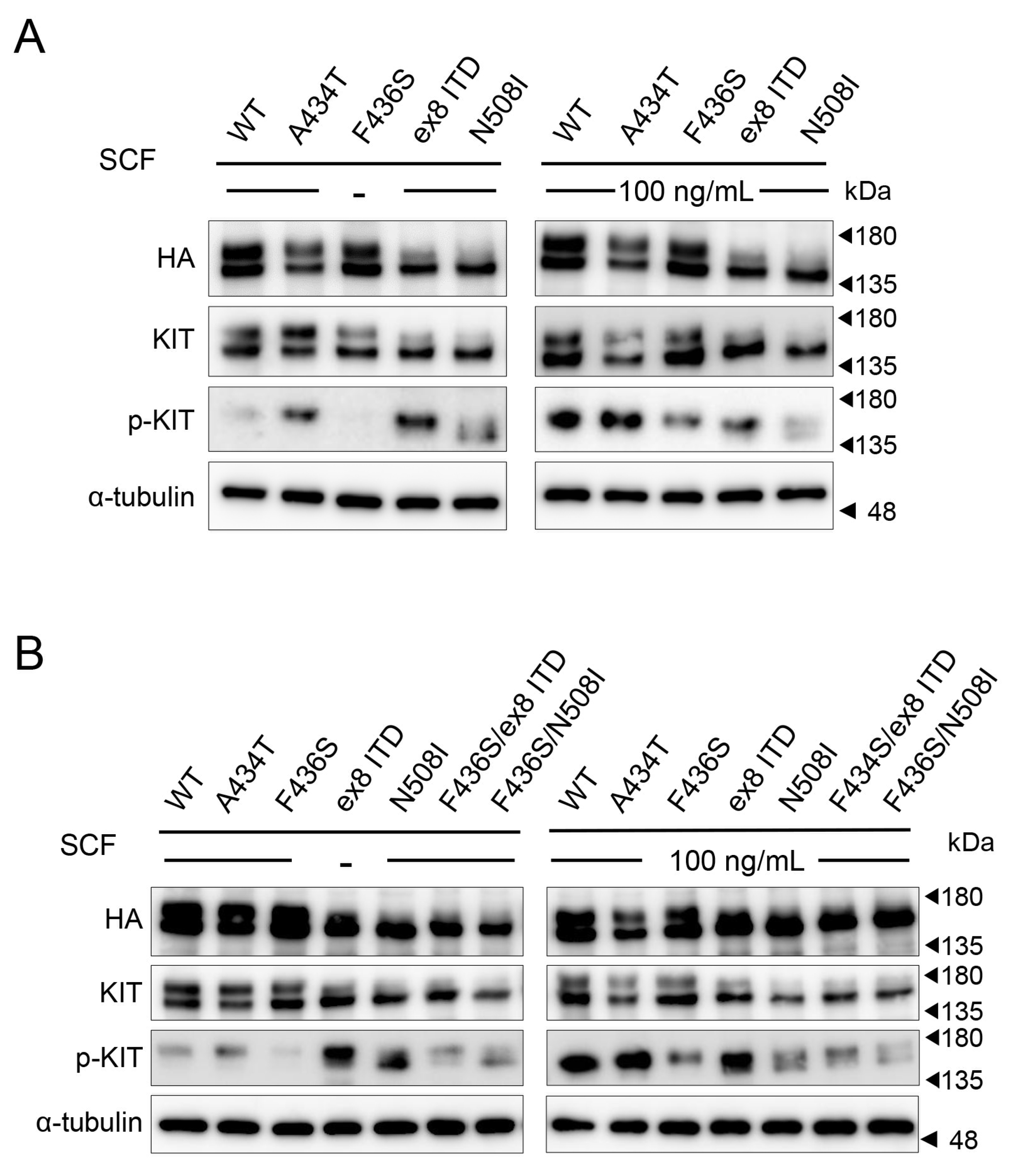
3.2. The Canine KIT Mutant of F433S Lacks Autophosphorylation Ability upon SCF Addition
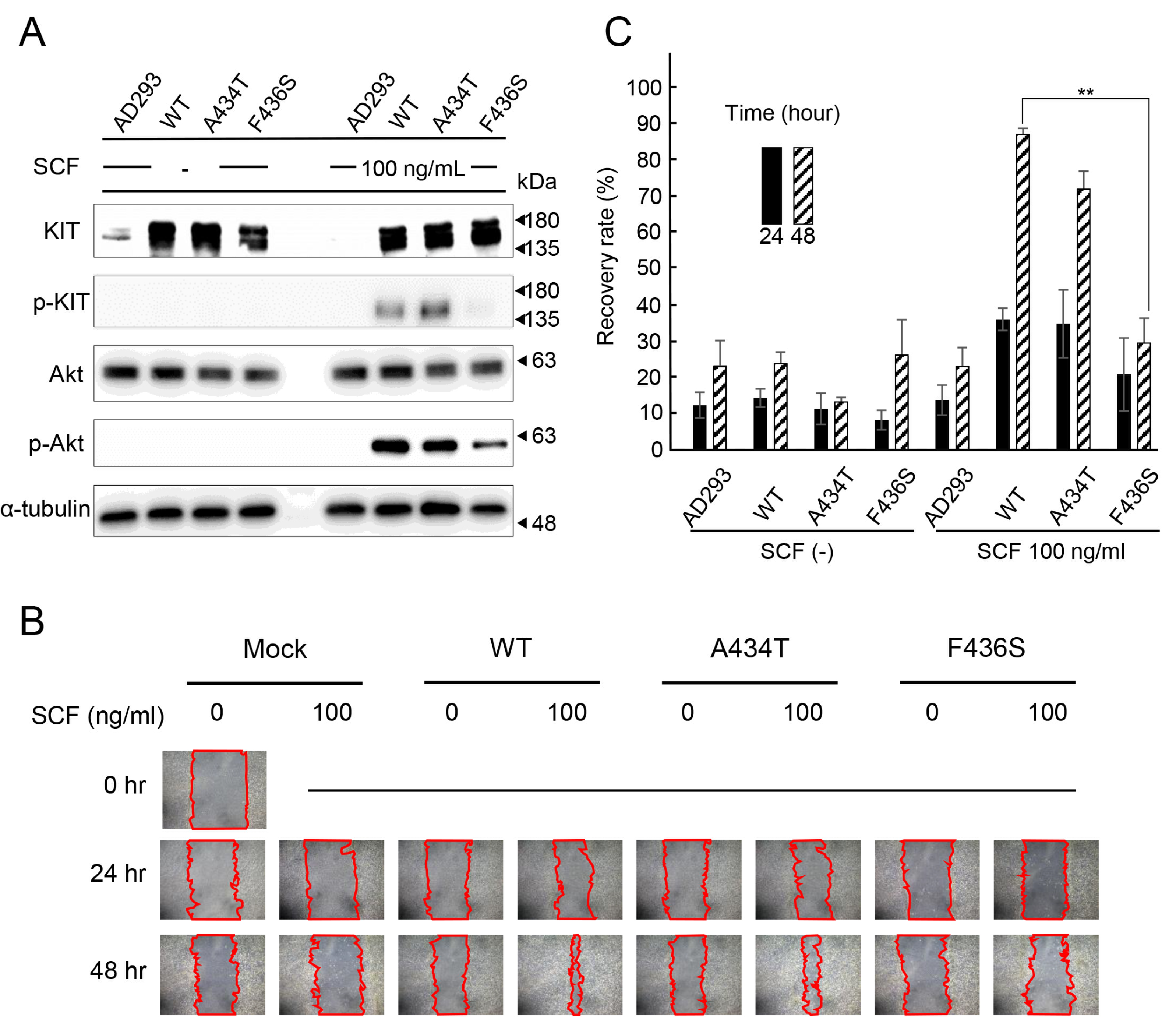
3.3. Suppression of Cell Proliferation and Migration by KIT F436S Mutant
3.4. The F436S Mutation Did Not Affect Protein Maturation or Subcellular Localization of the Canine KIT
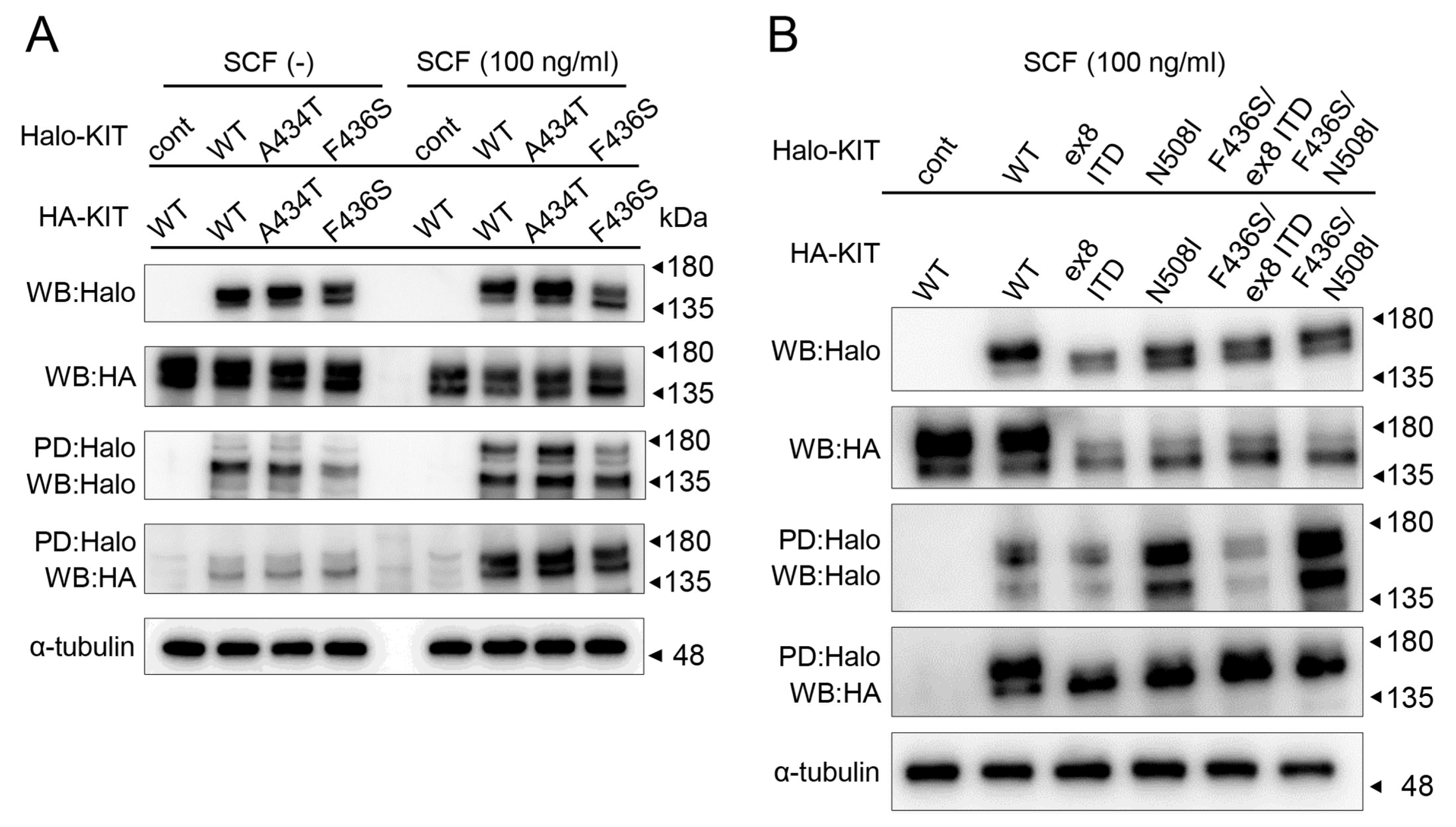
3.5. Loss of F433S Phosphorylation of KIT Did Not Affect Its Ability to Form Dimers
3.6. Effects of F436S Mutation on the Conformation of Canine KIT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hirota, S.; Isozaki, K.; Moriyama, Y.; Hashimoto, K.; Nishida, T.; Ishiguro, S.; Kawano, K.; Hanada, M.; Kurata, A.; Takeda, M.; et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science 1998, 279, 577–580. [Google Scholar] [CrossRef] [PubMed]
- Morini, M.; Gentilini, F.; Turba, M.E.; Gobbo, F.; Mandrioli, L.; Bettini, G. Mutational Analysis of c-kit and PDGFRA in Canine Gastrointestinal Stromal Tumors (GISTs). Vet. Sci. 2022, 9, 376. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y.; Ullrich, A. Growth factor receptor tyrosine kinases. Annu. Rev. Biochem. 1988, 57, 443–478. [Google Scholar] [CrossRef]
- Lennartsson, J.; Ronnstrand, L. Stem cell factor receptor/c-kit: From basic science to clinical implications. Physiol. Rev. 2012, 92, 1619–1649. [Google Scholar] [CrossRef]
- Tsai, M.; Valent, P.; Galli, S.J. KIT as a master regulator of the mast cell lineage. J. Allergy Clin. Immunol. 2022, 149, 1845–1854. [Google Scholar] [CrossRef]
- Sheikh, E.; Tran, T.; Vranic, S.; Levy, A.; Bonfil, R.D. Role and significance of c-kit receptor tyrosine kinase in cancer: A review. Bosn. J. Basic Med. Sci. 2022, 22, 683–698. [Google Scholar] [CrossRef]
- Downing, S.; Chien, M.B.; Kass, P.H.; Moore, P.E.; London, C.A. Prevalence and importance of internal tandem duplications in exons 11 and 12 of c-kit in mast cell tumors of dogs. Am. J. Vet. Res. 2002, 63, 1718–1723. [Google Scholar] [CrossRef]
