Towards Automated Chicken Monitoring: Dataset and Machine Learning Methods for Visual, Noninvasive Reidentification
Simple Summary
Abstract
1. Introduction
1.1. Background
1.2. Motivation
1.3. Contributions
- i
- We present a summary of animal re-ID studies and give a comprehensive overview of publicly available datasets.
- ii
- We address the existing gap and introduce the first publicly available dataset for chicken re-ID: Chicks4FreeID. The dataset supports closed -and open-set re-ID, as well as semantic and instance segmentation tasks. We make this thoroughly documented dataset freely accessible to the research community and the public.
- iii
- We evaluate a species-agnostic state-of-the-art model on our dataset through two experiments. In the first experiment, we test the model using its frozen weights, which were not trained on chicken data. In the second experiment, we fine-tune the model to adapt it specifically to our dataset.
- iv
- We train two feature extractors from scratch in a standard supervised manner and test them on our dataset. Both models are based on transformer architectures.
- v
- We perform additional one-shot experiments with all previously mentioned models.
- vi
- Lastly, we make all associated code publicly available to ensure transparency and facilitate further research.
2. Related Work
2.1. Re-ID
2.2. State of the Art
2.3. Datasets
3. The Chicks4FreeID Dataset
3.1. Overview
3.2. Collection
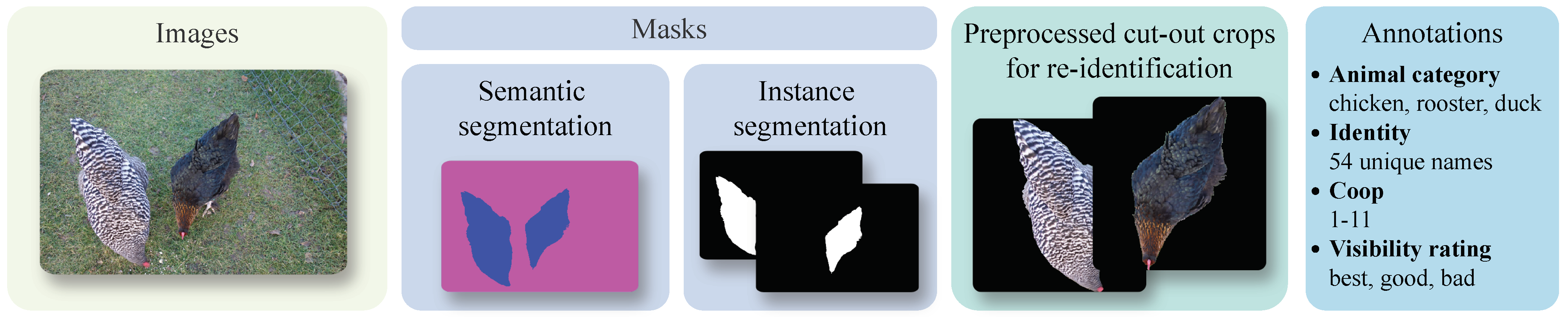
3.3. Annotation
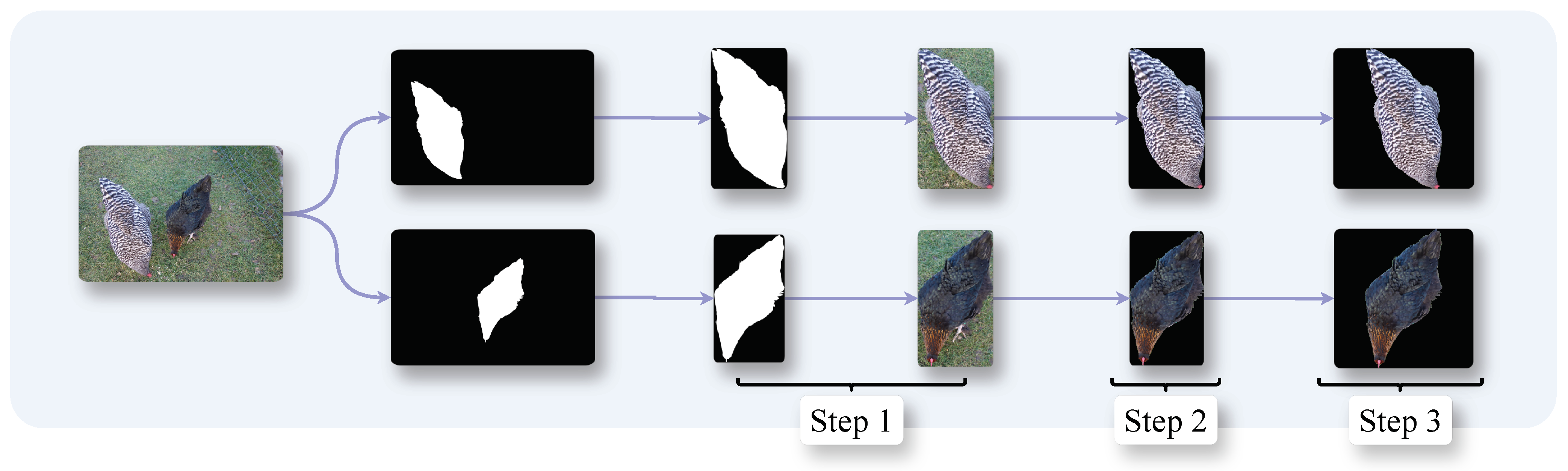
3.4. Preprocessing
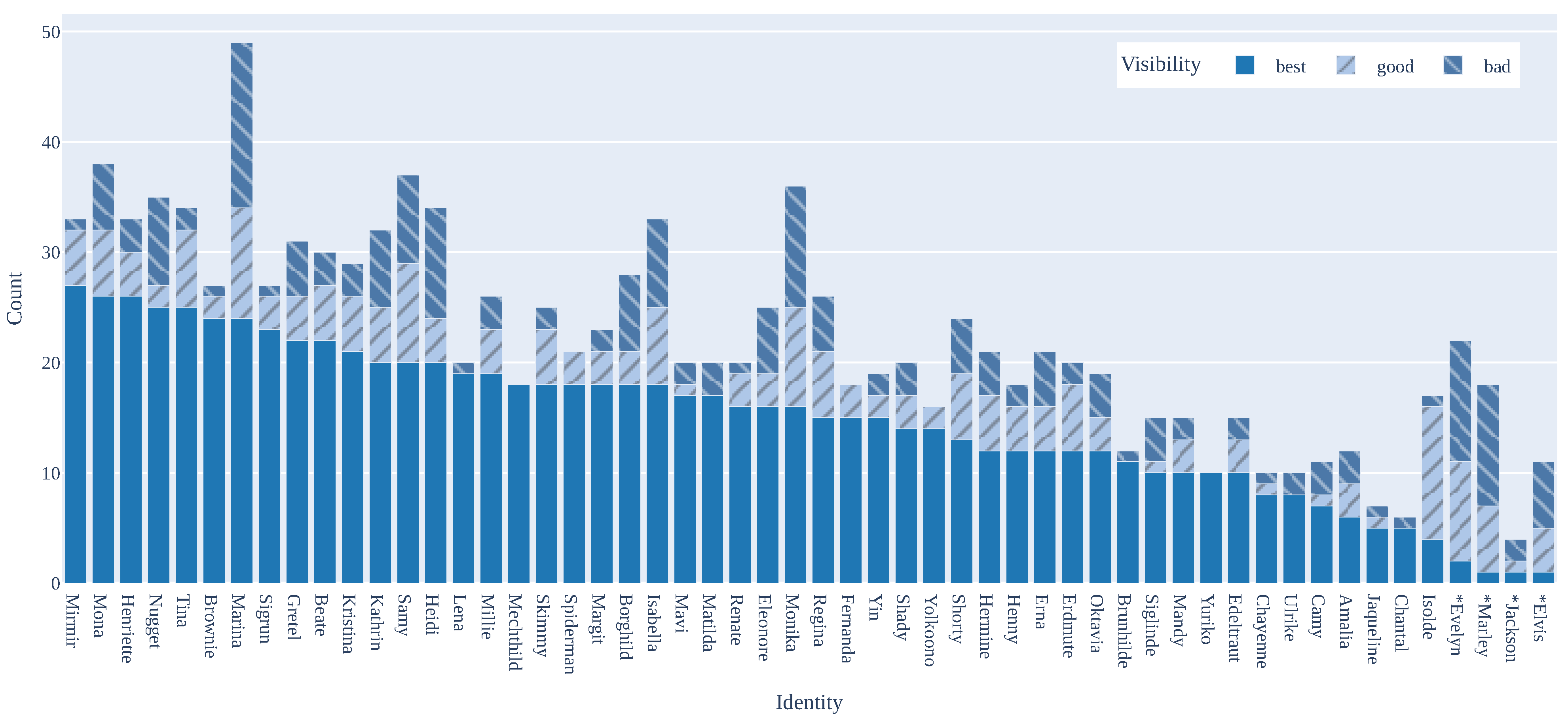
3.5. Dataset Statistics
4. Materials and Methods
4.1. Hardware
4.2. Data
4.3. Augmentation
4.4. Feature Extractors
4.5. Classifiers
4.6. Evaluation Metrics
5. Experiments
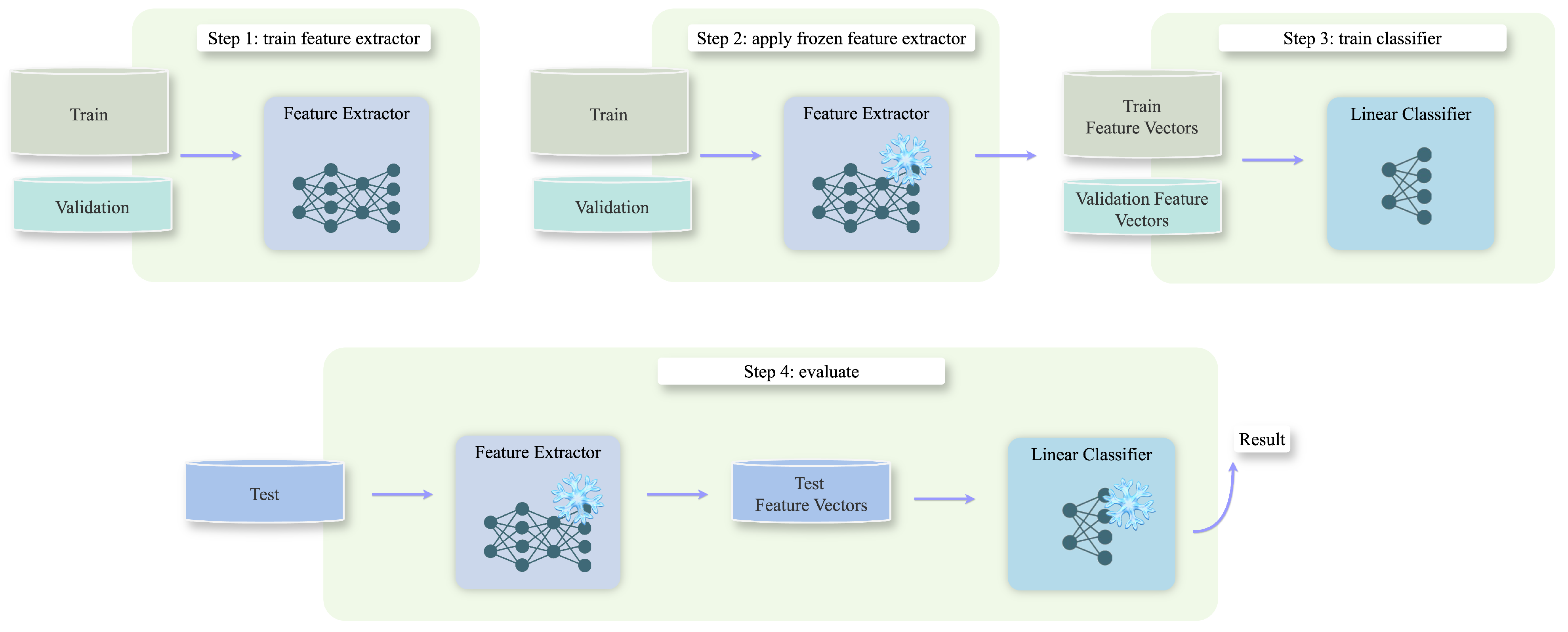
5.1. Domain Transfer Experiment
5.2. Standard Supervised Learning Experiment
5.3. One-Shot Experiment
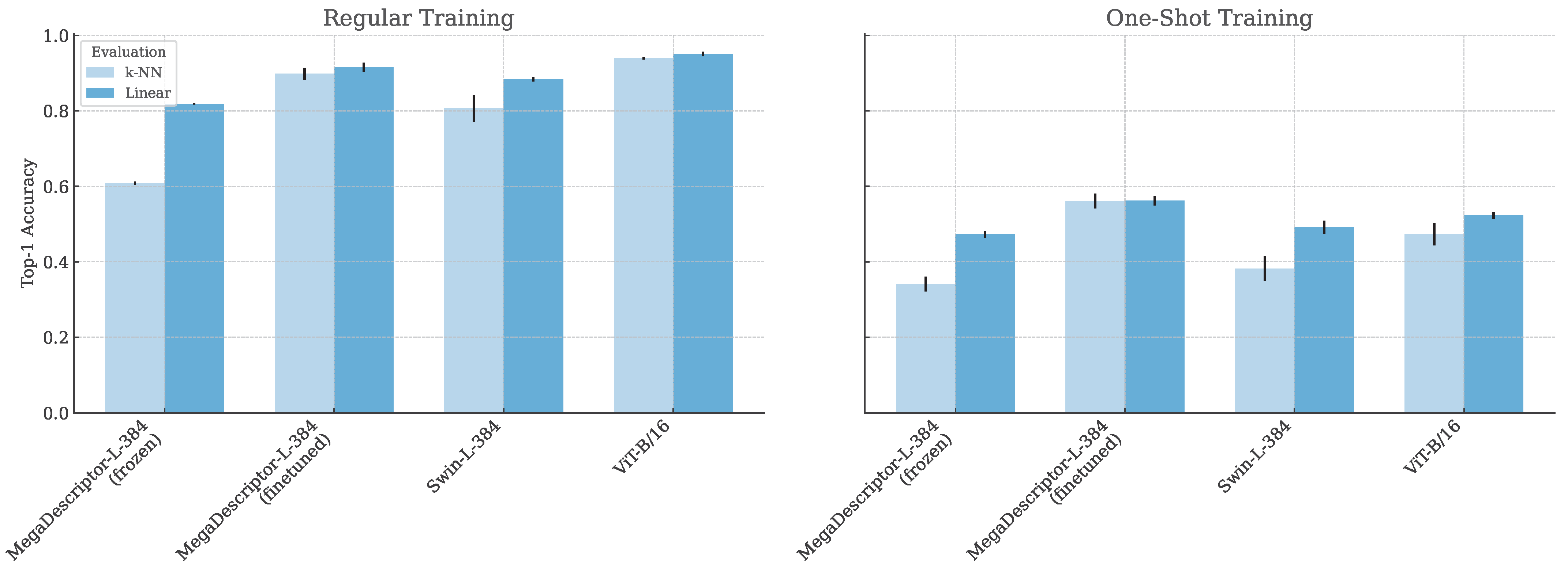
6. Results and Discussion
7. Conclusions
7.1. Findings
7.2. Limitations and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| k-NN | k-nearest neighbor |
| mAP | mean average precision |
| re-ID | reidentification |
Appendix A. Annotations

