Comparative Analysis of Genetic Structure and Diversity in Larimichthys polyactis, Larimichthys crocea, and Their Reciprocal Hybrids Based on Microsatellite Loci
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Microsatellite Loci Screening and Analysis
2.3. Data Processing and Analysis
3. Results
3.1. Genetic Diversity
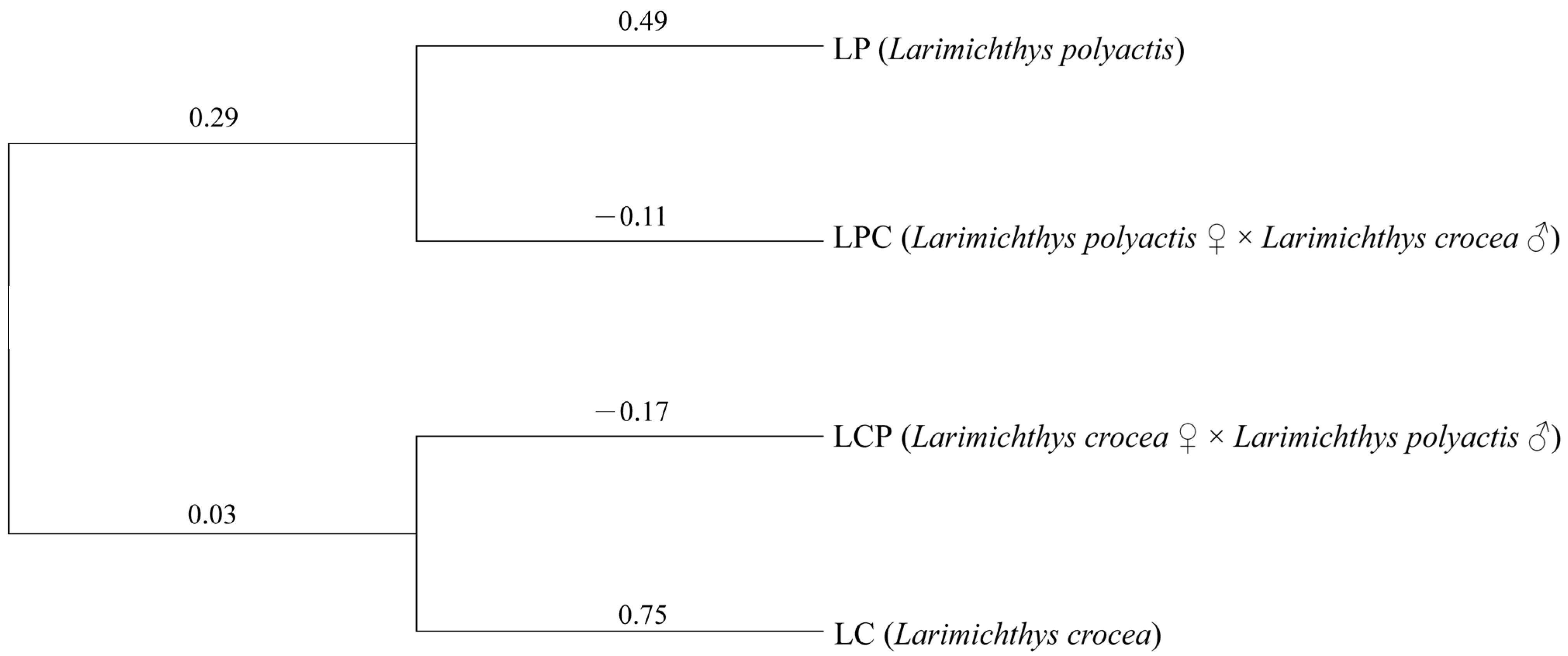
3.2. Genetic Differentiation
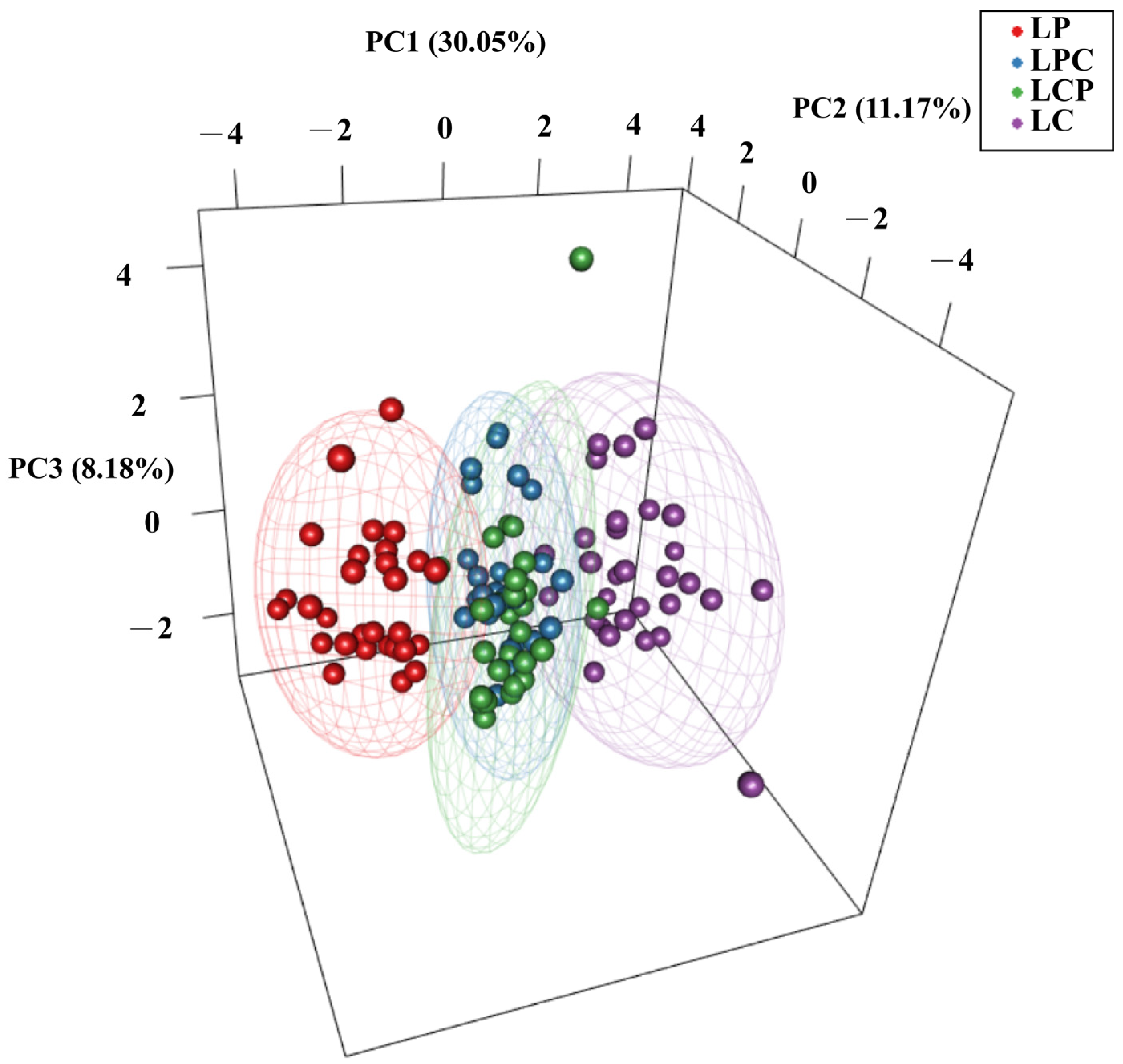
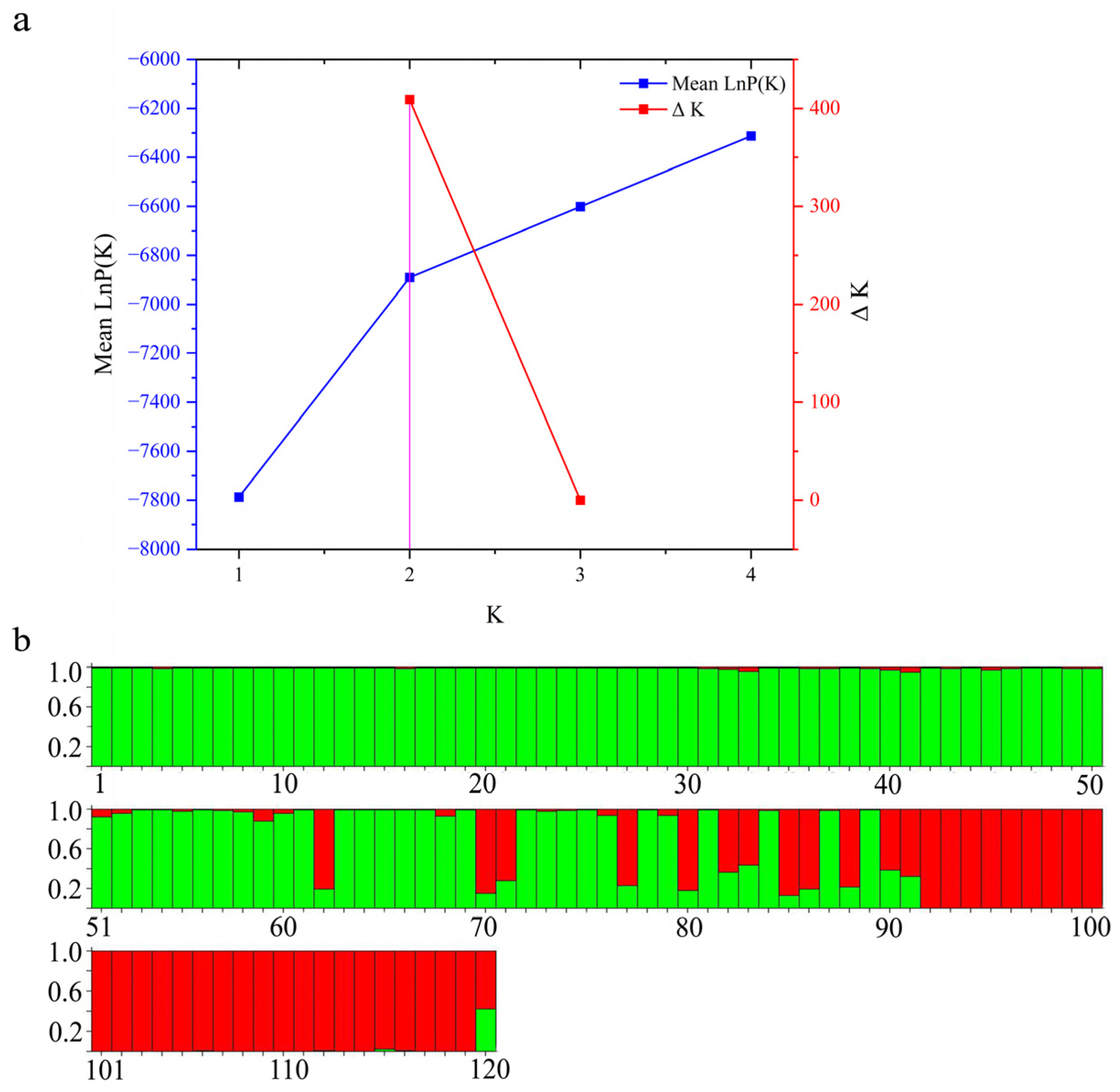
3.3. Population Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lou, B.; Zhan, W.; Chen, Y.R.; Liu, F.; Wang, L.G.; Xu, D.D.; Mao, G.M. Studies on techniques of the artificial breeding of Larimichthys polyactis. J. Zhejiang Ocean Univ. 2016, 35, 361–365. [Google Scholar]
- Liu, D.J.; Feng, Z.J.; Zheng, Z.Y.; Weng, Z.C.; Shu, Y.Z. Studies on the artificial propagation of thelarge yellow croake, Pseudosciaena crocea (Richardson). J. Fujian Norm. Univ. 1991, 7, 71–79. [Google Scholar]
- Cheng, Q.Q.; Chen, W.M.; Ma, L. Genetic diversity and population structure of small yellow croaker (Larimichthys polyactis) in the Yellow and East China seas based on microsatellites. Aquat. Living Resour. 2019, 32, 16. [Google Scholar] [CrossRef]
- Xu, Z.; Dou, S.Z.; Ding, S.X.; Liu, J.X. Temporal genetic stability despite decades of overexploitation for large yellow croaker in the East China sea. Front. Mar. Sci. 2022, 9, 861840. [Google Scholar] [CrossRef]
- Guo, D.D.; Liu, F.; Niu, B.L.; Lou, B. Genetic diversity of wild and cultured populations of little yellow croaker (Larimichthys polyactis) based on mitochondrial Cyt b gene and D-loop region. Acta Agric. Zhejiangensis 2022, 34, 1856–1865. [Google Scholar]
- Lei, F.L.; Chen, M.F.; Meng, Y.X.; Niu, S.F.; Wu, R.X.; Pan, Y. Analysis of COⅠ sequence variation in the wild and cultured populations of Larimichthys crocea. Guangxi Sci. 2023, 30, 794–803. [Google Scholar]
- Liu, Q.; Tang, B.J.; Liu, K.; Wang, L. Research progress on development and utilization of Larimichthys crocea germplasm resources based on CNKI database. Fish. Inf. Strat. 2022, 37, 54–60. [Google Scholar]
- Zhao, H.; Zou, H.F.; Lu, W.Q. Ploidy polymorphism and morphological variation among reciprocal hybrids of Pseudosciaena crocea (♀) × Miichthys miiuy (♂). Aquacult. Res. 2016, 47, 3390–3398. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Ralls, K.; Eldridge, M.; Dudash, M.R.; Fenster, C.B.; Lacy, R.C.; Sunnucks, P. Genetic Management of Fragmented Animal and Plant Populations; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Hughes, A.R.; Inouye, B.D.; Johnson, M.T.; Underwood, N.; Vellend, M. Ecological consequences of genetic diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef]