- London, C.A.; Galli, S.J.; Yuuki, T.; Hu, Z.Q.; Helfand, S.C.; Geissler, E.N. Spontaneous canine mast cell tumors express tandem duplications in the proto-oncogene c-kit. Exp. Hematol. 1999, 27, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, V.; George, S.; Cote, G.M. Molecular Advances in the Treatment of Advanced Gastrointestinal Stromal Tumor. Oncologist 2023, 28, 671–681. [Google Scholar] [CrossRef]
- Kobayashi, M.; Sugisaki, O.; Ishii, N.; Yamada, O.; Ito, K.; Kuroki, S.; Sasaki, Y.; Ono, K.; Washizu, T.; Bonkobara, M. Canine intestinal mast cell tumor with c-kit exon 8 mutation responsive to imatinib therapy. Vet. J. 2012, 193, 264–267. [Google Scholar] [CrossRef]
- Heinrich, M.C.; Maki, R.G.; Corless, C.L.; Antonescu, C.R.; Harlow, A.; Griffith, D.; Town, A.; McKinley, A.; Ou, W.B.; Fletcher, J.A.; et al. Primary and secondary kinase genotypes correlate with the biological and clinical activity of sunitinib in imatinib-resistant gastrointestinal stromal tumor. J. Clin. Oncol. 2008, 26, 5352–5359. [Google Scholar] [CrossRef] [PubMed]
- Zemke, D.; Yamini, B.; Yuzbasiyan-Gurkan, V. Characterization of an undifferentiated malignancy as a mast cell tumor using mutation analysis in the proto-oncogene c-kit. J. Vet. Diagn. Invest. 2001, 13, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Kobie, K.; Kawabata, M.; Hioki, K.; Tanaka, A.; Matsuda, H.; Mori, T.; Maruo, K. The tyrosine kinase inhibitor imatinib [STI571] induces regression of xenografted canine mast cell tumors in SCID mice. Res. Vet. Sci. 2007, 82, 239–241. [Google Scholar] [CrossRef] [PubMed]
- Isotani, M.; Ishida, N.; Tominaga, M.; Tamura, K.; Yagihara, H.; Ochi, S.; Kato, R.; Kobayashi, T.; Fujita, M.; Fujino, Y.; et al. Effect of tyrosine kinase inhibition by imatinib mesylate on mast cell tumors in dogs. J. Vet. Intern. Med. 2008, 22, 985–988. [Google Scholar] [CrossRef]