| Coop | Images | ID | Bad | Best | Good | Total | Coop | Images | ID | Bad | Best | Good | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 29 | Coop Total | 16 | 28 | 5 | 49 | ... | ... | Camy | 3 | 7 | 1 | 11 |
| #Unknown | 11 | 0 | 0 | 11 | Samy | 8 | 20 | 9 | 37 | ||||
| Chantal | 1 | 5 | 0 | 6 | Yin | 2 | 15 | 2 | 19 | ||||
| Chayenne | 1 | 8 | 1 | 10 | Yuriko | 0 | 10 | 0 | 10 | ||||
| Jaqueline | 1 | 5 | 1 | 7 | 7 | 42 | Coop Total | 1 | 42 | 5 | 48 | ||
| Mandy | 2 | 10 | 3 | 15 | Brownie | 1 | 24 | 2 | 27 | ||||
| 2 | 36 | Coop Total | 14 | 39 | 13 | 66 | Spiderman | 0 | 18 | 3 | 21 | ||
| #Unknown | 4 | 0 | 0 | 4 | 8 | 47 | Coop Total | 2 | 48 | 15 | 65 | ||
| Henny | 2 | 12 | 4 | 18 | Brunhilde | 1 | 11 | 0 | 12 | ||||
| Shady | 3 | 14 | 3 | 20 | Fernanda | 0 | 15 | 3 | 18 | ||||
| Shorty | 5 | 13 | 6 | 24 | Isolde | 1 | 4 | 12 | 17 | ||||
| 3 | 60 | Coop Total | 22 | 58 | 16 | 96 | Mechthild | 0 | 18 | 0 | 18 | ||
| #Unknown | 5 | 0 | 0 | 5 | 9 | 68 | Coop Total | 14 | 87 | 13 | 114 | ||
| Amalia | 3 | 6 | 3 | 12 | #Unknown | 1 | 0 | 0 | 1 | ||||
| Edeltraut | 2 | 10 | 3 | 15 | Mavi | 2 | 17 | 1 | 20 | ||||
| Erdmute | 2 | 12 | 6 | 20 | Mirmir | 1 | 27 | 5 | 33 | ||||
| Oktavia | 4 | 12 | 3 | 19 | Nugget | 8 | 25 | 2 | 35 | ||||
| Siglinde | 4 | 10 | 1 | 15 | Skimmy | 2 | 18 | 5 | 25 | ||||
| Ulrike | 2 | 8 | 0 | 10 | 10 | 140 | Coop Total | 57 | 189 | 36 | 282 | ||
| 4 | 26 | Coop Total | 7 | 29 | 5 | 41 | #Unknown | 23 | 0 | 0 | 23 | ||
| Hermine | 4 | 12 | 5 | 21 | Beate | 3 | 22 | 5 | 30 | ||||
| Matilda | 3 | 17 | 0 | 20 | Borghild | 7 | 18 | 3 | 28 | ||||
| 5 | 116 | Coop Total | 84 | 141 | 48 | 273 | Eleonore | 6 | 16 | 3 | 25 | ||
| #Unknown | 22 | 0 | 0 | 22 | Henriette | 3 | 26 | 4 | 33 | ||||
| Erna | 5 | 12 | 4 | 21 | Kristina | 3 | 21 | 5 | 29 | ||||
| Heidi | 10 | 20 | 4 | 34 | Margit | 2 | 18 | 3 | 23 | ||||
| Isabella | 8 | 18 | 7 | 33 | Millie | 3 | 19 | 4 | 26 | ||||
| Kathrin | 7 | 20 | 5 | 32 | Mona | 6 | 26 | 6 | 38 | ||||
| Marina | 15 | 24 | 10 | 49 | Sigrun | 1 | 23 | 3 | 27 | ||||
| Monika | 11 | 16 | 9 | 36 | 11 | 67 | Coop Total | 8 | 80 | 13 | 101 | ||
| Regina | 5 | 15 | 6 | 26 | Gretel | 5 | 22 | 4 | 31 | ||||
| Renate | 1 | 16 | 3 | 20 | Lena | 1 | 19 | 0 | 20 | ||||
| 6 | 46 | Coop Total | 16 | 52 | 12 | 80 | Tina | 2 | 25 | 7 | 34 | ||
