Simple Summary
In brief, the mitochondrial genome of L. ikeae has a total length of 16,498 bp and encodes 13 PCGs, 22 transfer RNA genes, two ribosomal RNA genes, and a D-loop (control) region. Gene rearrangement is not observed. The mitochondrial genome of L. ikeae exhibits an AT preference, with AT skew > 0 and GC skew < 0 across the entire genome. The phylogenetic relationships of Sicydiinae based on 13 mitochondrial PCG sequences are Sicydium + (Stiphodon + (Sicyopus + Lentipes)) + Sicyopterus, indicating that Sicydium, Sicyopterus, Lentipes, and Stiphodon are all monophyletic groups.
Abstract
We sequenced and analyzed the complete mitochondrial genome of Lentipes ikeae and explored the phylogenetic relationships among Sicydiinae based on mitochondrial genome sequences. The complete mitochondrial genome sequence of L. ikeae was determined using the Illumina HiSeq X Ten sequencing platform, and the gene structural characteristics and base composition were analyzed. Based on the mitochondrial genome sequences of 28 Sicydiinae species published in GenBank and mitochondrial protein-coding genes (PCGs), Acanthogobius flavimanus (Gobionellinae) was selected as an outgroup to construct phylogenetic trees of Sicydiinae using the maximum likelihood and Bayesian inference methods. The mitochondrial genome of L. ikeae (GenBank number: OP764680) has a total length of 16,498 bp and encodes 13 PCGs, 22 transfer RNA genes, two ribosomal RNA genes, and a D-loop (control) region. Gene rearrangement is not observed. The mitochondrial genome of L. ikeae exhibits an AT preference, with AT skew > 0 and GC skew < 0 across the entire genome. The phylogenetic relationships of Sicydiinae based on 13 mitochondrial PCG sequences are Sicydium + (Stiphodon + (Sicyopus + Lentipes)) + Sicyopterus, indicating that Sicydium, Sicyopterus, Lentipes, and Stiphodon are all monophyletic groups.
1. Introduction
The complexity of fishery ecosystems requires an accurate understanding of species. However, due to their different growth environments and developmental stages, aquatic organisms often exhibit different morphological characteristics, such as “the same species but different forms” and “different species and the same forms”, as well as commonly existing hidden taxonomic units, which complicates traditional morphological research. The morphology of gobies is diverse and widely distributed [1,2]. Their morphology rapidly differentiates and forms within an extremely short period of time, which is also common in other groups. It is difficult to reconstruct the phylogenetic relationships of gobies using general molecular markers. Therefore, it is extremely important to clarify the evolutionary relationships between them.
Lentipes ikeae Keith, Hubert, Busson & Hadiaty, 2014 belonging to the order Gobiiformes, family Gobiidae, subfamily Sicydiinae [1]. They are mainly distributed in Cisolok, Kab Sukabumi, Java, and Indonesia [1,3] and occur in fast-flowing mountain streams with high gradients and small yet clear, oxygen-rich streams with rocky bottoms, typically at an altitude of 310–488 m. L. ikeae is an omnivorous species that accepts frozen red worms and mainly eats shrimp and water fleas, and even algae when there are no other food sources. In its combat mode, it has bright colors and important ornamental value, and it is a water quality indicator species [2,4].
The mitochondrial genome can autonomously replicate under the control of nuclear genes, and it is characterized by maternal inheritance, a small size, and a fast evolution rate. Further, it can be easily PCR-amplified. With the continuous development of sequencing technology, mitochondrial genomes have been widely used to study the phylogeny of various fish species and serve as important markers for classification and molecular evolution [5,6,7]. In recent years, many researchers have used mitochondrial genome data to analyze the phylogeny of Sicydiinae [8,9,10]. The mitochondrial genome sequences of 28 species of Sicydiinae have been published in the GenBank database, and the most complete sequencing data cover five genera of Sicydiinae.
In this study, we sequenced and analyzed the complete mitochondrial genome sequence of L. ikeae and compared, analyzed the protein-coding gene (PCG) sequences in the mitochondrial genomes of 28 Sicydiinae species, and constructed a phylogenetic tree based on them. We systematically discuss the phylogenetic relationships among Sicydiinae, laying a foundation for future research on the phylogenetic relationships in this subfamily.