- Kobayashi, M.; Kuroki, S.; Tanaka, Y.; Moriya, Y.; Kozutumi, Y.; Uehara, Y.; Ono, K.; Tamura, K.; Washizu, T.; Bonkobara, M. Molecular changes associated with the development of resistance to imatinib in an imatinib-sensitive canine neoplastic mast cell line carrying a KIT c.1523A>T mutation. Eur. J. Haematol. 2015, 95, 524–531. [Google Scholar] [CrossRef]
- Kobayashi, M.; Kuroki, S.; Kurita, S.; Miyamoto, R.; Tani, H.; Tamura, K.; Bonkobara, M. A decrease in ubiquitination and resulting prolonged life-span of KIT underlies the KIT overexpression-mediated imatinib resistance of KIT mutation-driven canine mast cell tumor cells. Oncol. Rep. 2017, 38, 2543–2550. [Google Scholar] [CrossRef][Green Version]
- Frost, D.; Lasota, J.; Miettinen, M. Gastrointestinal stromal tumors and leiomyomas in the dog: A histopathologic, immunohistochemical, and molecular genetic study of 50 cases. Vet. Pathol. 2003, 40, 42–54. [Google Scholar] [CrossRef]
- Gregory-Bryson, E.; Bartlett, E.; Kiupel, M.; Hayes, S.; Yuzbasiyan-Gurkan, V. Canine and human gastrointestinal stromal tumors display similar mutations in c-kit exon 11. BMC Cancer 2010, 10, 559. [Google Scholar] [CrossRef]
- Hayes, S.; Yuzbasiyan-Gurkan, V.; Gregory-Bryson, E.; Kiupel, M. Classification of canine nonangiogenic, nonlymphogenic, gastrointestinal sarcomas based on microscopic, immunohistochemical, and molecular characteristics. Vet. Pathol. 2013, 50, 779–788. [Google Scholar] [CrossRef]
- Takanosu, M.; Amano, S.; Kagawa, Y. Analysis of c-kit exon 11 mutations in canine gastrointestinal stromal tumours. Vet. J. 2016, 207, 118–123. [Google Scholar] [CrossRef]