- Mallet, J. Hybrid speciation. Nature 2007, 446, 279–283. [Google Scholar] [CrossRef]
- Wang, S.; Tang, C.C.; Tao, M.; Qin, Q.B.; Zhang, C.; Luo, K.K.; Zhao, R.R.; Wang, J.; Ren, L.; Xiao, J.; et al. Establishment and application of distant hybridization technology in fish. Sci. China Life Sci. 2019, 62, 22–45. [Google Scholar] [CrossRef] [PubMed]
- Li, S.N.; Xie, L.H.; Xiao, J.; Yuan, L.J.; Zhou, T.; Luo, K.K.; Zhang, C.; Zhao, R.R.; Tao, M.; Liu, S.J. Diploid hybrid fish derived from the cross between female Bleeker’s yellow tail and male topmouth culter, two cyprinid fishes belonging to different subfamilies. BMC Genet. 2019, 20, 80. [Google Scholar] [CrossRef]
- Liu, Q.Z.; Wang, S.; Tang, C.C.; Tao, M.; Zhang, C.; Zhou, Y.; Qin, Q.B.; Luo, K.K.; Wu, C.; Hu, F.Z. The research advances in distant hybridization and gynogenesis in fish. Rev. Aquacult. 2025, 17, e12972. [Google Scholar] [CrossRef]
- Yang, C.H.; Dai, C.H.; Liu, Q.; Zhu, Y.T.; Huang, X.X.; Xu, X.W.; Zhou, Y.; Wang, S.; Liu, Q.F.; Liu, S.J. Different ploidy-level hybrids derived from female common carp × male topmouth culter. Aquaculture 2025, 594, 741366. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Chen, J.; Li, L.; Tao, M.; Zhang, C.; Qin, Q.B.; Xiao, J.; Liu, Y.; Liu, S.J. Research advances in animal distant hybridization. Sci. China Life Sci. 2014, 57, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Sarker, B.S.; Ali, A.; Rahman, S.S.; Alam, M.S.; Islam, M.S. Monogamous hybridization of Nile tilapia (Oreochromis niloticus) with Mozambique tilapia (O. mossambicus) results in unprecedented all-female F1 hybrid. Aquacult. Fish. 2024, 9, 871–1018. [Google Scholar] [CrossRef]
- Fan, J.J. Investigation on the Biological Characteristics of the Hybrid of Megalobrama amblycephala (♀) × Erythroculter mongolicus (♂). Master’s Thesis, Hunan Normal University, Changsha, China, 2020. [Google Scholar]
- Tao, Y.F.; Qiang, J.; Dagoudo, M.; Zhu, H.J.; Bao, J.W.; Ma, J.L.; Li, M.X.; Xu, P. Transcriptome profiling reveals differential expression of immune-related genes in gills of hybrid yellow catfish (Tachysurus fulvidraco ♀ × Pseudobagrus vachellii ♂) under hypoxic stress: Potential NLR-mediated immune response. Fish Shellfish Immun. 2021, 119, 409–419. [Google Scholar] [CrossRef]