2. Materials and Methods
2.1. Mitochondrial Genome Sequencing and Assembly
Lentipes ikeae was collected in July 2021 at the New World of Flowers, Birds, Fish, and Insects in Fangcun, Liwan District, Guangzhou City, Guangdong Province, China (23°3′47.69″ N, 113°12′19.42″ E). The fish specimens were soaked in anhydrous ethanol and stored at −20 °C in the fish specimen library of the Department of Ecology, Jinan University, Guangzhou City, Guangdong Province, China. Total genomic DNA was extracted from back muscle tissues using the improved CTAB extraction protocol [11] and sent to Biozero Biotechnology Co., Ltd. (Shanghai, China) for 350-bp small-fragment library construction and high-throughput sequencing. PE150 sequencing (paired-end 150 bp: we performed separate sequencing of DNA samples to generate two reads with a length of 150 bp) was conducted on the Illumina HiSeq X Ten sequencing platform and the sequencing data subjected to quality control; effective read segments larger than 10 GB were retained. The read segments from each sample were assembled de novo using SPAdes v3.0.0 to obtain the complete mitochondrial genome sequence [12,13].
2.2. Mitochondrial Genome Sequence Annotation and Comparative Genome Analysis
FASTA files of the L. ikeae mitochondrial genome sequence were imported into MITOS (http://mitos2.bioinf.uni-leipzig.de/index.py, accessed on 1 January 2022) for preliminary annotation [14]. PCG and ribosomal (r)RNA gene sequences were compared with those of related species and corrected using Genesis v4.8.5 [15]. tRNAscan-SE v2.0 was used to predict transfer (t)RNA genes and their secondary structures [16]. The L. ikeae mitochondrial genome’s AT content, AT skew, GC skew, and relative synonymous codon usage (RSCU) were calculated in PhyloSuite v1.2.1 [17]. Base composition skewness was calculated as follows: AT skew = (A − T)/(A + T), GC skew = (G − C)/(G + C).
2.3. Phylogenetic Analysis
To fully explore the evolutionary position of L. ikeae in Sicydiinae, phylogenetic analysis was conducted using mitochondrial sequence data from 28 representative Sicydiinae species reported in GenBank and the complete mitochondrial whole genome of L. ikeae. Acanthogobius flavimanus (GenBank accession MW271007; Gobionellinae) was selected as the outgroup; phylogenetic trees were constructed on the basis of a 13 PCG sequence dataset. For the PCGs, we used MAFFT for multiple sequence alignment [18] and Gblocks [19] to remove vacancies and fuzzy alignment sites. The multiple sequence alignment results of individual genes were combined using SequenceMatrix [20] to obtain a dataset of the 13 PCGs. Dataset saturation was evaluated using DAMBE v5.0 [21], which revealed that all datasets were non-saturated (Iss < lss.cSym or lss.cAsym, p < 0.05). We used PartitionFinder2 [22] to select the evolutionary model corresponding to the optimal dataset partition. Phylogenetic analysis was conducted on the PhyloSuite platform. A maximum likelihood (ML) phylogenetic tree was constructed using IQ-TREEv1.6.8 [23], with 50,000 bootstraps to evaluate the branching node reliability, and the bootstrap value of each branch was calculated. We used MrBayes v3.2.6 [24] to construct a Bayesian inference (BI) phylogenetic tree. Four independent Markov chains were set to run simultaneously for 200 million generations, with sampling once every 1000 generations. When the effective sampling size was ≥200 and the average standard deviation of the split frequency was ≤0.01, assuming that all runs had reached saturation and that the MrBayes results had converged, 25% of the aging samples were discarded, the remaining samples were used to construct a unified tree, and the Bayesian posterior probability values for each node were calculated. We used iTOL (https://itol.embl.de/, accessed on 1 January 2022) [25] for the beautification of the final phylogenetic tree.