| #Unknown | 3 | 0 | 0 | 3 | Yolkoono | 0 | 14 | 2 | 16 | ||||
| ... | ... | ... | ... | ... | ... | ... | Total | 677 | 50 | 241 | 793 | 181 | 1215 |
| Coop | ID | Category | Bad | Best | Good | Total |
|---|---|---|---|---|---|---|
| 4 | Coop Total | 22 | 3 | 15 | 40 | |
| Evelyn | Duck | 11 | 2 | 9 | 22 | |
| Marley | Duck | 11 | 1 | 6 | 18 | |
| 5 | Elvis | Rooster | 6 | 1 | 4 | 11 |
| 9 | Jackson | Rooster | 2 | 1 | 1 | 4 |
| Grand Total | 4 | 30 | 5 | 20 | 55 |
Appendix B. Plumage

| Plumage | ID | Coop | Bad | Best | Good | Total |
|---|---|---|---|---|---|---|
| Solid White | Total | 5 | 28 | 5 | 38 | |
| Chantal | 1 | 1 | 5 | 0 | 6 | |
| Chayenne | 1 | 1 | 8 | 1 | 10 | |
| Jaqueline | 1 | 1 | 5 | 1 | 7 | |
| Mandy | 1 | 2 | 10 | 3 | 15 | |
| Solid Black | Total | 5 | 39 | 21 | 65 | |
| Erdmute | 3 | 2 | 12 | 6 | 20 | |
| Ulrike | 3 | 2 | 8 | 0 | 10 | |
| Isolde | 8 | 1 | 4 | 12 | 17 | |
| Fernanda | 8 | 0 | 15 | 3 | 18 | |
| Shades of Gray | Total | 8 | 66 | 10 | 84 | |
| Erna | 5 | 5 | 12 | 4 | 21 | |
| Mavi | 9 | 2 | 17 | 1 | 20 | |
| Sigrun | 10 | 1 | 23 | 3 | 27 | |
| Yolkoono | 11 | 0 | 14 | 2 | 16 | |
| Shades of Orange | Total | 16 | 90 | 17 | 123 | |
| Henny | 2 | 2 | 12 | 4 | 18 | |
| Shady | 2 | 3 | 14 | 3 | 20 | |
| Shorty | 2 | 5 | 13 | 6 | 24 | |
| Brunhilde | 8 | 1 | 11 | 0 | 12 | |
| Mechthild | 8 | 0 | 18 | 0 | 18 | |
| Gretel | 11 | 5 | 22 | 4 | 31 | |
| Uniform Plumage | Grand Total | 34 | 223 | 53 | 310 | |
| Mixed Plumage | Grand Total | 207 | 570 | 128 | 905 |
Appendix C. Runtime

References
- Guhl, A.M.; Ortman, L.L. Visual Patterns in the Recognition of Individuals among Chickens. Condor 1953, 55, 287–298. [Google Scholar] [CrossRef]
- Andrew, W.; Hannuna, S.; Campbell, N.; Burghardt, T. Automatic individual holstein friesian cattle identification via selective local coat pattern matching in RGB-D imagery. In Proceedings of the 2016 IEEE International Conference on Image Processing (ICIP), Phoenix, AZ, USA, 25–28 September 2016; pp. 484–488. [Google Scholar] [CrossRef]
- Li, S.; Fu, L.; Sun, Y.; Mu, Y.; Chen, L.; Li, J.; Gong, H. Cow Dataset. 2021. Available online: https://doi.org/10.6084/m9.figshare.16879780 (accessed on 3 November 2024).