- Irie, M.; Takeuchi, Y.; Ohtake, Y.; Suzuki, H.; Nagata, N.; Miyoshi, T.; Kagawa, Y.; Yamagami, T. Imatinib mesylate treatment in a dog with gastrointestinal stromal tumors with a c-kit mutation. J. Vet. Med. Sci. 2015, 77, 1535–1539. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Elliott, J.W.; Swinbourne, F.; Parry, A.; Baines, L. Successful treatment of a metastatic, gastrointestinal stromal tumour in a dog with toceranib phosphate (Palladia). J. Small Anim. Pract. 2017, 58, 416–418. [Google Scholar] [CrossRef] [PubMed]
- Berger, E.P.; Johannes, C.M.; Jergens, A.E.; Allenspach, K.; Powers, B.E.; Du, Y.; Mochel, J.P.; Fox, L.E.; Musser, M.L. Retrospective evaluation of toceranib phosphate (Palladia(R)) use in the treatment of gastrointestinal stromal tumors of dogs. J. Vet. Intern. Med. 2018, 32, 2045–2053. [Google Scholar] [CrossRef] [PubMed]
- London, C.A.; Malpas, P.B.; Wood-Follis, S.L.; Boucher, J.F.; Rusk, A.W.; Rosenberg, M.P.; Henry, C.J.; Mitchener, K.L.; Klein, M.K.; Hintermeister, J.G.; et al. Multi-center, placebo-controlled, double-blind, randomized study of oral toceranib phosphate (SU11654), a receptor tyrosine kinase inhibitor, for the treatment of dogs with recurrent (either local or distant) mast cell tumor following surgical excision. Clin. Cancer Res. 2009, 15, 3856–3865. [Google Scholar] [CrossRef]
- Rassner, M.; Waldeck, S.; Follo, M.; Jilg, S.; Philipp, U.; Jolic, M.; Wehrle, J.; Jost, P.J.; Peschel, C.; Illert, A.L.; et al. Development of Highly Sensitive Digital Droplet PCR for Detection of cKIT Mutations in Circulating Free DNA That Mediate Resistance to TKI Treatment for Gastrointestinal Stromal Tumor (GIST). Int. J. Mol. Sci. 2023, 24, 5411. [Google Scholar] [CrossRef]
- Orfao, A.; Garcia-Montero, A.C.; Sanchez, L.; Escribano, L. Recent advances in the understanding of mastocytosis: The role of KIT mutations. Br. J. Haematol. 2007, 138, 12–30. [Google Scholar] [CrossRef]
