Identification and Analysis of Circular RNAs in Mammary Gland from Yaks Between Lactation and Dry Period
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animal and Sample Collection
2.3. RNA Extraction and Quality Control
2.4. Library Construction and Sequencing
2.5. Identification and Annotation of circRNAs
2.6. Differentially Expressed circRNAs and miRNAs
2.7. Target miRNAs and Genes Prediction and ceRNA Network Analysis
2.8. GO and KEGG Enrichment Analysis
2.9. Verification of Sequencing Data Using qRT-PCR
3. Results
3.1. RNA-Seq Data Analysis
3.2. Identification of circRNAs
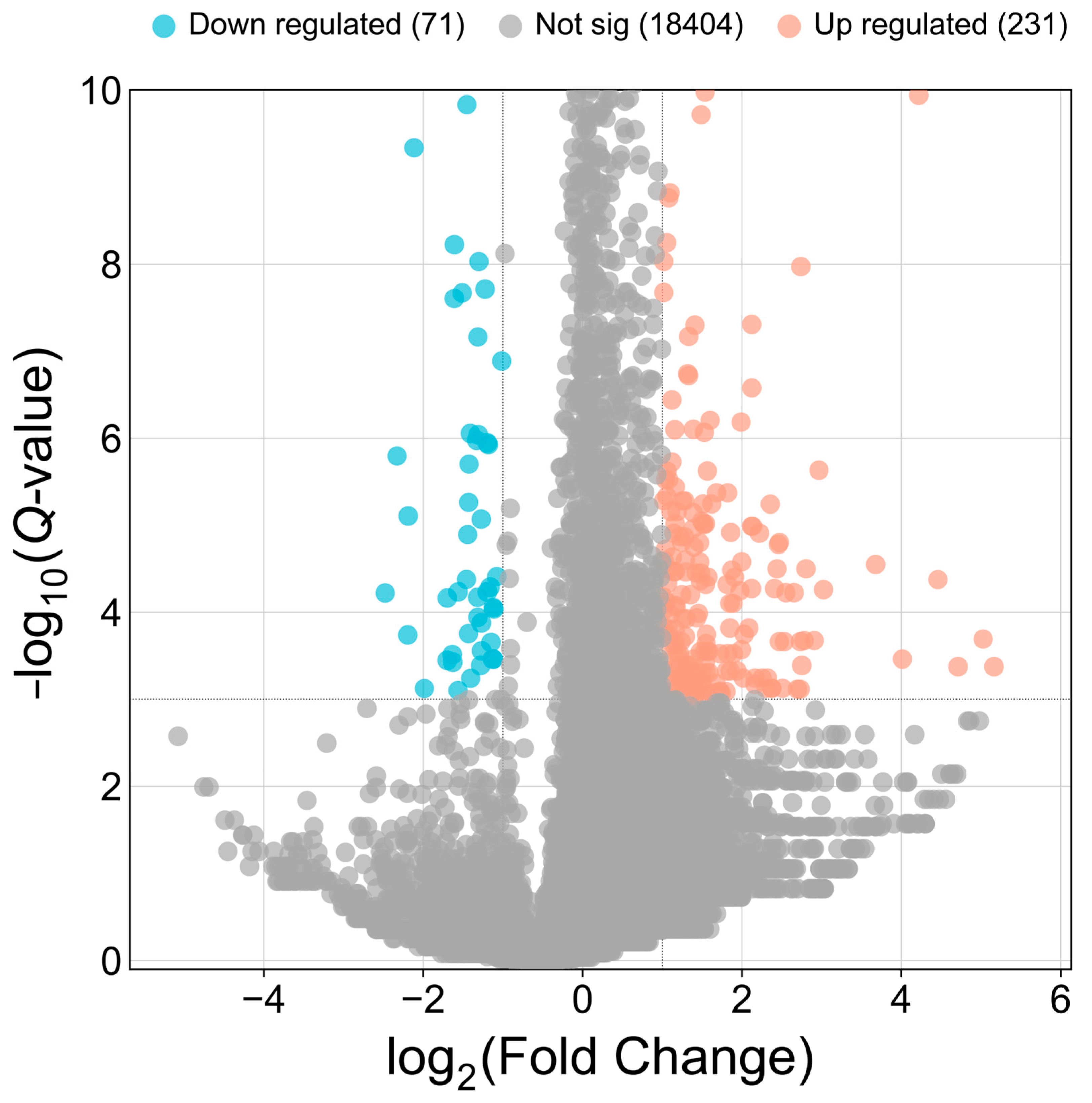
3.3. Differentially Expressed circRNAs Between DP and LP Groups
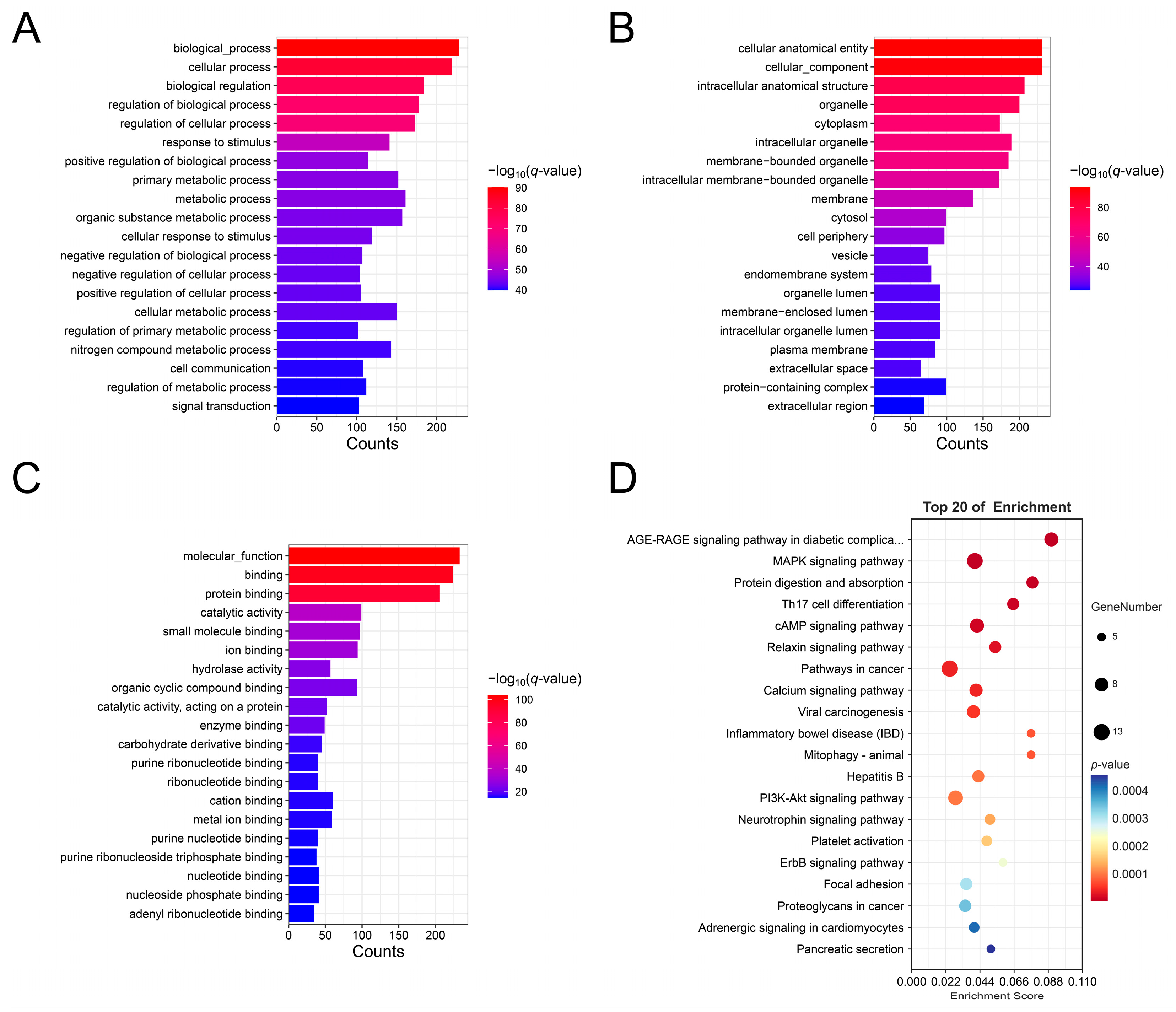
3.4. Functional Enrichment Analysis of Host Gene
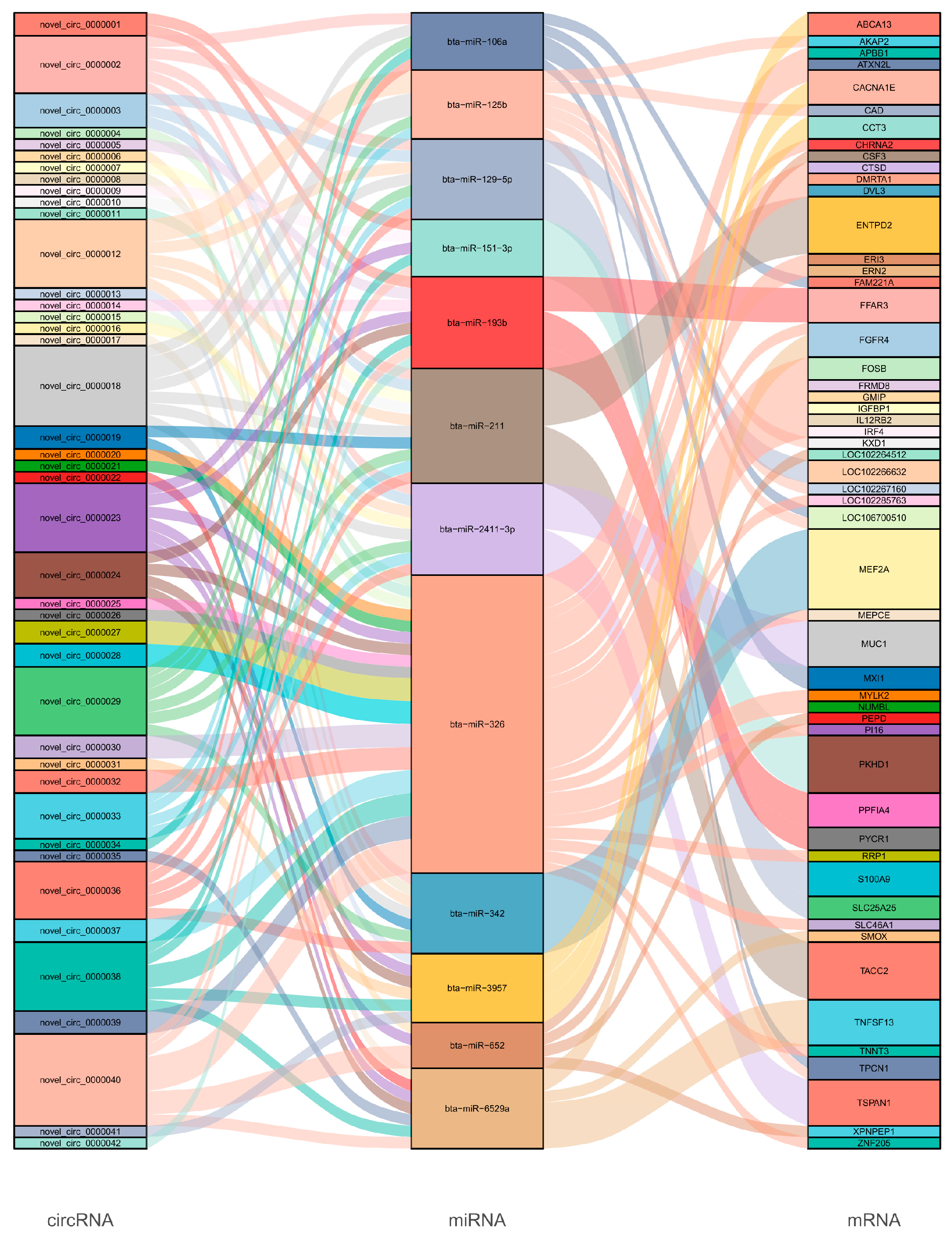
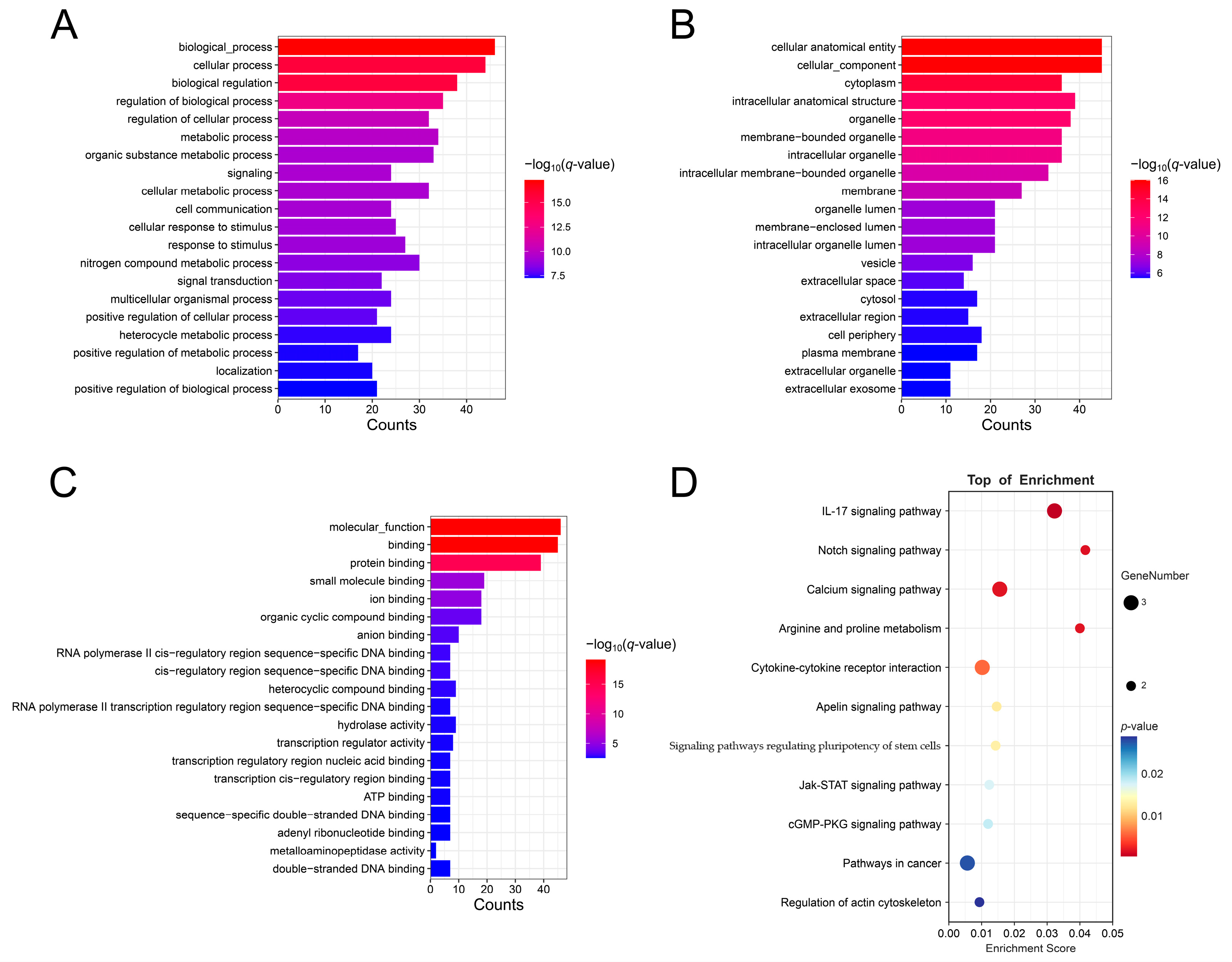
3.5. Construction of ceRNA Network
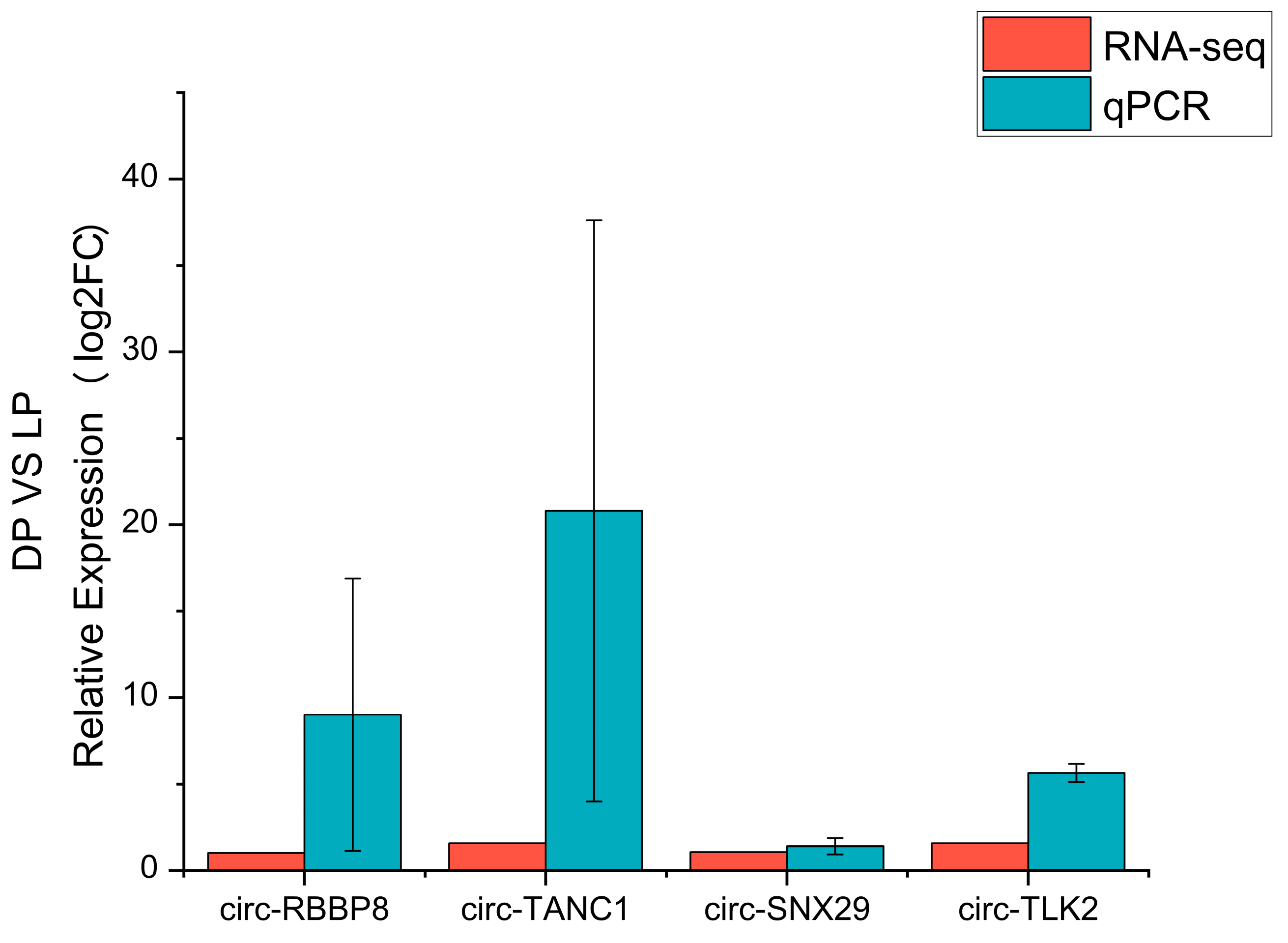
3.6. Real-Time Quantitative PCR Validation of Sequencing Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ding, L.; Wang, Y.; Kreuzer, M.; Guo, X.; Mi, J.; Gou, Y.; Shang, Z.; Zhang, Y.; Zhou, J.; Wang, H.; et al. Seasonal Variations in the Fatty Acid Profile of Milk from Yaks Grazing on the Qinghai-Tibetan Plateau. J. Dairy Res. 2013, 80, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Long, R.; Kreuzer, M.; Ding, L.; Shang, Z.; Zhang, Y.; Yang, Y.; Cui, G. Importance of Functional Ingredients in Yak Milk-Derived Food on Health of Tibetan Nomads Living Under High-Altitude Stress: A Review. Crit. Rev. Food Sci. Nutr. 2014, 54, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ma, Y.; Li, Q.; Wang, J.; Cheng, J.; Xue, J.; Shi, J. The Chemical Composition and Nitrogen Distribution of Chinese Yak (Maiwa) Milk. Int. J. Mol. Sci. 2011, 12, 4885–4895. [Google Scholar] [CrossRef]