- Andrew, W.; Greatwood, C.; Burghardt, T. Visual Localisation and Individual Identification of Holstein Friesian Cattle via Deep Learning. In Proceedings of the 2017 IEEE International Conference on Computer Vision Workshops (ICCVW), Venice, Italy, 22–29 October 2017; pp. 2850–2859. [Google Scholar] [CrossRef]
- Gao, J.; Burghardt, T.; Andrew, W.; Dowsey, A.W.; Campbell, N.W. Towards Self-Supervision for Video Identification of Individual Holstein-Friesian Cattle: The Cows2021 Dataset. arXiv 2021, arXiv:2105.01938. [Google Scholar]
- Andrew, W.; Gao, J.; Mullan, S.; Campbell, N.; Dowsey, A.W.; Burghardt, T. Visual identification of individual Holstein-Friesian cattle via deep metric learning. Comput. Electron. Agric. 2021, 185, 106133. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Q.; Da, C.; Zhou, L.; Li, L.; Jiancuo, S. YakReID-103: A Benchmark for Yak reidentification. In Proceedings of the 2021 IEEE International Joint Conference on Biometrics (IJCB), Shenzhen, China, 4–7 August 2021; pp. 1–8. [Google Scholar] [CrossRef]
- Chan, J.; Carrión, H.; Mégret, R.; Rivera, J.L.A.; Giray, T. Honeybee reidentification in Video: New Datasets and Impact of Self-supervision. In Proceedings of the 17th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2022)—Volume 5: VISAPP, INSTICC, Avenida de S. Francisco Xavier, Lote 7 Cv. C, Online, 6–8 February 2022; pp. 517–525. [Google Scholar] [CrossRef]
- Tausch, F.; Stock, S.; Fricke, J.; Klein, O. Bumblebee reidentification Dataset. In Proceedings of the 2020 IEEE Winter Applications of Computer Vision Workshops (WACVW), Snowmass Village, CO, USA, 1–5 March 2020; pp. 35–37. [Google Scholar] [CrossRef]
- Borlinghaus, P.; Tausch, F.; Rettenberger, L. A Purely Visual Re-ID Approach for Bumblebees (Bombus terrestris). Smart Agric. Technol. 2023, 3, 100135. [Google Scholar] [CrossRef]
- Kulits, P.; Wall, J.; Bedetti, A.; Henley, M.; Beery, S. ElephantBook: A Semi-Automated Human-in-the-Loop System for Elephant reidentification. In Proceedings of the 4th ACM SIGCAS Conference on Computing and Sustainable Societies, New York, NY, USA, 28 June–2 July 2021; pp. 88–98. [Google Scholar] [CrossRef]
- Moskvyak, O.; Maire, F.; Dayoub, F.; Armstrong, A.O.; Baktashmotlagh, M. Robust reidentification of Manta Rays from Natural Markings by Learning Pose Invariant Embeddings. In Proceedings of the 2021 Digital Image Computing: Techniques and Applications (DICTA), Gold Coast, Australia, 29 November–1 December 2021; pp. 1–8. [Google Scholar] [CrossRef]
- Wang, L.; Ding, R.; Zhai, Y.; Zhang, Q.; Tang, W.; Zheng, N.; Hua, G. Giant Panda Identification. IEEE Trans. Image Process. 2021, 30, 2837–2849. [Google Scholar] [CrossRef]
- He, Q.; Zhao, Q.; Liu, N.; Chen, P.; Zhang, Z.; Hou, R. Distinguishing Individual Red Pandas from Their Faces. In Pattern Recognition and Computer Vision; Lin, Z., Wang, L., Yang, J., Shi, G., Tan, T., Zheng, N., Chen, X., Zhang, Y., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 714–724. [Google Scholar]
- Vidal, M.; Wolf, N.; Rosenberg, B.; Harris, B.P.; Mathis, A. Perspectives on Individual Animal Identification from Biology and Computer Vision. Integr. Comp. Biol. 2021, 61, 900–916. [Google Scholar] [CrossRef]
- Neethirajan, S. ChickTrack—A quantitative tracking tool for measuring chicken activity. Measurement 2022, 191, 110819. [Google Scholar] [CrossRef]
- T. Psota, E.; Schmidt, T.; Mote, B.; C. Pérez, L. Long-Term Tracking of Group-Housed Livestock Using Keypoint Detection and MAP Estimation for Individual Animal Identification. Sensors 2020, 20, 3670. [Google Scholar] [CrossRef]
- Li, N.; Ren, Z.; Li, D.; Zeng, L. Review: Automated techniques for monitoring the behaviour and welfare of broilers and laying hens: Towards the goal of precision livestock farming. Animal 2020, 14, 617–625. [Google Scholar] [CrossRef]
- Food Traceability. European Commission—B-1049 Brussels. 2007. Available online: https://food.ec.europa.eu/system/files/2016-10/gfl_req_factsheet_traceability_2007_en.pdf (accessed on 13 August 2024).