- Guo, D.D.; Liu, F.; Niu, B.L.; Zhan, W.; Xie, Q.P.; Zhang, Y.; Lou, B. Establishment of diploid hybrid strains derived from female Larimichthys crocea × male Larimichthys polyactis and transmission of parental mtDNA in hybrid progenies. Aquaculture 2022, 561, 738693. [Google Scholar] [CrossRef]
- Liu, F.; Gao, S.B.; Liu, Y.Y.; Chu, T.Q.; Zhan, W.; Lou, B. Morphological characteristics comparison of Larimichthys polyactis, L. crocea and their hybrids (L. polyactis ♀ × L. crocea ♂). J. Shanghai Ocean Univ. 2020, 29, 189–198. [Google Scholar] [CrossRef]
- Liu, F.; Gao, S.B.; Zhan, W.; Chu, T.Q.; Lou, B. An analysis of nutritive composition of Larimichthys polyactis ♀ × Larimichthys crocea ♂ hybridmuscle and their parents. Period. Ocean Univ. China 2020, 50, 34–42. [Google Scholar] [CrossRef]
- Hartl, D.L.; Clark, A.G. Principles of Population Genetics; Sinauer Assoc Inc.: Sunderland, MA, USA, 1989. [Google Scholar]
- Bagshaw, A.T. Functional mechanisms of microsatellite DNA in eukaryotic genomes. Genome Biol. Evol. 2017, 9, 2428–2443. [Google Scholar] [CrossRef] [PubMed]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; de Freitas Munhoz, C. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Merritt, B.J.; Culley, T.M.; Avanesyan, A.; Stokes, R.; Brzyski, J. An empirical review: Characteristics of plant microsatellite markers that confer higher levels of genetic variation. Appl. Plant Sci. 2015, 3, 1500025. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.C.; Si, Z.G.; Li, G.L. Microsatellites and its application to the genetic diversity in fish. Biotechnology 2007, 17, 83–85. [Google Scholar] [CrossRef]
- Srivastava, S.; Mukherjee, S.; Pragya, P.; Burman, S.; Rana, M.; Kumar, R.; Katyayani, K.K.S.; Karnwal, A.; Kumar, S.; Shukla, M. Microsatellite markers for crop improvement: A review. J. Appl. Nat. Sci. 2023, 15, 1018–1035. [Google Scholar] [CrossRef]
- Wang, Y.J.; Sha, H.; Li, X.H.; Zhou, T.; Luo, X.Z.; Zou, G.W.; Chai, Y.; Liang, H.W. Microsatellite characteristics of silver carp (Hypophthalmichthys molitrix) genome and genetic diversity analysis in four cultured populations. Genes 2022, 13, 1267. [Google Scholar] [CrossRef]
- Li, Y.P.; Xu, F.; Wang, Y.M.; Lv, Y.Y.; Shi, J.R.; Xie, B.W.; Cai, W.Y.; Liu, D. Genetic diversity evaluation of two loach fishes and their artificial hybrid population based on 19 polymorphic microsatellite loci. Pak. J. Zool. 2023, 55, 1665–1675. [Google Scholar] [CrossRef]
