Simple Summary
Post-weaning diarrhea is a critical health and welfare issue in swine production systems as the transition from sow milk to solid feed is a stressful event that is accompanied by microbiome community disturbances that can require antimicrobial treatment or other individualized dietary interventions. This study investigated how feeding sows live yeast during gestation and lactation affects the growth and microbiomes of their offspring in the post-weaning period, without directly feeding live yeast to offspring. The offspring of yeast-fed sows displayed altered growth performance and gut microbiome composition when compared to offspring born to sows fed a control diet. The microbiomes of offspring born to yeast-fed sows contained more taxa associated with beneficial fermentative processes, indicating that the manipulation of offspring microbiomes through maternal diet is a viable method for conferring beneficial microbiome characteristics to offspring in swine systems.
Abstract
The supplementation of live yeast in pig diets is common in the post-weaning phase due to its prebiotic and probiotic effects, but little is known regarding the potential of feeding live yeast to gestating or lactating sows for transferring such benefits to their offspring through maternal programming. The objective of this study was to investigate the effects of live yeast supplementation in sow diets during late gestation and lactation on their reproductive performance and its impact on offspring performance and gut microbiomes in the post-weaning period. Three dietary treatments were imposed on 92 mixed-parity sows during late gestation and lactation based upon the inclusion level of live yeast in corn/soybean meal-based diets: Control (0% yeast), Low (0.1% yeast), and High (0.5% yeast). Nursery pigs in the Low group displayed the highest feed intake in the post-weaning period and greater total gain and average daily gain in comparison to pigs in the High group. The gut microbiomes of nursery pigs differed in composition according to maternal dietary treatment groups at days 4 and 28 post weaning, highlighting higher abundances of bacterial genera typically associated with fermentation roles in the gut microbiomes of offspring of yeast-fed sows. These results indicate that the supplementation of live yeast in sow diets, depending on the inclusion level, may result in beneficial performance and specific microbiome traits for their offspring in the post-weaning period.
1. Introduction
Maternal programming refers to the process by which maternal characteristics can predetermine or influence the growth and development of offspring. In swine production systems, several maternal factors have been associated previously with the growth performance, health status, and microbiome composition of offspring [1,2,3,4]. Due to the intrinsic ties between gut microbial communities and host health, maternal programming strategies that aim to target gut microbiome composition in their offspring represent a novel approach to favorably influence their growth and development. One of the most compelling factors influencing piglet performance and microbiomes in early life is maternal diet. Maternal diets during gestation and lactation can affect the initial seeding of piglet gut microbiomes early in life, potentially impacting their physiological performance.
Previous researchers demonstrated that the manipulation of maternal diets altered the composition of gut microbiomes in suckling pigs [5,6,7,8]. However, whether these changes persist past weaning into the nursery phase of production is unclear. The gut microbiome of newborn piglets diversifies quickly in the pre-weaning period, with increasing microbial diversity throughout each life phase in swine production systems [9,10,11,12]. A distinct shift in gut microbiome composition occurs at weaning in tandem with the shift from dietary oligosaccharides found in sow milk to more complex carbohydrates found in plant-based solid feed [1,13,14]. This shift is also accompanied by increased susceptibility to health challenges and post-weaning diarrhea (PWD) in the weeks immediately after weaning, which can decrease growth performance in mild cases and cause death in severe cases [15,16]. Consequently, the weaning and nursery phases of swine production are often the target of investigations involving dietary feed additives to prevent or ameliorate these challenges [17].
Supplementation with live yeast has been studied extensively in food animal nutrition due to its prebiotic and probiotic potential. Previous research reported the effects of live yeast supplementation on microbiome composition and favorable immune function in food animals [18,19,20]. Live yeast can act as a probiotic through various potential mechanisms including the competitive exclusion of pathogen growth, the production of antimicrobial peptides, and the activation of host immune pathways [18]. Beneficial prebiotic effects associated with live yeast are attributable to mannan oligosaccharides and β-glucans found in yeast cell walls [19,21,22] that can stimulate beneficial bacteria, eliciting a wide variety of immunogenic effects and resulting in increased growth performance [22,23,24,25,26]. The supplementation of diets for suckling and nursery pigs with live yeast supported increased average daily gain (ADG) and improved fecal consistency [27,28,29]. However, potential effects on post-weaning pig performance by supplementing only sow diets with live yeast have not been reported. Therefore, the objective of this study was to investigate the effects of feeding live yeast to gestating and lactating sows on their reproductive performance and growth performance of their offspring in lactation and throughout the nursery period. In addition, we studied the effects of maternal yeast feeding on the gut microbiome of weaned offspring.