- Dennis, R.L.; Fahey, A.G.; Cheng, H.W. Different Effects of Individual Identification Systems on Chicken Well-Being1. Poult. Sci. 2008, 87, 1052–1057. [Google Scholar] [CrossRef]
- Anderson, G.; Johnson, A.; Arguelles-Ramos, M.; Ali, A. Impact of Body-worn Sensors on Broiler Chicken Behavior and Agonistic Interactions. J. Appl. Anim. Welf. Sci. 2023, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Stadig, L.M.; Rodenburg, T.B.; Ampe, B.; Reubens, B.; Tuyttens, F.A. An automated positioning system for monitoring chickens’ location: Effects of wearing a backpack on behaviour, leg health and production. Appl. Anim. Behav. Sci. 2018, 198, 83–88. [Google Scholar] [CrossRef]
- Marino, L. Thinking chickens: A review of cognition, emotion, and behavior in the domestic chicken. Anim. Cogn. 2017, 20, 127–147. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Zhao, Y.; Wang, J.; Zheng, Z.; Feng, L.; Tang, J. MammalClub: An Annotated Wild Mammal Dataset for Species Recognition, Individual Identification, and Behavior Recognition. Electronics 2023, 12, 4506. [Google Scholar] [CrossRef]
- Ye, M.; Shen, J.; Lin, G.; Xiang, T.; Shao, L.; Hoi, S.C.H. Deep Learning for Person reidentification: A Survey and Outlook. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 2872–2893. [Google Scholar] [CrossRef]
- Witham, C.L. Automated face recognition of rhesus macaques. J. Neurosci. Methods 2018, 300, 157–165. [Google Scholar] [CrossRef]
- Freytag, A.; Rodner, E.; Simon, M.; Loos, A.; Kühl, H.S.; Denzler, J. Chimpanzee Faces in the Wild: Log-Euclidean CNNs for Predicting Identities and Attributes of Primates. In Pattern Recognition; Rosenhahn, B., Andres, B., Eds.; Springer: Cham, Switzerland, 2016; pp. 51–63. [Google Scholar]
- Lin, T.Y.; Kuo, Y.F. Cat Face Recognition Using Deep Learning; American Society of Agricultural and Biological Engineers (ASABE) Annual International Meeting: St. Joseph, MI, USA, 2018. [Google Scholar] [CrossRef]
- Dlamini, N.; Zyl, T.L.v. Automated Identification of Individuals in Wildlife Population Using Siamese Neural Networks. In Proceedings of the 2020 7th International Conference on Soft Computing and Machine Intelligence (ISCMI), Stockholm, Sweden, 14–15 November 2020; pp. 224–228. [Google Scholar] [CrossRef]
- Mougeot, G.; Li, D.; Jia, S. A Deep Learning Approach for Dog Face Verification and Recognition. In Proceedings of the PRICAI 2019: Trends in Artificial Intelligence, Cuvu, Fiji, 26–30 August 2019; Nayak, A.C., Sharma, A., Eds.; Springer International Publishing: Cham, Swtizerland, 2019; pp. 418–430. [Google Scholar] [CrossRef]
- Lamping, C.; Kootstra, G.; Derks, M. Transformer-Based Similarity Learning for Re-Identification of Chickens. Available online: https://doi.org/10.2139/ssrn.4886408 (accessed on 22 October 2024).
- Li, S.; Li, J.; Tang, H.; Qian, R.; Lin, W. ATRW: A Benchmark for Amur Tiger Re-identification in the Wild. In Proceedings of the 28th ACM International Conference on Multimedia, Seattle, WA, USA, 12–16 October 2020; pp. 2590–2598. [Google Scholar] [CrossRef]
- Parham, J.; Crall, J.; Stewart, C.; Berger-Wolf, T.; Rubenstein, D. Animal population censusing at scale with citizen science and photographic identification. Aaai Spring Symp. Tech. Rep. 2017, SS-17-01–SS-17-08, 37–44. [Google Scholar]
- Haurum, J.B.; Karpova, A.; Pedersen, M.; Bengtson, S.H.; Moeslund, T.B. reidentification of Zebrafish using Metric Learning. In Proceedings of the 2020 IEEE Winter Applications of Computer Vision Workshops (WACVW), Snowmass Village, CO, USA, 1–5 March 2020; pp. 1–11. [Google Scholar] [CrossRef]
- Lahiri, M.; Tantipathananandh, C.; Warungu, R.; Rubenstein, D.I.; Berger-Wolf, T.Y. Biometric animal databases from field photographs: Identification of individual zebra in the wild. In Proceedings of the 1st ACM International Conference on Multimedia Retrieval, Vancouver, BC, Canada, 30–31 October 2008. [Google Scholar] [CrossRef]
- Adam, L.; Čermák, V.; Papafitsoros, K.; Picek, L. SeaTurtleID2022: A long-span dataset for reliable sea turtle reidentification. In Proceedings of the 2024 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 3–8 January 2024; pp. 7131–7141. [Google Scholar] [CrossRef]
- Bouma, S.; Pawley, M.D.; Hupman, K.; Gilman, A. Individual Common Dolphin Identification via Metric Embedding Learning. In Proceedings of the 2018 International Conference on Image and Vision Computing New Zealand (IVCNZ), Auckland, New Zealand, 19–21 November 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Hughes, B.; Burghardt, T. Automated Visual Fin Identification of Individual Great White Sharks. Int. J. Comput. Vision 2017, 122, 542–557. [Google Scholar] [CrossRef]
- Bae, H.B.; Pak, D.; Lee, S. Dog Nose-Print Identification Using Deep Neural Networks. IEEE Access 2021, 9, 49141–49153. [Google Scholar] [CrossRef]
- Zuerl, M.; Dirauf, R.; Koeferl, F.; Steinlein, N.; Sueskind, J.; Zanca, D.; Brehm, I.; Fersen, L.v.; Eskofier, B. PolarBearVidID: A Video-Based reidentification Benchmark Dataset for Polar Bears. Animals 2023, 13, 801. [Google Scholar] [CrossRef]
- Clapham, M.; Miller, E.; Nguyen, M.; Darimont, C.T. Automated facial recognition for wildlife that lack unique markings: A deep learning approach for brown bears. Ecol. Evol. 2020, 10, 12883–12892. [Google Scholar] [CrossRef] [PubMed]
- Körschens, M.; Denzler, J. ELPephants: A Fine-Grained Dataset for Elephant reidentification. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision Workshop (ICCVW), Seoul, Republic of Korea, 27–28 October 2019; pp. 263–270. [Google Scholar] [CrossRef]
- Čermák, V.; Picek, L.; Adam, L.; Papafitsoros, K. WildlifeDatasets: An Open-Source Toolkit for Animal reidentification. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 3–8 January 2024; pp. 5953–5963. [Google Scholar]
- Wahltinez, O.; Wahltinez, S.J. An open-source general purpose machine learning framework for individual animal reidentification using few-shot learning. Methods Ecol. Evol. 2024, 15, 373–387. [Google Scholar] [CrossRef]
- Guo, S.; Xu, P.; Miao, Q.; Shao, G.; Chapman, C.A.; Chen, X.; He, G.; Fang, D.; Zhang, H.; Sun, Y.; et al. Automatic Identification of Individual Primates with Deep Learning Techniques. iScience 2020, 23, 32. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.C.; Silva, L.R.; Renna, F.; Brandl, H.B.; Renoult, J.P.; Farine, D.R.; Covas, R.; Doutrelant, C. Bird individualID. 2020. Available online: https://github.com/AndreCFerreira/Bird_individualID (accessed on 3 November 2024).
- Kuncheva, L.I.; Williams, F.; Hennessey, S.L.; Rodríguez, J.J. Animal-Identification-from-Video. 2022. Available online: https://github.com/LucyKuncheva/Animal-Identification-from-Video (accessed on 3 November 2024).
- Adam, L.; Čermák, V.; Papafitsoros, K.; Picek, L. SeaTurtleID. 2022. Available online: https://www.kaggle.com/datasets/wildlifedatasets/seaturtleid2022 (accessed on 3 November 2024).