- Wang, T.; Fang, M.Y.; Yang, Y.; Song, L.L.; Cai, C.Y.; Meng, Z.N.; Liu, X.C. Comparison of phenotypes and genetic characteristics between hybrids and parental generation of Epinephelus Fuscoguttatus (♀) × Epinephelus Polyphekadion (♂). Acta Hydrobiol. Sin. 2024, 48, 444–453. [Google Scholar] [CrossRef]
- Han, Z. Studies of Morphological Characteristics and Genetic Diversity on Populations of Small Yellow Croaker (Larimichthys polyactis) and the Difference Between Small Yellow Croaker and Large Yellow Croaker (Larimichthys crocea). Master’s Thesis, Ocean University of China, Qingdao, China, 2012. [Google Scholar]
- Lin, N.F.; Su, Y.Q.; Ding, S.X.; Wang, J. Cross-species amplification of microsatellite markers from Pseudosciaena crocea in Sciaenidae. J. Fish. Sci. China 2008, 15, 237–243. [Google Scholar]
- Liu, B.J. Population Genetic Structure and Local Adaptation of the Small Yellow Croaker (Larimichthys polyactis) and Japanese Eel (Anguilla japonica). Ph.D. Thesis, University of Chinese Academy of Sciences, Beijing, China, 2017. [Google Scholar]
- Wu, L.N.; Zhang, N.Y.; Sun, S.; Yuan, J.G.; Chen, J.; Li, M.M.; Lin, N.; You, Y.; Wang, W.J.; Ding, S.X. Application of microsatellite markers for evaluating the effect of restocking enhancement in Larimichthys crocea. J. Fish. Sci. China 2021, 28, 1100–1108. [Google Scholar] [CrossRef]
- Xie, F.A.; Zhao, R.P.; He, Q.; Mao, J.L.; Wang, Y.F.; Jiang, L.H.; Wansuk, S.; Chen, Y.J. Development and validation of microsatellite markers derived from the genome DNA sequence of Larimichthys crocea. J. Zhejiang Ocean Univ. 2020, 39, 394–400. [Google Scholar]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Rousset, F. Genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Li, X. Introduction to a population genetic software-GENALEX 6. Chin. Wild Plant Resour. 2008, 27, 59–62. [Google Scholar]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Zhang, Y.R.; Li, B.Y.; Zheng, G.D.; Zou, S.M. Microsatellite genetic structure analysis of two successive generations of gynogenetic populations of Megalobrama amblycephala “Pujiang No.2”. J. Fish. Sci. China 2022, 29, 643–652. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Liu, J.X. StructureSelector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Wu, Y.P.; Tian, Y.S.; Wang, L.N.; Li, Z.T.; Zhang, J.J.; Li, L.L.; Li, Z.Q.; Chen, S.; Ma, W.H.; Wang, Q.B.; et al. Genetic diversity analysis of Epinephelus fuscoguttatus (♀) and E. tukula (♂) hybrids. Prog. Fish. Sci. 2021, 42, 25–32. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. Evolution and the Genetics of Populations, Volume 4: Variability Within and Among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1984. [Google Scholar]