2. Materials and Methods
2.1. Animals, Housing, and Diets
Mixed-parity, crossbred sows (n = 92; Topigs NorsvinTN 70 females) from three contemporary farrowing groups were used in this experiment. Sows were assigned based on parity to one of three dietary treatments which included: 0 (Control, C), 0.1% (Low, L) or 0.5% (High, H) of a live yeast additive (Saccharomyces cerevisiae Sc47, Lesaffre Yeast Corporation, Milwaukee, WI, USA). The diets were mixed using pre-blends of live yeast to ensure a uniform distribution of the yeast product throughout the final diets. Pre-blends for the Control group did not include the yeast feed additive. Dietary treatments were imposed from approximately day 85 of gestation (Table 1) and continued through lactation (Table 2) until piglets were weaned (at about 19 days old). Sows were fed their assigned gestation diet (1.8 kg/head/day) from day 85 to day 110 of gestation on an as-fed basis. At day 111 of gestation, sows were fed their assigned lactation diet (3.6 kg/head/day) until parturition. Sows were allowed ad libitum access to their assigned lactation diet beginning the first day postpartum until piglets were weaned, with diets formulated to meet or exceed nutrient recommendations set by the National Research Council [30] for gestating and lactating sows. Metabolizable energy (ME) was calculated from the ME density of the ingredients [30].

Table 1.
Ingredient composition and calculated energy and nutrient content of sow gestation diets (as-fed basis).

Table 2.
Ingredient composition and calculated energy and nutrient content of sow lactation diets (as-fed basis).
Sows were split into 3 contemporary farrowing groups containing about 30 sows each, with dietary treatment groups assigned randomly within each farrowing group. Across all 3 farrowing groups, treatments were balanced for sow parity. The sows were moved from group pens in a straw-bedded hoop gestation barn on day 78 of gestation to individual stalls with slatted floors in a conventional confinement farrowing barn. Litter size, piglets born live per litter, and stillborn piglets were recorded at birth for all sows prior to cross-fostering. Sow body weight, backfat depth at the last rib, and body condition were recorded on day 85 of gestation, the day prior to the expected farrowing date, and on the day before weaning. Body condition was scored using a caliper device at the last rib as previously described [31]. Parity, gestation length, and lactation length were recorded for all sows. Cross-fostering was allowed only within dietary treatment groups, excluding focal sows that were selected for further microbiome and immune analyses. Piglets born to focal sows were not cross-fostered. Piglets were processed and weighed within 24 h of birth. Piglet processing included tail docking, iron injections, clipping needle teeth, and the castration of males. Piglets were weighed individually within 24 h of birth and the day before weaning.
Pigs from the third farrowing group (n = 240; 80 per maternal dietary treatment) were selected randomly for further performance data collection throughout the nursery period. The pigs were balanced by litter, body weight, and sex during nursery allotment. At weaning, the pigs were moved to a research nursery barn in groups based on the maternal dietary treatment group. The treatment groups were kept separate from each other by maintaining one empty pen between the treatment groups. Pigs were housed in pens according to the maternal dietary treatment (10 pigs/pen; 8 pens per maternal dietary treatment). The pigs were provided 0.28 m2 of floor space per pig. Each pen was equipped with a stainless-steel feeder with five feeding spaces, a water cup (Drink-o-Mat, Vittetoe Inc., Keota, IA, USA), and slotted plastic flooring over a manure pit. The pigs were weighed individually at the end of each of the 4 dietary phases (d 4, d 14, d 28, d 42). All nursery pigs were allowed ad libitum access to water and a common four-phase feeding program that was devoid of a yeast feed additive. The phase one diet was a proprietary pelleted diet, with antibiotics excluded (First Feed®, VitaPlus Corp., Madison, WI, USA), and was fed from d 0 to d 4. The phase two diet was fed from d 4 to d 14 and was a corn and soybean meal-based proprietary blend (Launch®, VitaPlus Corp., Madison, WI, USA). Phases 3 and 4 were corn and soybean meal-based (Table 3) and were fed from days 14 to 28 post weaning and days 28–42 post weaning, respectively.