- Wahltinez, O. Sea Star Re-ID. 2023. Available online: https://lila.science/sea-star-re-id-2023/ (accessed on 3 November 2024).
- Me, W. Beluga ID. info@wildme.org. 2022. Available online: https://lila.science/datasets/beluga-id-2022/ (accessed on 3 November 2024).
- Cheeseman, T.; Southerland, K.; Reade, W.; Howard, A. Happywhale—Whale and Dolphin Identification. 2022. Available online: https://kaggle.com/competitions/happy-whale-and-dolphin (accessed on 3 November 2024).
- Nepovinnykh, E. SealID. Lappeenranta University of Technology, School of Engineering Science Yhteiset. 2022. Available online: https://doi.org/10.23729/0f4a3296-3b10-40c8-9ad3-0cf00a5a4a53 (accessed on 3 November 2024).
- Papafitsoros, K.; Adam, L.; Čermák, V.; Picek, L. SeaTurtleID. 2022. Available online: https://www.kaggle.com/datasets/wildlifedatasets/seaturtleidheads (accessed on 3 November 2024).
- Watch, W.T.; Conservation, L.O. Turtle Recall: Conservation Challenge. 2022. Available online: https://zindi.africa/competitions/turtle-recall-conservation-challenge/data (accessed on 3 November 2024).
- Trotter, C.; Atkinson, G.; Sharpe, M.; Richardson, K.; McGough, A.S.; Wright, N.; Burville, B.; Berggren, P. The Northumberland Dolphin Dataset 2020; Newcastle University: Newcastle upon Tyne, UK, 2020; Available online: https://doi.org/10.25405/data.ncl.c.4982342 (accessed on 3 November 2024).
- Humpback Whale Identification, Kaggle. 2018. Available online: https://kaggle.com/competitions/humpback-whale-identification (accessed on 3 November 2024).
- Khan, C.B.; Shashank; Kan, W. Right Whale Recognition, Kaggle. 2015. Available online: https://kaggle.com/competitions/noaa-right-whale-recognition (accessed on 3 November 2024).
- Holmberg, J.; Norman, B.; Arzoumanian, Z. Whale Shark ID. info@wildme.org. 2020. Available online: https://lila.science/datasets/whale-shark-id (accessed on 3 November 2024).
- Gao, J.; Burghardt, T.; Andrew, W.; Dowsey, A.W.; Campbell, N.W. Cows2021. 2021. Available online: https://doi.org/10.5523/bris.4vnrca7qw1642qlwxjadp87h7 (accessed on 3 November 2024).
- Andrew, W.; Gao, J.; Mullan, S.; Campbell, N.; Dowsey, A.W.; Burghardt, T. OpenCows2020. 2020. Available online: https://doi.org/10.5523/bris.10m32xl88x2b61zlkkgz3fml17 (accessed on 3 November 2024).
- Andrew, W.; Greatwood, C.; Burghardt, T. AerialCattle2017, University of Bristol. 2017. Available online: https://doi.org/10.5523/bris.3owflku95bxsx24643cybxu3qh (accessed on 3 November 2024).
- Andrew, W.; Greatwood, C.; Burghardt, T. FriesianCattle2017, University of Bristol. 2017. Available online: https://doi.org/10.5523/bris.2yizcfbkuv4352pzc32n54371r (accessed on 3 November 2024).
- Andrew, W.; Hannuna, S.; Campbell, N.; Burghardt, T. FriesianCattle2015, University of Bristol. 2016. Available online: https://doi.org/10.5523/bris.wurzq71kfm561ljahbwjhx9n3 (accessed on 3 November 2024).
- Kern, D.; Schiele, T.; Klauck, U.; Ingabire, W. Chicks4FreeID. 2024. Available online: https://huggingface.co/datasets/dariakern/Chicks4FreeID (accessed on 3 November 2024).
- Lu, W.; Zhao, Y.; Wang, J.; Zheng, Z.; Feng, L.; Tang, J. MammalClub. 2023. Available online: https://github.com/WJ-0425/MammalClub (accessed on 3 November 2024).
- He, Z.; Qian, J.; Yan, D.; Wang, C.; Xin, Y. Animal reidentification Algorithm for Posture Diversity. In Proceedings of the ICASSP 2023—2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Rhodes Island, Greece, 4–10 June 2023; pp. 1–5. [Google Scholar] [CrossRef]
- He, Z. Multi-Pose Dog Dataset. 2023. Mendeley Data, V1. Available online: https://data.mendeley.com/datasets/v5j6m8dzhv/1 (accessed on 3 November 2024).
- Dataset for: PolarBearVidID: A Video-Based Re-Identification Benchmark Dataset for Polar Bears. Available online: https://zenodo.org/records/7564529 (accessed on 3 November 2024).
- Kuncheva, L.I.; Williams, F.; Hennessey, S.L.; Rodríguez, J.J. A Benchmark Database for Animal reidentification and Tracking. In Proceedings of the 2022 IEEE 5th International Conference on Image Processing Applications and Systems (IPAS), Genova, Italy, 5–7 December 2022; Volume 5, pp. 1–6. [Google Scholar] [CrossRef]
- Trust, B.P.C. Hyiena ID. Panthera Pardus CSV Custom Export. Retrieved from African Carnivore Wildbook 2022-04-28. 2022. Available online: https://lila.science/datasets/hyena-id-2022/ (accessed on 3 November 2024).
- Trust, B.P.C. Leopard ID. Panthera Pardus CSV Custom Export. Retrieved from African Carnivore Wildbook 2022-04-28. 2022. Available online: https://lila.science/datasets/leopard-id-2022/ (accessed on 3 November 2024).
- Nepovinnykh, E.; Eerola, T.; Biard, V.; Mutka, P.; Niemi, M.; Kunnasranta, M.; Kälviäinen, H. SealID: Saimaa Ringed Seal reidentification Dataset. Sensors 2022, 22, 7602. [Google Scholar] [CrossRef]
- Papafitsoros, K.; Adam, L.; Čermák, V.; Picek, L. SeaTurtleID: A novel long-span dataset highlighting the importance of timestamps in wildlife re-identification. arXiv 2022, arXiv:2211.10307. [Google Scholar] [CrossRef]
- Li, S.; Fu, L.; Sun, Y.; Mu, Y.; Chen, L.; Li, J.; Gong, H. Individual dairy cow identification based on lightweight convolutional neural network. PLoS ONE 2021, 16, 13. [Google Scholar] [CrossRef]
- Miele, V.; Dussert, G.; Spataro, B.; Chamaillé-Jammes, S.; Allainé, D.; Bonenfant, C. Revisiting animal photo-identification using deep metric learning and network analysis. Methods Ecol. Evol. 2021, 12, 863–873. [Google Scholar] [CrossRef]
- Miele, V.; Dussert, G.; Spataro, B.; Chamaillé-Jammes, S.; Allainé, D.; Bonenfant, C. Giraffe Dataset. 2020. Available online: https://plmlab.math.cnrs.fr/vmiele/animal-reid/ (accessed on 3 November 2024).
- Wang, L.; Ding, R.; Zhai, Y.; Zhang, Q.; Tang, W.; Zheng, N.; Hua, G. iPanda-50. 2021. Available online: https://github.com/iPandaDateset/iPanda-50 (accessed on 3 November 2024).