- Xie, Z.G. The cross breeding and genetic analysis of hybrids of Pseduosciaena crocea ♀ and Miichthys miiuy ♂. Master’s Thesis, Hunan Agricultural University, Changsha, China, 2006. [Google Scholar]
- Takezaki, N.; Nei, M. Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 1996, 144, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Ruane, J. A critical review of the value of genetic distance studies in conservation of animal genetic resources. J. Anim. Breed. Genet. 2001, 116, 317–323. [Google Scholar] [CrossRef]
- Chan, K.M.; Levin, S.A. Leaky prezygotic isolation and porous genomes: Rapid introgression of maternally inherited DNA. Evolution 2005, 59, 720–729. [Google Scholar] [CrossRef]
- Tiffin, P.; Olson, M.S.; Moyle, L.C. Asymmetrical crossing barriers in angiosperms. Proc. R. Soc. Lond. B 2001, 268, 861–867. [Google Scholar] [CrossRef]
- Hamilton, J.A.; Miller, J.M. Adaptive introgression as a resource for management and genetic conservation in a changing climate. Conserv. Biol. 2015, 30, 33–41. [Google Scholar] [CrossRef]
| Locus | Reference | Repeat Motif | Primer Sequence (5′–3′) | Tm/°C | Allele Size | Fluorescent Labeling | Null Allele Frequency |
|---|---|---|---|---|---|---|---|
| L1 | Xie et al. 2020 [36] | (TGAT)5 | F: TGTAGATCGGATGCCAGTTG R: TTCATGAAACATGCAGAGGG | 55 | 231~271 | FAM | 0.04112 |
| L2 | Xie et al. 2020 [36] | (ATAG)12 | F: GGCAGCGGTGACATTATTCT R: AACTCACCGCAGAAACTGAAA | 56 | 261~352 | HEX | 0.00008 |
| L3 | Xie et al. 2020 [36] | (AGAT)9 | F: CACAGCCCACTGATGATGTC R: ATCCTCCCCCATACAAGTCC | 55 | 274~415 | FAM | −0.01262 |
| L4 | Wu et al. 2021 [35] | (ACAT)9 | F: CACAGCCTTTCTTTGGAATCA R: CACTGTCACTTTTGCTGTATGGA | 56 | 176~252 | HEX | −0.01312 |
| L5 | Xie et al. 2020 [36] | (GACA)5 | F: TTAGGCGATCACCAAAGTCA R: TTCAGTTTTCTGCTGGTTTCTG | 55 | 235~249 | HEX | 0.01281 |
| L6 | Xie et al. 2020 [36] | (CCTG)7 | F: AAACTCACGACCGGAACAAC R: TGTAGCTGAACGCTCATTGG | 56 | 239~263 | HEX | −0.09336 |
| L7 | EF635869 | (TC)9(CTT)6 | F: CATCTCCCCCACTCATATCG R: TTCAGACTGCTGCCCTGTC | 56 | 281~304 | FAM | −0.04372 |
| L8 | Xie et al. 2020 [36] | (TATT)5 | F: CAATTCAAACACCGTCCTGA R: GTTTCCTGTGAATCGCCTGT | 55 | 254~288 | HEX | −0.01393 |
| L9 | EF635876 | (CT)8 | F: CTTTGCTGTGAGGCTTTTCC R: TCGCAGACAGAATCTCCAAG | 57 | 213~264 | HEX | 0.08007 |
| L10 | KF805068 | (AG)11 | F: CTTCAACATTTCCTCCATTT R: GTGTTCAGGACTGCGTATTT | 52 | 152~166 | HEX | 0.00609 |
| L11 | HQ678309 | (AG)6 | F: AGCCTACAGGTGAATGAGTG R: GCTTGGGTCTGAGGTTGC | 55 | 209~256 | HEX | 0.05032 |