3. Results
3.1. Mitochondrial Genome Structure
The complete mitochondrial genome of L. ikeae encodes 37 genes (13 PCGs, 22 tRNA genes, and two rRNA genes) and a circular DNA molecule composed of a control region, with a total length of 16,498 bp (Figure 1). The difference in the mitochondrial genome length between L. ikeae and the other three species (16,496–16,499 bp) in this genus is very small. One PCG and eight tRNA genes are located on the light (L-)strand, and 28 genes on the heavy (H-)strand. Gene rearrangement is not observed. The L. ikeae mitochondrial genome genes are tightly arranged, and there are six gene overlaps and 11 gene intervals with lengths of 1–7 bp. There is a 34-bp gap between the tRNA-Asn and tRNA-Cys genes (Table 1). Among the 13 PCGs, only COI has the start codon GTG, whereas the other genes have the ATG start codon. Except for ATP6, COII, COIII, Cytb, ND3, and ND4, which have an incomplete termination codon T or TA, the other PCGs have a TAA/TAG termination codon. The 16S rRNA and 12S rRNA genes are 1682 and 850 bp, respectively, in size. The non-coding D-loop region is located between the tRNA-Pro and tRNA-Phe genes and has a length of 843 bp.
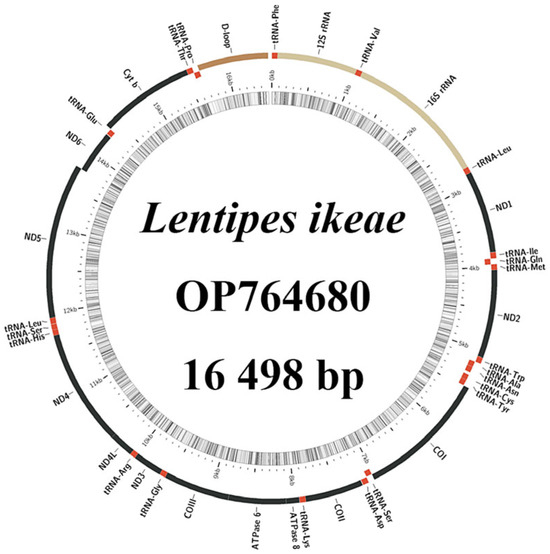
Figure 1.
Lentipes ikeae mitochondrial genome structure.

Table 1.
Organization of the Lentipes ikeae mitochondrial genome.
3.2. Mitochondrial Genome Base Preference and Relative Synonymous Codon Usage Frequency
The complete mitochondrial genome of L. ikeae has a clear AT preference, with total AT content of 54.9%. The mitochondrial genomes of the other three species in this genus also have a clear AT preference (54.9–55.0%). The AT content of the PCGs and tRNA and rRNA genes is 54.3%, 55.4%, and 54.9%, respectively, indicating that different positions in the mitochondrial genome of L. ikeae have different base usage preferences. The value of the AT skew of the mitochondrial genome is 0.035 and that of GC skew is −0.264, indicating that the mitochondrial genome of L. ikeae preferentially uses A and C bases. We found significant differences in the AT content of different coding sites in the PCGs, with the AT content being significantly higher in the second and third sites than in the first site (Table 2). The high AT content was also reflected in the use of relatively synonymous codons in the PCGs. Codons rich in G and C bases, such as CCG, ACG, GCG, and UCG, had RSCU values below 1 and a relatively low usage frequency, whereas codons rich in A and T bases, such as ACA and CAA, were the most commonly used (Figure 2). The RSCU values of different codons were found to greatly vary, with a preference for the use of codons rich in A and T bases and codons ending with A and T bases in the PCGs of the L. ikeae mitochondrial genome.

Table 2.
Nucleotide composition of the Lentipes ikeae mitochondrial genome.
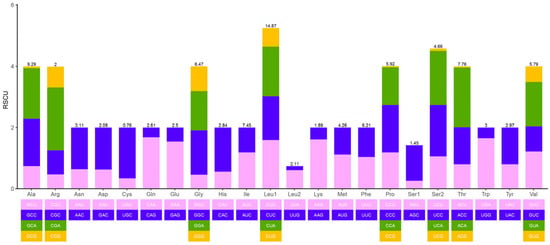
Figure 2.