- Haurum, J.B.; Karpova, A.; Pedersen, M.; Bengtson, S.H.; Moeslund, T.B. AAU Zebrafish Re-Identification Dataset. 2020. Available online: https://www.kaggle.com/datasets/aalborguniversity/aau-zebrafish-reid (accessed on 3 November 2024).
- Guo, S.; Xu, P.; Miao, Q.; Shao, G.; Chapman, C.A.; Chen, X.; He, G.; Fang, D.; Zhang, H.; Sun, Y.; et al. AFD. Mendeley Data, Version 2. 2020. Available online: https://doi.org/10.17632/z3x59pv4bz.2 (accessed on 3 November 2024).
- Li, S.; Li, J.; Tang, H.; Qian, R.; Lin, W. ATRW (Amur Tiger Re-identification in the Wild). 2020. Available online: https://lila.science/datasets/atrw (accessed on 3 November 2024).
- Dlamini, N.; Zyl, T.L.v. Lion Face Dataset. Mara Masia Project, Kenya. 2020. Available online: https://github.com/tvanzyl/wildlife_reidentification/ (accessed on 3 November 2024).
- Trotter, C.; Atkinson, G.; Sharpe, M.; Richardson, K.; McGough, A.S.; Wright, N.; Burville, B.; Berggren, P. NDD20: A large-scale few-shot dolphin dataset for coarse and fine-grained categorisation. arXiv 2020, arXiv:2005.13359. [Google Scholar]
- Dlamini, N.; Zyl, T.L.v. Nyala Dataset. South African Nature Reserves. 2020. Available online: https://github.com/tvanzyl/wildlife_reidentification/ (accessed on 3 November 2024).
- Ferreira, A.C.; Silva, L.R.; Renna, F.; Brandl, H.B.; Renoult, J.P.; Farine, D.R.; Covas, R.; Doutrelant, C. Deep learning-based methods for individual recognition in small birds. Methods Ecol. Evol. 2020, 11, 1072–1085. [Google Scholar] [CrossRef]
- Mougeot, G.; Li, D.; Jia, S. Dog Face Dataset. 2019. Available online: https://github.com/GuillaumeMougeot/DogFaceNet (accessed on 3 November 2024).
- Lin, T.Y.; Kuo, Y.F. Cat Individual Images. 2018. Available online: https://www.kaggle.com/datasets/timost1234/cat-individuals (accessed on 3 November 2024).
- Schneider, J.; Murali, N.; Taylor, G.; Levine, J. Can Drosophila melanogaster tell who’s who? PLoS ONE 2018, 13, e0205043. [Google Scholar] [CrossRef]
- Dataset for: Can Drosophila Melanogaster Tell Who’s Who? Available online: https://borealisdata.ca/dataset.xhtml?persistentId=doi:10.5683/SP2/JP4WDF (accessed on 3 November 2024).
- Witham, C.L. MacaqueFaces. 2018. Available online: https://github.com/clwitham/MacaqueFaces (accessed on 3 November 2024).
- Parham, J.; Crall, J.; Stewart, C.; Berger-Wolf, T.; Rubenstein, D. Great Zebra and Giraffe Count ID. info@wildme.org. 2017. Available online: https://lila.science/datasets/great-zebra-giraffe-id (accessed on 3 November 2024).
- Freytag, A.; Rodner, E.; Simon, M.; Loos, A.; Kühl, H.S.; Denzler, J. Chimpanzee Faces in the Wild. Acknowledgements: Tobias Deschner, Laura Aporius, Karin Bahrke, Zoo Leipzig. 2016. Available online: https://github.com/cvjena/chimpanzee_faces (accessed on 3 November 2024).
- Holmberg, J.; Norman, B.; Arzoumanian, Z. Estimating population size, structure, and residency time for whale sharks Rhincodon typus through collaborative photo-identification. Endanger. Species Res. 2009, 7, 39–53. [Google Scholar] [CrossRef]
- Labelbox. “Labelbox”. 2024. Available online: https://labelbox.com (accessed on 3 November 2024).
- Cubuk, E.D.; Zoph, B.; Shlens, J.; Le, Q. RandAugment: Practical Automated Data Augmentation with a Reduced Search Space. In Advances in Neural Information Processing Systems; Larochelle, H., Ranzato, M., Hadsell, R., Balcan, M., Lin, H., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2020; Volume 33, pp. 18613–18624. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. 2021. Available online: https://openreview.net/forum?id=YicbFdNTTy (accessed on 8 May 2012).
- TorchVision: PyTorch’s Computer Vision Library. 2016. Available online: https://github.com/pytorch/vision (accessed on 3 November 2024).
- Lightly (Software Version 1.5.2). 2020. Available online: https://github.com/lightly-ai/lightly/blob/master/CITATION.cff (accessed on 3 November 2024).
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 11–17 October 2021; pp. 9992–10002. [Google Scholar] [CrossRef]
- Wightman, R. PyTorch Image Models. 2019. Available online: https://github.com/rwightman/pytorch-image-models (accessed on 3 November 2024).
- Deng, J.; Guo, J.; Xue, N.; Zafeiriou, S. ArcFace: Additive Angular Margin Loss for Deep Face Recognition. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–19 June 2019. [Google Scholar]
- Čermák, V.; Picek, L.; Adam, L.; Papafitsoros, K. MegaDescriptor-L-384. 2024. Available online: https://huggingface.co/BVRA/MegaDescriptor-L-384 (accessed on 3 November 2024).