| L12 | Xie et al. 2020 [36] | (TGAA)5 | F: ATAGCTGTCTCCATGCCCAC R: AAAATTGACCTCCAGCCAAA | 55 | 215~235 | HEX | 0.12789 |
| L13 | KC773866 | (TG)11 | F: AAAGCCTCCGTCAAGCAC R: CGTATTCAAACCAGCACA | 53 | 175~203 | FAM | −0.03206 |
| L14 | EF635877 | (CTT)6 | F: CCTCCTCACCTGCTAACT R: AACAAACGAAGCCCAACT | 53 | 353~402 | HEX | −0.05425 |
| Locus | K | PIC | ||||||
|---|---|---|---|---|---|---|---|---|
| LP | LPC | LCP | LC | LP | LPC | LCP | LC | |
| L1 | 28 | 21 | 28 | 17 | 0.94 | 0.91 | 0.95 | 0.88 |
| L2 | 40 | 29 | 43 | 23 | 0.97 | 0.95 | 0.97 | 0.86 |
| L3 | 45 | 29 | 40 | 18 | 0.97 | 0.95 | 0.96 | 0.90 |
| L4 | 36 | 33 | 41 | 23 | 0.96 | 0.95 | 0.97 | 0.92 |
| L5 | 13 | 22 | 23 | 18 | 0.82 | 0.94 | 0.94 | 0.89 |
| L6 | 6 | 13 | 21 | 12 | 0.63 | 0.85 | 0.93 | 0.86 |
| L7 | 33 | 21 | 34 | 20 | 0.96 | 0.91 | 0.95 | 0.91 |
| L8 | 34 | 19 | 29 | 12 | 0.96 | 0.92 | 0.93 | 0.84 |
| L9 | 25 | 21 | 35 | 17 | 0.94 | 0.92 | 0.96 | 0.90 |
| L10 | 18 | 15 | 12 | 9 | 0.90 | 0.89 | 0.85 | 0.77 |
| L11 | 28 | 22 | 30 | 13 | 0.95 | 0.93 | 0.95 | 0.85 |
| L12 | 21 | 12 | 23 | 17 | 0.92 | 0.88 | 0.93 | 0.90 |
| L13 | 29 | 27 | 30 | 27 | 0.95 | 0.94 | 0.95 | 0.94 |
| L14 | 28 | 25 | 30 | 23 | 0.94 | 0.94 | 0.96 | 0.90 |
| Total | 384 | 309 | 419 | 249 | - | - | - | - |
| Mean | 27.43 | 22.07 | 29.93 | 17.79 | 0.91 | 0.92 | 0.94 | 0.88 |
| Locus | Na | Ne | I | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LP | LPC | LCP | LC | LP | LPC | LCP | LC | LP | LPC | LCP | LC | |
| L1 | 13 | 8 | 10 | 8 | 4.32 | 2.64 | 5.13 | 3.61 | 1.85 | 1.39 | 1.83 | 1.53 |
| L2 | 26 | 14 | 23 | 12 | 17.82 | 7.35 | 12.59 | 2.77 | 3.05 | 2.24 | 2.81 | 1.61 |
| L3 | 22 | 10. | 17 | 10 | 16.22 | 6.32 | 8.22 | 4.57 | 2.91 | 2.06 | 2.45 | 1.80 |
| L4 | 14 | 13 | 18 | 10 | 7.89 | 7.50 | 10.71 | 3.61 | 2.30 | 2.22 | 2.60 | 1.72 |
| L5 | 7 | 10 | 7 | 6 | 2.25 | 6.32 | 5.23 | 2.85 | 1.16 | 2.09 | 1.77 | 1.26 |
| L6 | 3 | 5 | 7 | 4 | 1.07 | 3.06 | 3.10 | 2.13 | 0.17 | 1.33 | 1.38 | 0.97 |
| L7 | 11 | 10 | 13 | 10 | 7.23 | 4.33 | 5.33 | 4.44 | 2.13 | 1.79 | 2.06 | 1.82 |
| L8 | 11 | 7 | 10 | 5 | 6.57 | 2.82 | 2.80 | 2.52 | 2.11 | 1.36 | 1.51 | 1.13 |
| L9 | 11 | 9 | 12 | 10 | 4.75 | 4.47 | 5.75 | 4.60 | 1.88 | 1.71 | 2.04 | 1.86 |
| L10 | 6 | 4 | 5 | 4 | 2.20 | 2.46 | 2.65 | 1.79 | 1.15 | 1.05 | 1.11 | 0.82 |
| L11 | 9 | 9 | 9 | 4 | 5.47 | 3.79 | 4.63 | 2.03 | 1.88 | 1.61 | 1.79 | 0.85 |
| L12 | 5 | 3 | 7 | 4 | 3.24 | 2.87 | 3.46 | 2.48 | 1.35 | 1.08 | 1.41 | 1.04 |
| L13 | 13 | 10 | 13 | 9 | 8.87 | 6.02 | 6.98 | 4.68 | 2.34 | 1.98 | 2.13 | 1.77 |