- Richert, M.M.; Schwertfeger, K.L.; Ryder, J.W.; Anderson, S.M. An Atlas of Mouse Mammary Gland Development. J. Mammary Gland. Biol. Neoplasia 2000, 5, 227–241. [Google Scholar] [CrossRef]
- Inman, J.L.; Robertson, C.; Mott, J.D.; Bissell, M.J. Mammary Gland Development: Cell Fate Specification, Stem Cells and the Microenvironment. Development 2015, 142, 1028–1042. [Google Scholar] [CrossRef]
- Hennighausen, L.; Robinson, G.W. Information Networks in the Mammary Gland. Nat. Rev. Mol. Cell Biol. 2005, 6, 715–725. [Google Scholar] [CrossRef]
- Watson, C.J.; Khaled, W.T. Mammary Development in the Embryo and Adult: A Journey of Morphogenesis and Commitment. Development 2008, 135, 995–1003. [Google Scholar] [CrossRef]
- Sanger, H.L.; KLOTZt, G.; RIESNERt, D.; Gross, H.J. Viroids Are Single-Stranded Covalently Closed Circular RNA Molecules Existing as Highly Base-Paired Rod-like Structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef]
- Kolakofsky, D. Isolation and Characterization of Sendai Virus DI-RNAs. Cell 1976, 8, 547–555. [Google Scholar] [CrossRef]
- Rong, D.; Sun, H.; Li, Z.; Liu, S.; Dong, C.; Fu, K.; Tang, W.; Cao, H. An Emerging Function of circRNA-miRNAs-mRNA Axis in Human Diseases. Oncotarget 2017, 8, 73271–73281. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs Are a Large Class of Animal RNAs with Regulatory Potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient microRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhao, Z.; Shi, Y.; Luo, J.; Chen, T.; Xi, Q.; Zhang, Y.; Sun, J. CircEZH2 Regulates Milk Fat Metabolism through miR-378b Sponge Activity. Animals 2022, 12, 718. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hou, J.; Zhang, M.; Seleh-Zo, E.; Wang, J.; Cao, B.; An, X. Circ-016910 Sponges miR-574-5p to Regulate Cell Physiology and Milk Synthesis via MAPK and PI3K/AKT–mTOR Pathways in GMECs. J. Cell. Physiol. 2020, 235, 4198–4216. [Google Scholar] [CrossRef]