RSCU in PCGs in the mitochondrial genome of Lentipes ikeae. Different colors correspond to different third codons.
3.3. Phylogenetic Relationships
The topological structures of the ML and BI trees based on the 13 PCG dataset were basically the same (Figure 3), with both trees supporting that Sicydium, Sicyopterus, Lentipes, and Stiphodon are monophyletic groups, and Lentipes, Sicyopus, and Stiphodon are sister groups. Sicyopus and Lentipes first form a sister group and then form a branch with Stiphodon. L. ikeae and (Lentipes palawanirufus + Lentipes kijimuna) first form a branch and then cluster with Lentipes bunagaya, supporting the taxonomic status of L. ikeae. The phylogenetic relationships among Sicydiinae are as follows: Sicydium + (Stiphodon + (Sicyopus + Lentipes)) + Sicyopterus.
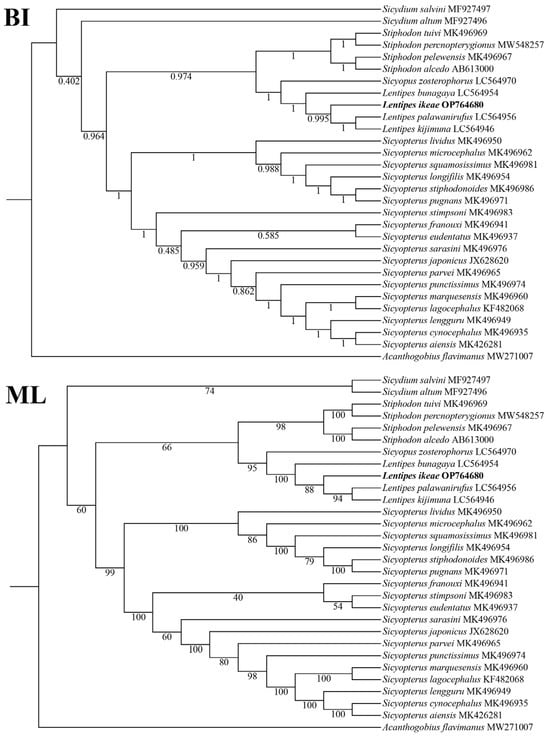
Figure 3.
Phylogenetics relationships among Sicydiinae species based on nucleotide sequences of 13 mitochondrial PCGs.
4. Discussion
The mitochondrial genome of L. ikeae has a total length of 16,498 bp and encodes 22 tRNA genes, 13 PCGs, two rRNAs, and one D-loop. Other Lentipes mitochondrial genomes have similar structures [4]. The A+T base content (54.9%) is greater than that of G+C bases (45.1%), revealing an AT preference, as observed in other fish, such as Oliotius Kottelat [26] and Rhinogobius [27].
In the mitochondrial genome of L. ikeae, PCGs are present on both the H- and L-strands. Brown [28] reported that the PCGs on the H-strand are incompletely protected because the H-strand is often in a hydrolyzed, single-stranded state. Like in Oliotius Kottelat [26], the majority of PCGs in the L. ikeae mitochondrial genome are located on the H-strand, rendering them prone to hydrolysis and oxidation. The ND6 gene located on the L-strand is substantially more stable, reflecting the diversity and importance of ND6 genes.
L. ikeae has 22 tRNA genes, ranging in length from 67 bp to 76 bp and totaling 15,57 bp, similar to Rhinogobius [27] and Lentipes [4]. The mitochondrial genome of L. ikeae contains two rRNAs, 16S rRNA and 12S rRNA, which are not isolated or overlapping with adjacent genes. Repetitive sequences, partially inserted sequences, and sequences contained in the mitochondrial genome are all characteristics of species evolution [5].