Table 3.
Ingredient composition and calculated energy and nutrient content of phase 3 and phase 4 nursery diets.
2.2. Microbiome Profiling Methods
Fecal samples were collected from all 240 pigs at d 4 and 28 (n = 480) post weaning to coincide with the end of dietary phase 1 and phase 3, respectively, and to profile gut microbiome composition without interference from transitions between dietary phases. Only samples from pigs that did not receive antibiotic treatment or experience adverse health conditions were used for microbiome profiling. In total, 180 samples (60/maternal dietary treatment group) from each time-point (n = 360 total) were selected randomly for microbiome analysis.
All fecal samples for microbiome analyses were collected with sterile cotton swabs and sterile collection tubes. Fecal swabs were collected from pigs by inserting the cotton swab tips just inside the rectum. All samples were immediately placed on dry ice after collection and then stored at −80 °C until DNA extraction. DNA was extracted from the swabs using PowerSoil Pro DNA extraction kits (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Negative controls, consisting of a sterile blank cotton swab, were created for each individual extraction kit and set of reagents.
Sequence data were generated through targeting the V4 variable region of the 16S rRNA bacterial gene on the MiSeq sequencing platform using the primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) and dual-indexing library preparation [32]. Raw sequence data were processed using a custom bioinformatics pipeline to remove primer sequences and perform quality control and filtering, as previously described [2]. Filtered sequence data were then processed using the QIIME2 pipeline [33] and its DADA2 plugin [34]. The Silva database [35] was used to taxonomically identify amplicon sequence variants (ASVs) and estimate their abundances in each sample.
An analysis of the processed 16S rRNA sequence data was performed using various packages in the R statistical software (version 4.2.2) [36]. Sequence data from negative controls generated for each extraction kit were used to screen for potential contamination using the R microDecon package [37]. ASVs that were identified as contaminants were filtered out of the corresponding sequence datasets, and potential sequencing artifacts were removed using the labdsv package by eliminating ASVs present in fewer than 3 samples and with fewer than 5 reads across all the dataset [38]. Beta diversity and principal coordinate analyses were performed using the vegan package [39]. The ANCOMBC package [40] was used to identify discriminant genera among the dietary treatment groups. Taxa were considered statistically significant discriminators for the High and Low treatments if q < 0.05 after false discovery rate (FDR) adjustments and if they showed log-fold change values in relative abundance of at least (+/−) 0.5 compared to the Control group.
2.3. Statistical Analyses
Generalized linear mixed models were built for all performance comparisons using the R lme4 package [41]. Models for sows included treatment and farrowing room as fixed effects and farrowing group, parity, and litter sire as random effects. Models for piglets in the pre-weaning period included maternal treatment, farrowing room, and sex as fixed effects and farrowing group, sire breed of sow, and maternal parity as random effects. Models in the nursery period considered treatment, sex, and time as fixed effects and sire breed of sow, maternal parity, and pen as random effects. A p-value of <0.05 was considered a significant result. The Tukey–Kramer post hoc test for multiple comparisons was used to differentiate among treatment means. The standard error was reported as a pooled standard error for each comparison. All statistical analyses for comparisons of alpha and beta diversity were based on nonparametric Kruskal–Wallis tests and permutational multivariate analyses of variance (PERMANOVA).
3. Results
3.1. Sow and Piglet Performance
Sow parity, gestation length, and lactation length did not differ among the dietary treatment groups (Table 4). Dietary treatment had no effect on the total feed intake or average daily feed intake (ADFI) during lactation. Sow body weight, backfat depth at the last rib, and caliper score were unaffected by yeast supplementation at each sampling time-point (Table 5). Similarly, dietary treatment had no effect on the total piglets born, liveborn piglets, stillborn piglets per litter, or the pre-weaning mortality of piglets (Table 6). The average birth weight, weaning weight, and average daily gain (ADG) did not differ among piglets born to sows fed different dietary treatments (Table 7).

Table 4.