- Li, X.; Wu, Y.; Wang, Y.; Yang, X.; Gao, R.; Lu, Q.; Lv, X.; Chen, Z. The Molecular Mechanism of circRNA-11228/miR-103/INSIG1 Pathway Regulating Milk Fat Synthesis in Bovine Mammary Epithelial Cells. Agriculture 2024, 14, 538. [Google Scholar] [CrossRef]
- Elnour, I.E.; Wang, X.; Zhansaya, T.; Akhatayeva, Z.; Khan, R.; Cheng, J.; Hung, Y.; Lan, X.; Lei, C.; Chen, H. Circular RNA circMYL1 Inhibit Proliferation and Promote Differentiation of Myoblasts by Sponging miR-2400. Cells 2021, 10, 176. [Google Scholar] [CrossRef]
- Wang, X.; Cao, X.; Dong, D.; Shen, X.; Cheng, J.; Jiang, R.; Yang, Z.; Peng, S.; Huang, Y.; Lan, X.; et al. Circular RNA TTN Acts As a miR-432 Sponge to Facilitate Proliferation and Differentiation of Myoblasts via the IGF2/PI3K/AKT Signaling Pathway. Mol. Ther. Nucleic Acids 2019, 18, 966–980. [Google Scholar] [CrossRef]
- Zhang, X.-O.; Wang, H.-B.; Zhang, Y.; Lu, X.; Chen, L.-L.; Yang, L. Complementary Sequence-Mediated Exon Circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet.journal 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Pearson, W.R.; Wood, T.; Zhang, Z.; Miller, W. Comparison of DNA Sequences with Protein Sequences. Genomics 1997, 46, 24–36. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate Alignment of Transcriptomes in the Presence of Insertions, Deletions and Gene Fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Salzberg, S.L. TopHat-Fusion: An Algorithm for Discovery of Novel Fusion Transcripts. Genome Biol. 2011, 12, R72. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA Targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Kruger, J.; Rehmsmeier, M. RNAhybrid: microRNA Target Prediction Easy, Fast and Flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The Role of Site Accessibility in microRNA Target Recognition. Nat. Genet. 2007, 39, 1278–1284. [Google Scholar] [CrossRef]
- Wu, X.; Zhou, X.; Xiong, L.; Pei, J.; Yao, X.; Liang, C.; Bao, P.; Chu, M.; Guo, X.; Yan, P. Transcriptome Analysis Reveals the Potential Role of Long Non-Coding RNAs in Mammary Gland of Yak During Lactation and Dry Period. Front. Cell Dev. Biol. 2020, 8, 579708. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent Prioritization and Exploratory Visualization of Biological Functions for Gene Enrichment Analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. G:Profiler—Interoperable Web Service for Functional Enrichment Analysis and Gene Identifier Mapping (2023 Update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an Enhanced Web Interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of Housekeeping Genes for Gene Expression Studies in Human Reticulocytes Using Real-Time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Zi, X.-D.; Luo, B.; Xia, W.; Zheng, Y.-C.; Xiong, X.-R.; Li, J.; Zhong, J.-C.; Zhu, J.-J.; Zhang, Z.-F. Characterization of Transcriptional Complexity during Pre-Implantation Development of the Yak (Bos Grunniens) Using RNA-Seq. Reprod. Domest. Anim. 2018, 53, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Guo, X.; Yongfu, L.; Wang, T.; Bao, P.; Chu, M.; Wu, X.; Yan, P.; Liang, C. Identification of circRNA-Associated ceRNA Networks in the Longissimus Dorsi of Yak Under Different Feeding Systems. BMC Vet. Res. 2024, 20, 67. [Google Scholar] [CrossRef] [PubMed]