In the phylogenetic trees, L. ikeae clustered together with three other species in the genus, Lentipes palawanirufus, Lentipes kijimuna, and Lentipes bunagaya, and showed the closest genetic relationship with Sicyopus zosterophorus. The monophyletic nature of the four genera Sicydium, Sicyopterus, Lentipes, and Stiphodon was well supported. Related genes in mitochondrial genomes are currently widely used in phylogenetic research and species classification. Kim et al. [29] constructed a phylogenetic tree based on mitochondrial 12S rRNA genes to study the evolutionary status of Acanthogobius hasta. With the widespread use of molecular biology methods, domestic and foreign researchers are increasingly studying the phylogenetics of Gobiidae fish. Agorreta et al. [30] analyzed the phylogenetic relationships of 222 European Gobioidei fish species. Thacker et al. [31] confirmed that Trachinoidei is the sister lineage of Gobioidei and studied the systematic distribution of Gobioidei in Acanthomorpha [32]. In general, the evolutionary speed of mitochondrial genes is greater than that of species, and different geographical environments and lifestyle habits can lead to DNA variations in species. To fully unravel the evolution of the L. ikeae mitochondrial genome, further research on the mitochondrial genes is needed.
Author Contributions
C.-H.S.: investigation, formal analysis, writing—original draft, writing—review and editing. Y.-L.G.: funding acquisition, writing—review and editing. D.-W.L.: formal analysis, writing—review and editing. H.-W.D.: writing—review and editing. C.-H.L.: conceptualization, writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
The present study was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD); the Response of Plankton Community Dynamics to Eutrophication in Beijiang and Dongjiang River Basins (PM-zx703-202204-176); and the study on extinction process and driving factors of harmful algae in Beijiang and Dongjiang rivers (PM-zx421-202209-346).
Institutional Review Board Statement
All specimens in this study were collected in accordance with Chinese laws. The collection and sampling of the specimens were reviewed and approved by the Animal Ethics Committee of Nanjing Forestry University (Approval Code: NFU2018-2023187, Approval Date: 2022.07.01). All experiments were conducted with respect for animal welfare and care. The study complied with the CBD and Nagoya protocols and with the ARRIVE guidelines (https://arriveguidelines.org, accessed on 1 January 2022).
Informed Consent Statement
Not applicable.
Data Availability Statement
The complete mitochondrial genome sequence and annotations of Lentipes ikeae are available in GenBank with accession number OP764680.
Acknowledgments
We are very grateful to the editor and reviewers for critically evaluating the manuscript and providing constructive comments for its improvement.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Keith, P.; Hadiaty, R.K.; Hubert, N.; Busson, F.; Lord, C. Three new species of Lentipes from Indonesia (Gobiidae). Cybium Rev. Int. Ichtyol. 2014, 38, 133–146. [Google Scholar]
- Dahruddin, H.; Hutama, A.; Busson, F.; Sauri, S.; Hanner, R.; Keith, P.; Hadiaty, R.; Hubert, N. Revisiting the ichthyodiversity of Java and Bali through DNA barcodes: Taxonomic coverage, identification accuracy, cryptic diversity and identification of exotic species. Mol. Ecol. Resour. 2017, 17, 288–299. [Google Scholar] [CrossRef]
- Hadiaty, R.K.; Rahardjo, M.F.; Allen, G.R. Small islands and coral reef Ichthyofaunas as the basic data for the fisheries resource management. J. Iktiologi Indones. 2019, 19, 167–186. [Google Scholar] [CrossRef]
- Maeda, K.; Kobayashi, H.; Palla, H.P.; Shinzato, C.; Koyanagi, R.; Montenegro, J.; Nagano, A.J.; Saeki, T.; Kunishima, T.; Mukai, T.; et al. Do colour-morphs of an amphidromous goby represent different species? Taxonomy of Lentipes (Gobiiformes) from Japan and Palawan, Philippines, with phylogenomic approaches. Syst. Biodivers. 2021, 19, 1080–1112. [Google Scholar] [CrossRef]