Sow parity, gestation and lactation length, and lactation feed consumption for each dietary treatment group.

Table 5.
Sow body weight, backfat depth, and caliper scores for each dietary treatment.

Table 6.
Effect of dietary yeast treatments on litter size and piglet mortality.

Table 7.
Effect of maternal dietary yeast on piglet body weight and pre-weaning growth rate.
3.2. Performance and Gut Microbiome Composition of Nursery Pigs
Seven pigs were removed from the study due to death or adverse health conditions. Due to an outbreak of suspected Streptococcus suis, which is endemic in the research swine herd, and observed poor overall pig health throughout the nursery barn around day 16 of the experiment, all pigs received amoxicillin continuously through drinking water from days 16 to 20 of the experiment.
Nursery pigs born to sows fed the high-yeast diet gained less weight throughout the entire nursery period when compared to those born to sows fed the low-yeast diet. Total weight gain did not differ between nursery pigs born to sows in the Control and the Low groups (Figure 1a). Similarly, the overall ADG for the entire nursery period was higher for pigs born to sows fed the low-yeast diet when compared to pigs born to sows fed the high-yeast diet (Figure 1b). However, nursery pigs belonging to the Low treatment group displayed higher ADFI than both the Control or High groups throughout the nursery period (Figure 1c). Gain-to-feed ratios did not differ among the treatment groups (Figure 1d).

Figure 1.
Effects of maternal dietary yeast treatment on nursery pig body weight (a), ADG (b), ADFI (c), and G:F ratios (d). ADG was calculated individually, while ADFI was calculated on a pen basis. G:F ratios were calculated by dividing individual ADG by pen ADFI. Error bars represent standard error pooled across treatment groups for each comparison. Differing letter superscripts denote differences (p < 0.05) among treatment groups in repeated measures models.
To visualize and explore potential differences in the microbial community composition of nursery pig gut microbiomes, the top 20 genera in pigs from each treatment group were identified at each sampling time-point and displayed in Figure 2. The relative abundances of each genus were averaged for each maternal treatment group at each time-point. The top 20 genera were consistent across maternal treatment groups at both days 4 (Figure 2a) and 28 (Figure 2b) post weaning. However, differences in the relative abundances of several taxa were observed among the maternal treatment groups (Kruskal–Wallis, p < 0.05).
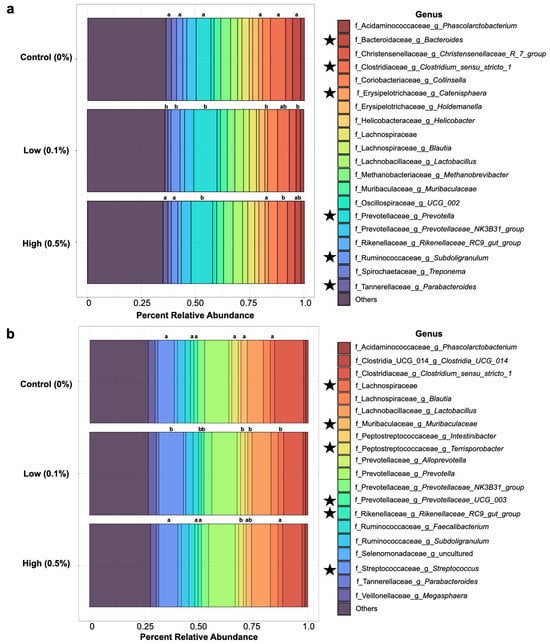
Figure 2.
The top 20 most abundant genera in nursery pig gut microbiomes at day 4 post weaning (a) and day 28 post weaning (b), averaged by maternal dietary treatment. Abundance is expressed as percent relative abundance as a proportion of the total. Listed taxa are identified at the genus level or represent an unidentified genus within the listed family. Star symbols next to the legend represent genera that were identified as different in terms of their abundances among treatment groups. Significance is denoted above each corresponded colored bar by letter superscripts, with differing letters representing differences (p < 0.05) in relative abundances.