- La, Y.; Ma, X.; Bao, P.; Chu, M.; Yan, P.; Liang, C.; Guo, X. Genome-Wide Landscape of mRNAs, lncRNAs, and circRNAs during Testicular Development of Yak. Int. J. Mol. Sci. 2023, 24, 4420. [Google Scholar] [CrossRef]
- Liang, Y.; Gao, Q.; Wang, H.; Guo, M.; Arbab, A.A.I.; Nazar, M.; Li, M.; Yang, Z.; Karrow, N.A.; Mao, Y. Identification and Characterization of Circular RNAs in Mammary Tissue from Holstein Cows at Early Lactation and Non-Lactation. Biomolecules 2022, 12, 478. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, H.; Hickford, J.G.H.; Hao, Z.; Gong, H.; Hu, J.; Liu, X.; Li, S.; Shen, J.; Ke, N.; et al. Identification and Characterization of Circular RNAs in Mammary Gland Tissue from Sheep at Peak Lactation and during the Nonlactating Period. J. Dairy Sci. 2021, 104, 2396–2409. [Google Scholar] [CrossRef]
- Xuan, R.; Wang, J.; Li, Q.; Wang, Y.; Du, S.; Duan, Q.; Guo, Y.; He, P.; Ji, Z.; Chao, T. Identification and Characterization of circRNAs in Non-Lactating Dairy Goat Mammary Glands Reveal Their Regulatory Role in Mammary Cell Involution and Remodeling. Biomolecules 2023, 13, 860. [Google Scholar] [CrossRef]
- Ma, D.; Zhao, Y.; Yu, S.; Zhang, H.; Cheng, M.; Cao, H.; Li, Q.; Min, L. CircRNA as CeRNA Mediated by microRNA May Be Involved in Goat Lactation. Small Rumin. Res. 2019, 171, 63–72. [Google Scholar] [CrossRef]
- Gonzalez-Iglesias, A.E.; Jiang, Y.; Tomić, M.; Kretschmannova, K.; Andric, S.A.; Zemkova, H.; Stojilkovic, S.S. Dependence of Electrical Activity and Calcium Influx-Controlled Prolactin Release on Adenylyl Cyclase Signaling Pathway in Pituitary Lactotrophs. Mol. Endocrinol. 2006, 20, 2231–2246. [Google Scholar] [CrossRef]
- Chiba, T.; Maeda, T.; Sanbe, A.; Kudo, K. Serotonin Suppresses β-Casein Expression via PTP1B Activation in Human Mammary Epithelial Cells. Biochem. Biophys. Res. Commun. 2016, 473, 323–328. [Google Scholar] [CrossRef]