- Krimmer, S.G.; Bertoletti, N.; Suzuki, Y.; Katic, L.; Mohanty, J.; Shu, S.; Lee, S.; Lax, I.; Mi, W.; Schlessinger, J. Cryo-EM analyses of KIT and oncogenic mutants reveal structural oncogenic plasticity and a target for therapeutic intervention. Proc. Natl. Acad. Sci. USA 2023, 120, e2300054120. [Google Scholar] [CrossRef]
- Amagai, Y.; Matsuda, A.; Jung, K.; Oida, K.; Jang, H.; Ishizaka, S.; Matsuda, H.; Tanaka, A. A point mutation in the extracellular domain of KIT promotes tumorigenesis of mast cells via ligand-independent auto-dimerization. Sci. Rep. 2015, 5, 9775. [Google Scholar] [CrossRef]
- Munday, J.S.; Löhr, C.V.; Kiupel, M. Tumors in Domestic Animals, 5th ed.; Wiley: Hoboken, NJ, USA, 2017; pp. 499–601. ISBN 978-1-11918-120-0. [Google Scholar]
- Yamada, O.; Kobayashi, M.; Sugisaki, O.; Ishii, N.; Ito, K.; Kuroki, S.; Sasaki, Y.; Isotani, M.; Ono, K.; Washizu, T.; et al. Imatinib elicited a favorable response in a dog with a mast cell tumor carrying a c-kit c.1523A>T mutation via suppression of constitutive KIT activation. Vet. Immunol. Immunopathol. 2011, 142, 101–106. [Google Scholar] [CrossRef]
- Maeda, M.; Ochiai, K.; Michishita, M.; Morimatsu, M.; Sakai, H.; Kinoshita, N.; Sakaue, M.; Onozawa, E.; Azakami, D.; Yamamoto, M.; et al. In vitro anticancer effects of alpelisib against PIK3CA-mutated canine hemangiosarcoma cell lines. Oncol. Rep. 2022, 47, 84. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef] [PubMed]
- Ning, Z.Q.; Li, J.; McGuinness, M.; Arceci, R.J. STAT3 activation is required for Asp(816) mutant c-kit induced tumorigenicity. Oncogene 2001, 20, 4528–4536. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Isotani, M.; Tamura, K.; Yagihara, H.; Hikosaka, M.; Ono, K.; Washizu, T.; Bonkobara, M. Identification of a c-kit exon 8 internal tandem duplication in a feline mast cell tumor case and its favorable response to the tyrosine kinase inhibitor imatinib mesylate. Vet. Immunol. Immunopathol. 2006, 114, 168–172. [Google Scholar] [CrossRef]