The overall gut microbiome community composition differed among nursery pigs based on their maternal treatment group at day 4 post weaning (PERMANOVA; R2 = 0.02, p = 0.001; Figure 3a). Differences in pairwise PERMANOVA comparisons between the control group and the yeast-fed groups for microbiome community composition were also observed, with the largest distinctions observed for the Low group (Control vs. Low: R2 = 0.02, p = 0.001; Control vs. High: R2 = 0.01, p = 0.03, Low vs. High: R2 = 0.02, p = 0.008). At day 28 post weaning, the effect (R2) of the maternal diet on pig gut microbiome composition increased slightly (PERMANOVA; R2 = 0.04, p = 0.001; Figure 3b), with significant pairwise differences between the Control and the High/Low groups maintained as well (Control vs. Low: R2 = 0.05, p = 0.001; Control vs. High: R2 = 0.02, p = 0.001, Low vs. High: R2 = 0.03, p = 0.001).
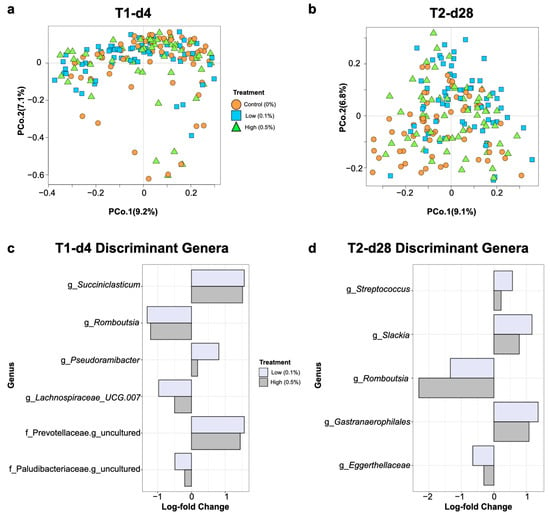
Figure 3.
Nursery pig gut microbiome composition at the ASV level at days 4 (a) and 28 (b) post weaning according to Bray–Curtis dissimilarities. Each shape represents an individual pig, with colors and shapes denoting maternal dietary treatments. Discriminant genera among treatments for nursery pigs were identified by ANCOMBC analysis at each sampling time-point (c,d). X axis values represent log-fold changes (q < 0.05) in relative abundances of listed genera or an unidentified genus within the listed family. Positive and negative log-fold changes indicate enriched or depleted taxa in low- or high-yeast treatments, respectively, compared with the control.
Multivariate ordination of microbial composition appeared to be constrained by unknown factors, particularly at day 4 post weaning as seen in Figure 3a. Hence, the analysis of compositions of microbiomes with bias correction (ANCOMBC) method [40] was implemented for reducing the effects of inherent biases introduced by amplicon sequencing at each time-point. This method revealed that the gut microbiomes of nursery pigs born to yeast-fed sows displayed differential abundances of several genera in comparison to those born to Control sows at days 4 and 28 post weaning, regardless of the yeast inclusion rate. The genera Romboutsia, Lachnospiraceae UCG.007, and an unidentified genus in the family Paludibacteraceae were identified as discriminant genera for the Control group at day 4 post weaning, while the genera Pseudoramibacter, Succiniclasticum, and an unidentified genus in the family Prevotellaceae were identified as discriminant genera (q < 0.05) for yeast-fed offspring (Figure 3c). At day 28 post weaning, the gut microbiomes of the offspring of yeast-fed sows were discriminated by higher abundances (q < 0.05) of the genera Streptococcus, Slackia, and Gastranaerophilales, while the offspring of Control sows were discriminated by higher abundances (q < 0.05) of the genus Romboutsia and an unidentified genus in the family Eggerthellaceae (Figure 3d).
4. Discussion
Here, we show that maternal programming based on the live yeast supplementation of sow diets during late gestation and lactation is associated with differences in growth performance and gut microbiome composition of offspring after weaning. In the post-weaning period, nursery pigs born to sows fed the low (0.1%)-yeast diet displayed greater overall ADFI when compared to pigs in both other maternal dietary treatment groups. Pigs in the Low group also displayed greater body weight gain and ADG in the nursery compared to pigs raised by sows fed the high (0.5%)-yeast diet, suggesting that higher inclusion rates of live yeast are not necessarily more beneficial.