- Chen, Z.; Zhou, J.; Wang, M.; Liu, J.; Zhang, L.; Loor, J.J.; Liang, Y.; Wu, H.; Yang, Z. Circ09863 Regulates Unsaturated Fatty Acid Metabolism by Adsorbing miR-27a-3p in Bovine Mammary Epithelial Cells. J. Agric. Food Chem. 2020, 68, 8589–8601. [Google Scholar] [CrossRef]
- Chen, Z.; Cao, X.; Lu, Q.; Zhou, J.; Wang, Y.; Wu, Y.; Mao, Y.; Xu, H.; Yang, Z. Circ01592 Regulates Unsaturated Fatty Acid Metabolism through Adsorbing miR-218 in Bovine Mammary Epithelial Cells. Food Funct. 2021, 12, 12047–12058. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Wang, M.; Zhang, X.; Ha, S.; Wang, C.; Ao, C.; Wang, H. Arginine levels affect growth and CSN3 gene expression of dairy cows mammary epithelial cells in vitro. Chin. J. Anim. Nutr. 2012, 24, 852–858. [Google Scholar]
- Wang, M.; Xu, B.; Wang, H.; Bu, D.; Wang, J.; Loor, J.-J. Effects of Arginine Concentration on the In Vitro Expression of Casein and mTOR Pathway Related Genes in Mammary Epithelial Cells from Dairy Cattle. PLoS ONE 2014, 9, e95985. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Bazer, F.W.; Hu, J.; Johnson, G.A.; Spencer, T.E. Polyamine Synthesis from Proline in the Developing Porcine Placenta1. Biol. Reprod. 2005, 72, 842–850. [Google Scholar] [CrossRef] [PubMed]
- Da Rosa, F.T.; Moreira, C.G.A.; Barbero, M.M.D.; Hurtado Lugo, N.A.; de Camargo, G.M.F.; Aspicueta Borquis, R.R.; de Oliveira, H.N.; Boligon, A.A.; de Vargas, L.; Moreira, H.L.M.; et al. Associations between MUC1 Gene Polymorphism and Resistance to Mastitis, Milk Production and Fertility Traits in Murrah Water Buffaloes. J. Appl. Anim. Res. 2020, 48, 151–155. [Google Scholar] [CrossRef]