| L14 | 11 | 9 | 12 | 8 | 6.34 | 6.04 | 7.83 | 2.94 | 2.03 | 1.98 | 2.23 | 1.40 |
| Mean | 11.57 b | 8.64 ab | 11.64 b | 7.43 a | 6.73 b | 4.71 ab | 6.03 b | 3.22 a | 1.88 b | 1.71 ab | 1.94 b | 1.40 a |
| Locus | Ho | He | Hm | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LP | LPC | LCP | LC | LP | LPC | LCP | LC | LP | LPC | LCP | LC | |
| L1 | 0.67 | 0.67 | 0.73 | 0.57 | 0.77 | 0.62 | 0.81 | 0.72 | 0.33 | 0.33 | 0.27 | 0.43 |
| L2 | 0.70 | 1.00 | 1.00 | 0.67 | 0.94 | 0.86 | 0.92 | 0.64 | 0.30 | 0.00 | 0.00 | 0.33 |
| L3 | 0.97 | 0.93 | 0.97 | 0.67 | 0.94 | 0.84 | 0.88 | 0.78 | 0.03 | 0.07 | 0.03 | 0.33 |
| L4 | 0.83 | 1.00 | 0.97 | 0.67 | 0.87 | 0.87 | 0.91 | 0.72 | 0.17 | 0.00 | 0.03 | 0.33 |
| L5 | 0.33 | 0.90 | 0.90 | 0.63 | 0.56 | 0.84 | 0.81 | 0.65 | 0.67 | 0.10 | 0.10 | 0.37 |
| L6 | 0.07 | 1.00 | 1.00 | 0.43 | 0.06 | 0.67 | 0.68 | 0.53 | 0.93 | 0.00 | 0.00 | 0.57 |
| L7 | 0.80 | 0.97 | 1.00 | 0.77 | 0.86 | 0.77 | 0.81 | 0.78 | 0.20 | 0.03 | 0.00 | 0.23 |
| L8 | 0.73 | 0.87 | 0.87 | 0.37 | 0.85 | 0.65 | 0.64 | 0.60 | 0.27 | 0.13 | 0.13 | 0.63 |
| L9 | 0.77 | 0.67 | 0.90 | 0.27 | 0.79 | 0.78 | 0.83 | 0.78 | 0.23 | 0.33 | 0.10 | 0.73 |
| L10 | 0.47 | 0.73 | 0.87 | 0.10 | 0.55 | 0.59 | 0.62 | 0.44 | 0.53 | 0.27 | 0.13 | 0.90 |
| L11 | 0.47 | 0.63 | 0.90 | 0.50 | 0.82 | 0.74 | 0.78 | 0.51 | 0.53 | 0.37 | 0.10 | 0.50 |
| L12 | 0.23 | 0.53 | 0.60 | 0.43 | 0.69 | 0.65 | 0.71 | 0.60 | 0.77 | 0.47 | 0.40 | 0.57 |
| L13 | 0.87 | 1.00 | 0.97 | 0.77 | 0.89 | 0.83 | 0.86 | 0.79 | 0.13 | 0.00 | 0.03 | 0.23 |
| L14 | 0.87 | 1.00 | 1.00 | 0.73 | 0.84 | 0.83 | 0.87 | 0.66 | 0.13 | 0.00 | 0.00 | 0.27 |
| Mean | 0.63 a | 0.85 b | 0.90 b | 0.54 a | 0.74 ab | 0.75 ab | 0.79 b | 0.66 a | 0.37 b | 0.15 a | 0.10 a | 0.46 b |
| Locus | Fis | HWE | ||||||
|---|---|---|---|---|---|---|---|---|
| LP | LPC | LCP | LC | LP | LPC | LCP | LC | |
| L1 | 0.13 | −0.07 | 0.09 | 0.22 | 0.184 NS | 0.959 NS | 0.039 * | 0.365 NS |
| L2 | 0.26 | −0.16 | −0.09 | −0.04 | 0.000 *** | 0.001 *** | 0.708 NS | 0.655 NS |
| L3 | −0.03 | −0.11 | −0.10 | 0.15 | 0.712 NS | 0.028 * | 0.984 NS | 0.000 *** |
| L4 | 0.05 | −0.15 | −0.07 | 0.08 | 0.489 NS | 0.000 *** | 0.816 NS | 0.001 *** |
| L5 | 0.40 | −0.07 | −0.11 | 0.02 | 0.043 * | 0.063 NS | 0.001 *** | 0.002 ** |
| L6 | −0.03 | −0.49 | −0.48 | 0.18 | 0.998 NS | 0.001 *** | 0.000 *** | 0.058 NS |
| L7 | 0.07 | −0.26 | −0.23 | 0.01 | 0.985 NS | 0.059 NS | 0.777 NS | 0.025 * |
| L8 | 0.13 | −0.34 | −0.35 | 0.39 | 0.003 ** | 0.349 NS | 0.946 NS | 0.000 *** |