Maternal dietary yeast supplementation did not result in alterations in sow or piglet performance in the pre-weaning period. Similarly, researchers utilizing live yeast products in sow feed in late gestation and lactation observed no effects on sow or piglet performance but did note increased levels of IgG in the colostrum and milk of yeast-fed sows [42,43]. Contrary to these results, other researchers supplementing sow diets with live yeast observed improved sow reproductive performance [44]. The variation in the reported benefits of maternal yeast supplementation in swine systems highlights the need for further research in this area, as well as inherent challenges due to variability among different strains of yeast products.
Previously, researchers demonstrated increased ADG of pigs in the post-weaning period when they were directly fed the same yeast product as used in the study reported herein [28]. The direct supplementation of nursery pigs with the same live yeast product used in our experiment also increased ADG and ADFI in the post-weaning period [45,46]. Similarly, directly feeding other yeast products to nursery pigs increases ADG [29]. Thus, one might easily surmise that feeding live yeast or yeast products directly to nursery pigs would improve growth performance. However, in this experiment, we documented the altered growth performance of nursery pigs through maternal programing, that is, when yeast was included only in maternal diets as opposed to yeast inclusion directly in nursery pig diets. This observation demonstrates an important carryover effect from feeding yeast to dams during gestation and lactation on the post-weaning performance of pigs.
We show that feeding live yeast to sows altered the post-weaning growth performance of their offspring and elicited changes in the gut microbial communities of nursery pigs. The overall structure of the gut microbiome community at days 4 and 28 post weaning differed based upon the maternal dietary treatment, although the magnitude of effects for these comparisons was small. Notably, at day 4 post weaning, pigs born to yeast-fed sows were discriminated by greater abundances of the genus Pseudoramibacter which has been associated with the digestion of fermentable carbohydrates and subsequently short-chain fatty acid (SCFA) production [47,48]. The offspring of yeast-fed sows were also discriminated by an unidentified genus in the family, Prevotellaceae, and the genus Succiniclasticum both of which harbor SCFA production capabilities [49,50,51]. The gut microbiomes of offspring from yeast-fed sows also harbored greater abundances of the genera Prevotella in comparison to the offspring of sows fed the Control diet, which underscores a potential of maternal supplementation with live yeast to impact the fermentative landscape of their offspring gut microbiomes. Specifically, pigs raised by sows fed the low-yeast diet harbored significantly decreased abundances of the genera Catenisphaera and Bacteroides compared with those raised by the Control sows. High abundances of some taxa from the genus Bacteroides in pre-weaning and weaning transition periods have been linked to incidences of post-weaning diarrhea [52]. Increased abundances of these taxa are also associated with the prevalence of enterotoxigenic Escherichia coli infections and post-weaning diarrhea [53].
At day 28 post weaning, pigs born to yeast-fed sows were discriminated by greater abundances of the genera Streptococcus, Slackia, and Gastranaerophilales, compared with the offspring of sows fed the control diet. Although the genera identified as discriminant taxa among the maternal treatment groups differed on days 4 and 28 post weaning, observed differences can likely be attributed to the natural diversification of pig gut microbiomes with increasing age [54,55]. Higher abundances of the genus Slackia at the end of the nursery phase have previously been positively associated with an increased growth performance [54]. Likewise, the genus Streptococcus was significantly more abundant in the gut microbiomes of pigs in the Low yeast group at day 28 post weaning compared to the gut microbiomes of pigs raised by the Control and High sows. Although this genus is composed of commensal species endemic to pig gut microbiomes across life phases [54], it is also composed of species that are potentially pathogenic in swine systems, such as Streptococcus suis [56,57]. However, previous research investigating post-weaning diarrhea noted that the proliferation of pathogenic taxa such as Escherichia coli was associated with marked reductions in abundances of the genus Streptococcus [58,59,60]. Our findings combined with those of previous researchers suggest that feeding yeast to sows may reduce the abundances of potentially pathogenic taxa in the gut microbiomes of their offspring and promote the growth of gut bacteria with potentially beneficial fermentative roles in the post-weaning period, though the exact mechanism is unknown.