- Yonezawa, T.; Haga, S.; Kobayashi, Y.; Katoh, K.; Obara, Y. Short-Chain Fatty Acid Signaling Pathways in Bovine Mammary Epithelial Cells. Regul. Pept. 2009, 153, 30–36. [Google Scholar] [CrossRef]
- Gerlando, R.D.; Sutera, A.M.; Mastrangelo, S.; Tolone, M.; Portolano, B.; Sottile, G.; Bagnato, A.; Strillacci, M.G.; Sardina, M.T. Genome-Wide Association Study between CNVs and Milk Production Traits in Valle Del Belice Sheep. PLoS ONE 2019, 14, e0215204. [Google Scholar] [CrossRef]
- Dai, W.; Zou, Y.; White, R.R.; Liu, J.; Liu, H. Transcriptomic Profiles of the Bovine Mammary Gland during Lactation and the Dry Period. Funct. Integr. Genom. 2018, 18, 125–140. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, F.; Song, Y.; Sheng, X.; Ren, F.; Xiong, K.; Chen, L.; Zhang, H.; Liu, D.; Lengner, C.J.; et al. Numb and Numbl Act to Determine Mammary Myoepithelial Cell Fate, Maintain Epithelial Identity, and Support Lactogenesis. FASEB J. 2016, 30, 3474–3488. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, Y.; Wang, K.; Zhang, Z.; Han, M.; Li, G.; Zhang, B.; Yang, Y.; Loor, J.J.; Yang, Z.; et al. CircRNA-02191 Regulating Unsaturated Fatty Acid Synthesis by Adsorbing miR-145 to Enhance CD36 Expression in Bovine Mammary Gland. Int. J. Biol. Macromol. 2023, 244, 125306. [Google Scholar] [CrossRef]
- Chen, Z.; Lu, Q.; Liang, Y.; Cui, X.; Wang, X.; Mao, Y.; Yang, Z. Circ11103 Interacts with miR-128/PPARGC1A to Regulate Milk Fat Metabolism in Dairy Cows. J. Agric. Food Chem. 2021, 69, 4490–4500. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, Y.; Yang, S.; Gao, R.; Lv, X.; Yang, Z.; Jiao, P.; Zhang, N.; Loor, J.J.; Chen, Z. m6A Methylation Mediates the Function of the circRNA-08436/miR-195/ELOVL6 Axis in Regards to Lipid Metabolism in Dairy Goat Mammary Glands. Animals 2024, 14, 1715. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Ma, L.; Liu, Y.; He, Y.; Li, G.; An, X.; Cao, B. CircRNA-006258 Sponge-Adsorbs miR-574-5p to Regulate Cell Growth and Milk Synthesis via EVI5L in Goat Mammary Epithelial Cells. Genes 2020, 11, 718. [Google Scholar] [CrossRef] [PubMed]