| L9 | 0.03 | 0.14 | −0.09 | 0.66 | 0.828 NS | 0.436 NS | 0.092 NS | 0.000 *** |
| L10 | 0.14 | −0.24 | −0.39 | 0.77 | 0.286 NS | 0.000 *** | 0.000 *** | 0.000 *** |
| L11 | 0.43 | 0.14 | −0.15 | 0.01 | 0.000 *** | 0.000 *** | 0.000 *** | 0.000 *** |
| L12 | 0.66 | 0.18 | 0.16 | 0.27 | 0.000 *** | 0.000 *** | 0.880 NS | 0.019 * |
| L13 | 0.02 | −0.20 | −0.13 | 0.02 | 0.905 NS | 0.354 NS | 0.199 NS | 0.603 NS |
| L14 | −0.03 | −0.20 | −0.15 | −0.11 | 0.117 NS | 0.000 *** | 0.036 * | 0.002 ** |
| NHW | - | - | - | - | 5 | 8 | 6 | 10 |
| Mean | 0.16 | −0.13 | −0.15 | 0.19 | - | - | - | - |
| Source of Variation | d.f. | Source of Squares | Variance Components | Percentage of Variation (%) | F-Statistic |
|---|---|---|---|---|---|
| Among populations | 3 | 70.387 | 0.25705 | 3.76 | FIS = 0.223 *** FST = 0.038 *** FIT = 0.252 *** |
| Among Individuals/ within populations | 116 | 932.550 | 1.46336 | 21.42 | |
| Within individuals | 120 | 613.500 | 5.11250 | 74.82 | |
| Total | 239 | 1616.438 | 6.83292 | 100 |
| LP | LPC | LCP | LC | |
|---|---|---|---|---|
| LP | 0.430 | 0.440 | 1.861 | |
| LPC | 0.031 *** | 0.357 | 0.729 | |
| LCP | 0.029 *** | 0.019 *** | 0.633 | |
| LC | 0.074 *** | 0.053 *** | 0.041 *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Guo, D.; Xie, Q.; Wei, F.; Jiang, L.; Liu, F.; Ye, T.; Lou, B. Comparative Analysis of Genetic Structure and Diversity in Larimichthys polyactis, Larimichthys crocea, and Their Reciprocal Hybrids Based on Microsatellite Loci. Animals 2025, 15, 1360. https://doi.org/10.3390/ani15101360
Wang Z, Guo D, Xie Q, Wei F, Jiang L, Liu F, Ye T, Lou B. Comparative Analysis of Genetic Structure and Diversity in Larimichthys polyactis, Larimichthys crocea, and Their Reciprocal Hybrids Based on Microsatellite Loci. Animals. 2025; 15(10):1360. https://doi.org/10.3390/ani15101360
Chicago/Turabian StyleWang, Zehui, Dandan Guo, Qingping Xie, Fuliang Wei, Lin Jiang, Feng Liu, Ting Ye, and Bao Lou. 2025. "Comparative Analysis of Genetic Structure and Diversity in Larimichthys polyactis, Larimichthys crocea, and Their Reciprocal Hybrids Based on Microsatellite Loci" Animals 15, no. 10: 1360. https://doi.org/10.3390/ani15101360
APA StyleWang, Z., Guo, D., Xie, Q., Wei, F., Jiang, L., Liu, F., Ye, T., & Lou, B. (2025). Comparative Analysis of Genetic Structure and Diversity in Larimichthys polyactis, Larimichthys crocea, and Their Reciprocal Hybrids Based on Microsatellite Loci. Animals, 15(10), 1360. https://doi.org/10.3390/ani15101360