5. Study Limitations
From these data, we cannot say with certainty that the supplementation of live yeast in sow diets caused the direct transmission of beneficial microbes from sows to piglets or whether observed differences in post-weaning gut microbiome composition were driven by some other mechanism(s). Additionally, the access of suckling piglets to sow feed was limited but not entirely prevented by the design of sow feeders, so piglets may have consumed small amounts of sow feed. The antibiotic treatment of pigs to maintain pig health and welfare between the sampling time-points may have also influenced nursery pig gut microbiome community composition and diversity. While the adverse health event and subsequent administration of oral antibiotic interfered with attempts to ensure antimicrobials did not alter observations of microbial patterns, this intervention mimics a common scenario and management practice in commercial swine productions systems. Future research in this area should profile markers of inflammation and cytokines in piglets in the pre-weaning period, as well as during the weaning transition period, to determine whether maternal live yeast supplementation has immunomodulatory effects that could potentially improve health status in the post-weaning period. Additionally, future studies should attempt to elucidate the mechanism(s) behind the carryover effects of maternal programming observed in this study, which remain unclear.
6. Conclusions
The supplementation of sow diets during gestation and lactation with 0.1% of live yeast increased the post-weaning ADFI of offspring. Nursery pigs raised by sows fed 0.1% yeast gained more weight and had greater ADG compared to those raised by sows fed 0.5% yeast. The performance of sows and suckling pigs was not affected by feeding live yeast to sows. Live yeast supplementation in sow diets was associated with greater abundances of several genera linked to fermentation and SCFA production, particularly at day 4 post weaning. These results indicate that maternal programming through live yeast supplementation at an inclusion rate of 0.1% in sow diets is a viable method for promoting the growth of potentially beneficial bacteria in piglet microbiomes that persist into the post-weaning period, though these effects must be balanced with observations of increased feed intake in the post-weaning period.
Author Contributions
Conceptualization, K.L., L.J.J., P.E.U. and A.G.; methodology, K.L., L.J.J., P.E.U. and A.G.; data curation, K.L., L.J.J. and P.E.U.; formal analysis, K.L.; writing—original draft preparation, K.L.; writing—review and editing, K.L., L.J.J., P.E.U. and A.G.; funding acquisition, L.J.J., P.E.U. and A.G. All authors have read and agreed to the published version of the manuscript.
Funding
This study was partially supported by funds from the Agricultural Research, Education, Extension and Technology Transfer (AGREETT) and MNDrive Global Food Ventures Programs, both from the University of Minnesota. Partial funding was supplied by Lesaffre Yeast Corporation, Milwaukee, WI, USA.
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Animal Care and Use Committee of the University of Minnesota (protocol # 2009-38494A, approved 26 October 2021).
Informed Consent Statement
The sows involved in this research were owned by the University of Minnesota and were housed at the West Central Research and Outreach Center in Morris, MN, USA.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to a temporary embargo held by the first author while their doctoral dissertation is finalized.
Acknowledgments
We appreciate the technical advice of Joe Loughmiller.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Guevarra, R.B.; Lee, J.H.; Lee, S.H.; Seok, M.-J.; Kim, D.W.; Kang, B.N.; Johnson, T.J.; Isaacson, R.E.; Kim, H.B. Piglet gut microbial shifts early in life: Causes and effects. J. Anim. Sci. Biotechnol. 2019, 10, 1. [Google Scholar] [CrossRef]