- Boore, L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, V.; Rego, N.; Naya, H.; García, B. First complete mitochondrial genome of the South American annual fish Austrolebias charrua (Cyprinodontiformes: Rivulidae): Peculiar features among cyprinodontiforms mitogenomes. BMC Genom. 2015, 16, 879. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.P.; Miya, M.; Mabuchi, K.; Nichida, M. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 2016, 17, 719. [Google Scholar] [CrossRef] [PubMed]
- Thacker, C.E. Molecular phylogeny of the gobioid fishes (Teleostei: Perciformes: Gobioidei). Mol. Phylogenet. Evol. 2003, 26, 354–368. [Google Scholar] [CrossRef]
- Keith, P.; Lord, C.; Lorion, J.; Watanabe, J.S.; Couloux, A.; Dettai, A. Phylogeny and biogeography of Sicydiinae (Teleostei: Gobiidae) inferred from mitochondrial and nuclear genes. Marine Biol. 2011, 158, 311–326. [Google Scholar] [CrossRef]
- Taillebois, L.; Castelin, M.; Lord, C.; Chabarria, R.; Dettaï, A.; Keith, P. New Sicydiinae phylogeny (Teleostei: Gobioidei) inferred from mitochondrial and nuclear genes: Insights on systematics and ancestral areas. Mol. Phylogenet. Evol. 2014, 70, 260–271. [Google Scholar] [CrossRef]
- Cota-Sánchez, J.H.; Remarchuk, K.; Ubayasena, K. Ready-to-use DNA extracted with a CTAB method adapted for herbarium specimens and mucilaginous plant tissue. Plant Mol. Biol. Rep. 2006, 24, 161–167. [Google Scholar] [CrossRef]
- Sun, C.H.; Liu, H.Y.; Min, X.; Lu, C.H. Mitogenome of the little owl Athene noctua and phylogenetic analysis of Strigidae. Int. J. Biol. Macromol. 2020, 151, 924–931. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, S.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. TRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jacovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.H. DAMBE5: A comprehensive software package for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2013, 30, 1720–1728. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3 2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Sun, C.H.; Sun, P.Y.; Lao, Y.L.; Wu, T.; Zhang, Y.N.; Huang, Q.; Zhang, Q. Mitogenome of a monotypic genus, Oliotius Kottelat, 2013 (Cypriniformes: Cyprinidae): Genomic characterization and phylogenetic position. Gene 2023, 851, 147035. [Google Scholar] [CrossRef]
- Wang, D.; Dai, C.; Li, Q.; Li, Y.; Liu, Z. Complete mitochondrial genome and phylogenic analysis of Rhinogobius cliffordpopei (Perciformes, Gobiidae). Mitochondrial DNA Part B Resour. 2019, 4, 2473–2474. [Google Scholar] [CrossRef]
- Brown, W.M.; Prager, E.M.; Wang, A.; Wilson, A.C. Mitochondrial DNA sequences of primates: Tempo and mode of evolution. J. Mol. Evol. 1982, 18, 225–239. [Google Scholar] [CrossRef]
- Kim, I.C.; Kweon, H.S.; Kim, Y.J.; Kim, C.B.; Gye, M.C.; Lee, W.O.; Lee, Y.S.; Lee, J.S. The complete mitochondrial genome of the javeline goby Acanthogobius hasta (Perciformes, Gobiidae) and phylogenetic considerations. Gene 2004, 336, 147–153. [Google Scholar] [CrossRef]
- Agorreta, A.; San Mauro, D.S.; Schliewen, U.; Van Tassell, J.L.; Kovačić, M.; Zardoya, R.; Rüber, L. Molecular phylogenetics of Gobioidei and phylogenetic placement of European gobies. Mol. Phylogenet. Evol. 2013, 69, 619–633. [Google Scholar] [CrossRef]
- Thacker, C.E.; Satoh, T.P.; Katayama, E.; Harrington, R.C.; Eytan, R.I.; Near, T.J. Molecular phylogeny of Percomorpha resolves Trichonotus as the sister lineage to Gobioidei (Teleostei: Gobiiformes) and confirms the polyphyly of Trachinoidei. Mol. Phylogenet. Evol. 2015, 93, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Thacker, C.E. Phylogeny of Gobioidei and placement within Acanthomorpha, with a new classification and investigation of diversification and character evolution. Copeia 2009, 2009, 93–104. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).