- Oquab, M.; Darcet, T.; Moutakanni, T.; Vo, H.; Szafraniec, M.; Khalidov, V.; Fernandez, P.; Haziza, D.; Massa, F.; El-Nouby, A.; et al. DINOv2: Learning Robust Visual Features without Supervision. arXiv 2024, arXiv:2304.07193. [Google Scholar]
- Radford, A.; Kim, J.W.; Hallacy, C.; Ramesh, A.; Goh, G.; Agarwal, S.; Sastry, G.; Askell, A.; Mishkin, P.; Clark, J.; et al. Learning Transferable Visual Models From Natural Language Supervision. PMLR 2021, 139, 8748–8763. [Google Scholar]
- Wu, Z.; Xiong, Y.; Yu, S.X.; Lin, D. Unsupervised Feature Learning via Non-parametric Instance Discrimination. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 3733–3742. [Google Scholar] [CrossRef]
- Detlefsen, N.S.; Borovec, J.; Schock, J.; Jha, A.H.; Koker, T.; Liello, L.D.; Stancl, D.; Quan, C.; Grechkin, M.; Falcon, W. TorchMetrics—Measuring Reproducibility in PyTorch. J. Open Source Softw. 2022, 7, 4101. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.P.; Sander, J.; Xu, X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. In Proceedings of the 2nd International Conference on Knowledge Discovery and Data Mining (KDD), Portland, OR, USA, 2–4 August 1996; pp. 226–231. [Google Scholar]
- Schubert, E.; Sander, J.; Ester, M.; Kriegel, H.P.; Xu, X. DBSCAN revisited, revisited: Why and how you should (still) use DBSCAN. Acm Trans. Database Syst. (Tods) 2017, 42, 19:1–19:21. [Google Scholar] [CrossRef]
- Nielsen, F. Introduction to HPC with MPI for Data Science; Hierarchical Clustering; Springer: Berlin/Heidelberg, Germany, 2016; Chapter 8; pp. 195–211. [Google Scholar]
| Year | Publ. | Dataset | IDs | Species | Annot. | Avail. at |
|---|---|---|---|---|---|---|
| ours | Chicks4FreeID | 50, 2, 2 | chicken, duck, rooster | 1215, 40, 15 | [64] | |
| 2024 | [36] | SeaTurtleID2022 | 438 | sea turtle | 8729 | [48] |
| 2023 | [24] | Mammal Club (IISD) | 218 | 11 terrestrial mammal species * | 33,612 | [65] |
| 2023 | [66] | Multi-pose dog dataset | 192 | dog | 1657 | [67] |
| 2023 | [40] | PolarBearVidID | 13 | polar bear * | 138,363 | [68] |
| 2023 | [44] | Sea Star Re-ID | 39, 56 | common starfish, Australian cushion star | 1204, 983 | [49] |
| 2022 | [69] | Animal-Identification-from-Video | 58, 26, 9 | pigeon *, pig *, Koi fish * | 12,671, 6184, 1635 | [47] |
| 2022 | n.a. | Beluga ID | 788 | beluga whale | 5902 | [50] |
| 2022 | n.a. | Happywhale | 15,587 | 30 different species of whales and dolphins | 51,033 | [51] |
| 2022 | n.a. | Hyiena ID | 256 | spotted hyena | 3129 | [70] |
| 2022 | n.a. | Leopard ID | 430 | African leopard | 6805 | [71] |
| 2022 | [72] | SealID | 57 | Saimaa ringed seal | 2080 | [52] |
| 2022 | [73] | SeaTurtleIDHeads | 400 | sea turtle | 7774 | [53] |
| 2022 | n.a. | Turtle Recall | 100 | sea turtle | 2145 | [54] |
| 2021 | [74] | Cow Dataset | 13 | cow | 3772 | [3] |
| 2021 | [5] | Cows2021 | 182 | Holstein-Friesian cattle * | 13,784 | [59] |
| 2021 | [75] | Giraffe Dataset | 62 | giraffe | 624 | [76] |
| 2021 | [13] | iPanda-50 | 50 | giant panda | 6874 | [77] |
| 2020 | [34] | AAU Zebrafish Dataset | 6 | zebrafish * | 6672 | [78] |
| 2020 | [45] | Animal Face Dataset | 1040 | 41 primate species | 102,399 | [79] |
| 2020 | [32] | ATRW | 92 | Amur tiger * | 3649 | [80] |
| 2020 | [29] | Lion Face Dataset | 94 | lion | 740 | [81] |
| 2020 | [82] | NDD20 | 44, 82 | bottlenose and white-beaked dolphin, white-beaked dolphin (underwater) * | 2201, 2201 | [55] |
| 2020 | [29] | Nyala Data | 237 | nyala | 1942 | [83] |
| 2020 | [6] | OpenCows2020 | 46 | Holstein-Friesian cattle * | 4736 | [60] |
| 2019 | [84] | Bird individualID | 30, 10, 10 | sociable weaver, great tit, zebra finch | 51,934 | [46] |
| 2019 | [30] | Dog Face Dataset | 1393 | dog | 8363 | [85] |
| 2018 | [28] | Cat Individual Images | 518 | cat | 13,536 | [86] |
| 2018 | [87] | Fruit Fly Dataset | 60 | fruit fly * | 2,592,000 | [88] |
| 2018 | n.a. | HumpbackWhaleID | 5004 | humpback whale | 15,697 | [56] |
| 2018 | [26] | MacaqueFaces | 34 | rhesus macaque * | 6280 | [89] |
| 2017 | [4] | AerialCattle2017 | 23 | Holstein-Friesian cattle * | 46,340 | [61] |
| 2017 | [4] | FriesianCattle2017 | 89 | Holstein-Friesian cattle * | 940 | [62] |
| 2017 | [33] | GZGC | 2056 | plains zebra and Masai giraffe | 6925 | [90] |
| 2016 | [27] | C-Tai | 78 | chimpanzee | 5078 | [91] |
| 2016 | [27] | C-Zoo | 24 | chimpanzee | 2109 | [91] |
| 2016 | [2] | FriesianCattle2015 | 40 | Holstein-Friesian cattle * | 377 | [63] |
| 2015 | n.a. | Right Whale Recognition | 447 | North Atlantic right whale | 4544 | [57] |
| 2011 | [35] | StripeSpotter | 45 | plains and Grevy’s zebra | 820 | [35] |
| 2009 | [92] | Whale Shark ID | 543 | whale shark | 7693 | [58] |
| Feature Extractor | Training | Epochs | Classifier | mAP | Top-1 | Top-5 |
|---|---|---|---|---|---|---|
| MegaDescriptor [43] | pretrained, frozen | - | k-NN | |||
| MegaDescriptor [43] | pretrained, frozen | - | linear | |||
| MegaDescriptor [43] | pretrained, fine-tuned | 200 | k-NN | |||
| MegaDescriptor [43] | pretrained, fine-tuned | 200 | linear |
| Feature Extractor | Training | Epochs | Classifier | mAP | Top-1 | Top-5 |
|---|---|---|---|---|---|---|
| Swin Transformer [98] | from scratch | 200 | k-NN | |||
| Swin Transformer [98] | from scratch | 200 | linear | |||
| Vision Transformer [95] | from scratch | 200 | k-NN | |||
| Vision Transformer [95] | from scratch | 200 | linear |
| Feature Extractor | Training | Epochs | Classifier | mAP | Top-1 | Top-5 |
|---|---|---|---|---|---|---|
| MegaDescriptor [43] | pretrained, frozen | - | k-NN | |||
| MegaDescriptor [43] | pretrained, frozen | - | linear | |||
| MegaDescriptor [43] | pretrained, fine-tuned | 200 | k-NN | |||
| MegaDescriptor [43] | pretrained, fine-tuned | 200 | linear | |||
| Swin Transformer [98] | from scratch | 200 | k-NN | |||
| Swin Transformer [98] | from scratch | 200 | linear | |||
| Vision Transformer [95] | from scratch | 200 | k-NN | |||
| Vision Transformer [95] | from scratch | 200 | linear |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kern, D.; Schiele, T.; Klauck, U.; Ingabire, W. Towards Automated Chicken Monitoring: Dataset and Machine Learning Methods for Visual, Noninvasive Reidentification. Animals 2025, 15, 1. https://doi.org/10.3390/ani15010001
Kern D, Schiele T, Klauck U, Ingabire W. Towards Automated Chicken Monitoring: Dataset and Machine Learning Methods for Visual, Noninvasive Reidentification. Animals. 2025; 15(1):1. https://doi.org/10.3390/ani15010001
Chicago/Turabian StyleKern, Daria, Tobias Schiele, Ulrich Klauck, and Winfred Ingabire. 2025. "Towards Automated Chicken Monitoring: Dataset and Machine Learning Methods for Visual, Noninvasive Reidentification" Animals 15, no. 1: 1. https://doi.org/10.3390/ani15010001
APA StyleKern, D., Schiele, T., Klauck, U., & Ingabire, W. (2025). Towards Automated Chicken Monitoring: Dataset and Machine Learning Methods for Visual, Noninvasive Reidentification. Animals, 15(1), 1. https://doi.org/10.3390/ani15010001