- Yang, Z.; Lasker, K.; Schneidman-Duhovny, D.; Webb, B.; Huang, C.C.; Pettersen, E.F.; Goddard, T.D.; Meng, E.C.; Sali, A.; Ferrin, T.E. UCSF Chimera, MODELLER, and IMP: An integrated modeling system. J. Struct. Biol. 2012, 179, 269–278. [Google Scholar] [CrossRef]
- Shen, H.; Nie, J.; Li, G.; Tian, H.; Zhang, J.; Luo, X.; Xu, D.; Sun, J.; Zhang, D.; Zhang, H.; et al. Stem cell factor restrains endoplasmic reticulum stress-associated apoptosis through c-kit receptor activation of JAK2/STAT3 axis in hippocampal neuronal cells. PLoS ONE 2024, 19, e0310872. [Google Scholar] [CrossRef]
- Hasegawa, K.; Zhao, Y.; Garbuzov, A.; Corces, M.R.; Neuhofer, P.; Gillespie, V.M.; Cheung, P.; Belk, J.A.; Huang, Y.H.; Wei, Y.; et al. Clonal inactivation of TERT impairs stem cell competition. Nature 2024, 632, 201–208. [Google Scholar] [CrossRef]
- Chen, X.; Meng, X.; Zhang, H.; Feng, C.; Wang, B.; Li, N.; Abdullahi, K.M.; Wu, X.; Yang, J.; Li, Z.; et al. Intestinal proinflammatory macrophages induce a phenotypic switch in interstitial cells of Cajal. J. Clin. Investig. 2020, 130, 6443–6456. [Google Scholar] [CrossRef]
- Zhou, S.; Abdihamid, O.; Tan, F.; Zhou, H.; Liu, H.; Li, Z.; Xiao, S.; Li, B. KIT mutations and expression: Current knowledge and new insights for overcoming IM resistance in GIST. Cell Commun. Signal. 2024, 22, 153. [Google Scholar] [CrossRef]
- Tabone-Eglinger, S.; Subra, F.; El Sayadi, H.; Alberti, L.; Tabone, E.; Michot, J.P.; Theou-Anton, N.; Lemoine, A.; Blay, J.Y.; Emile, J.F. KIT mutations induce intracellular retention and activation of an immature form of the KIT protein in gastrointestinal stromal tumors. Clin. Cancer Res. 2008, 14, 2285–2294. [Google Scholar] [CrossRef]
- Yuzawa, S.; Opatowsky, Y.; Zhang, Z.; Mandiyan, V.; Lax, I.; Schlessinger, J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell 2007, 130, 323–334. [Google Scholar] [CrossRef]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]