| Sample | Raw Reads | Clean Reads | Mapped Reads | Mapping Rate (%) | GC Content (%) | Q30 (%) |
|---|---|---|---|---|---|---|
| DP1 | 184,408,224 | 181,065,646 | 156,206,414 | 86.17 | 53.07 | 96.5 |
| DP2 | 155,782,622 | 152,534,698 | 131,660,504 | 86.32 | 53.23 | 96.53 |
| DP3 | 162,876,662 | 158,253,090 | 132,600,127 | 83.79 | 53.07 | 96.35 |
| LP1 | 160,155,930 | 156,985,398 | 135,755,839 | 86.48 | 53.17 | 96.53 |
| LP2 | 139,072,986 | 135,208,918 | 115,961,807 | 85.76 | 50.51 | 96.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Y.; Wu, X.; Meng, G.; Ma, X.; La, Y.; Bao, P.; Chu, M.; Yan, P. Identification and Analysis of Circular RNAs in Mammary Gland from Yaks Between Lactation and Dry Period. Animals 2025, 15, 89. https://doi.org/10.3390/ani15010089
Shi Y, Wu X, Meng G, Ma X, La Y, Bao P, Chu M, Yan P. Identification and Analysis of Circular RNAs in Mammary Gland from Yaks Between Lactation and Dry Period. Animals. 2025; 15(1):89. https://doi.org/10.3390/ani15010089
Chicago/Turabian StyleShi, Yilin, Xiaoyun Wu, Guangyao Meng, Xiaoming Ma, Yongfu La, Pengjia Bao, Min Chu, and Ping Yan. 2025. "Identification and Analysis of Circular RNAs in Mammary Gland from Yaks Between Lactation and Dry Period" Animals 15, no. 1: 89. https://doi.org/10.3390/ani15010089
APA StyleShi, Y., Wu, X., Meng, G., Ma, X., La, Y., Bao, P., Chu, M., & Yan, P. (2025). Identification and Analysis of Circular RNAs in Mammary Gland from Yaks Between Lactation and Dry Period. Animals, 15(1), 89. https://doi.org/10.3390/ani15010089