| Purpose | Base Sequence | |
|---|---|---|
| Forward | Reverse | |
| exon 8 | 5′-gtcctcttcaaactcaagaagg-3′ | 5′-gtagccaaaataatcctctc-3′ |
| exon 9 | 5′-gatggaatggacttaaaatcatg-3′ | 5′-gatggaatggacttaaaatcatg-3′ |
| exon 11del | 5′-catttgttctctaccctaagtgct-3′ | 5′-gtttccattgatctcctcaac-3′ |
| exon 11ins | 5′-cccatgtatgaagtacagtggaag-3′ | 5′-gttccctaaagtcattgttacacg-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shimakawa, K.; Doge, S.; Michishita, M.; Tanabe, E.; Tajima, T.; Kobayashi, M.; Bonkobara, M.; Watanabe, M.; Ochiai, K.; Tanaka, Y. A Canine c-kit Novel Mutation Isolated from a Gastrointestinal Stromal Tumor (GIST) Retains the Ability to Form Dimers but Lacks Autophosphorylation. Animals 2025, 15, 1444. https://doi.org/10.3390/ani15101444
Shimakawa K, Doge S, Michishita M, Tanabe E, Tajima T, Kobayashi M, Bonkobara M, Watanabe M, Ochiai K, Tanaka Y. A Canine c-kit Novel Mutation Isolated from a Gastrointestinal Stromal Tumor (GIST) Retains the Ability to Form Dimers but Lacks Autophosphorylation. Animals. 2025; 15(10):1444. https://doi.org/10.3390/ani15101444
Chicago/Turabian StyleShimakawa, Kei, So Doge, Masaki Michishita, Eri Tanabe, Tsuyoshi Tajima, Masato Kobayashi, Makoto Bonkobara, Masami Watanabe, Kazuhiko Ochiai, and Yoshikazu Tanaka. 2025. "A Canine c-kit Novel Mutation Isolated from a Gastrointestinal Stromal Tumor (GIST) Retains the Ability to Form Dimers but Lacks Autophosphorylation" Animals 15, no. 10: 1444. https://doi.org/10.3390/ani15101444
APA StyleShimakawa, K., Doge, S., Michishita, M., Tanabe, E., Tajima, T., Kobayashi, M., Bonkobara, M., Watanabe, M., Ochiai, K., & Tanaka, Y. (2025). A Canine c-kit Novel Mutation Isolated from a Gastrointestinal Stromal Tumor (GIST) Retains the Ability to Form Dimers but Lacks Autophosphorylation. Animals, 15(10), 1444. https://doi.org/10.3390/ani15101444