- Law, K.; Lozinski, B.; Torres, I.; Davison, S.; Hilbrands, A.; Nelson, E.; Parra-Suescun, J.; Johnston, L.; Gomez, A. Disinfection of Maternal Environments Is Associated with Piglet Microbiome Composition from Birth to Weaning. mSphere 2021, 6, e00663-21. [Google Scholar] [CrossRef]
- Liu, H.; Zeng, X.; Zhang, G.; Hou, C.; Li, N.; Yu, H.; Shang, L.; Zhang, X.; Trevisi, P.; Yang, F.; et al. Maternal milk and fecal microbes guide the spatiotemporal development of mucosa-associated microbiota and barrier function in the porcine neonatal gut. BMC Biol. 2019, 17, 106. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xu, J.; Ren, E.; Su, Y.; Zhu, W. Co-occurrence of early gut colonization in neonatal piglets with microbiota in the maternal and surrounding delivery environments. Anaerobe 2018, 49, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.A.; Davis, E.; Spencer, J.D.; Moser, R.; Rehberger, T. The effect of a Bacillus-based direct-fed microbial supplemented to sows on the gastrointestinal microbiota of their neonatal piglets. J. Anim. Sci. 2013, 91, 3390–3399. [Google Scholar] [CrossRef]
- Cheng, C.; Wei, H.; Xu, C.; Xie, X.; Jiang, S.; Peng, J. Maternal Soluble Fiber Diet during Pregnancy Changes the Intestinal Microbiota, Improves Growth Performance, and Reduces Intestinal Permeability in Piglets. Appl. Environ. Microbiol. 2018, 84, e01047-18. [Google Scholar] [CrossRef] [PubMed]
- Leblois, J.; Massart, S.; Li, B.; Wavreille, J.; Bindelle, J.; Everaert, N. Modulation of piglets’ microbiota: Differential effects by a high wheat bran maternal diet during gestation and lactation. Sci. Rep. 2017, 7, 7426. [Google Scholar] [CrossRef]
- Liu, B.; Zhu, X.; Cui, Y.; Wang, W.; Liu, H.; Li, Z.; Guo, Z.; Ma, S.; Li, D.; Wang, C.; et al. Consumption of Dietary Fiber from Different Sources during Pregnancy Alters Sow Gut Microbiota and Improves Performance and Reduces Inflammation in Sows and Piglets. mSystems 2021, 6, e00591-20. [Google Scholar] [CrossRef]
- Gaire, T.N.; Scott, H.M.; Noyes, N.R.; Ericsson, A.C.; Tokach, M.D.; Menegat, M.B.; Vinasco, J.; Roenne, B.; Ray, T.; Nagaraja, T.G.; et al. Age influences the temporal dynamics of microbiome and antimicrobial resistance genes among fecal bacteria in a cohort of production pigs. Anim. Microbiome 2023, 5, 2. [Google Scholar] [CrossRef]
- Kim, H.B.; Borewicz, K.; White, B.A.; Singer, R.S.; Sreevatsan, S.; Tu, Z.J.; Isaacson, R.E. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet. Microbiol. 2011, 153, 124–133. [Google Scholar] [CrossRef]
- Han, G.G.; Lee, J.-Y.; Jin, G.-D.; Park, J.; Choi, Y.H.; Kang, S.-K.; Chae, B.J.; Kim, E.B.; Choi, Y.-J. Tracing of the fecal microbiota of commercial pigs at five growth stages from birth to shipment. Sci. Rep. 2018, 8, 6012. [Google Scholar] [CrossRef]
- Ke, S.; Fang, S.; He, M.; Huang, X.; Yang, H.; Yang, B.; Chen, C.; Huang, L. Age-based dynamic changes of phylogenetic composition and interaction networks of health pig gut microbiome feeding in a uniformed condition. BMC Vet. Res. 2019, 15, 172. [Google Scholar] [CrossRef]
- Gaio, D.; DeMaere, M.Z.; Anantanawat, K.; Eamens, G.J.; Liu, M.; Zingali, T.; Falconer, L.; Chapman, T.A.; Djordjevic, S.P.; Darling, A.E. Community composition and development of the post-weaning piglet gut microbiome. bioRxiv 2020, 2020.07.20.211326. [Google Scholar] [CrossRef]
- Frese, S.A.; Parker, K.; Calvert, C.C.; Mills, D.A. Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome 2015, 3, 28. [Google Scholar] [CrossRef]
- Rhouma, M.; Fairbrother, J.M.; Beaudry, F.; Letellier, A. Post weaning diarrhea in pigs: Risk factors and non-colistin-based control strategies. Acta Vet. Scand. 2017, 59, 31. [Google Scholar] [CrossRef]
- Fairbrother, J.M.; Nadeau, É.; Gyles, C.L. Escherichia coli in postweaning diarrhea in pigs: An update on bacterial types, pathogenesis, and prevention strategies. Anim. Health Res. Rev. 2005, 6, 17–39. [Google Scholar] [CrossRef]
- Thacker, P.A. Alternatives to antibiotics as growth promoters for use in swine production: A review. J. Anim. Sci. Biotechnol. 2013, 4, 35. [Google Scholar] [CrossRef] [PubMed]
- Broadway, P.R.; Carroll, J.A.; Sanchez, N.C.B. Live Yeast and Yeast Cell Wall Supplements Enhance Immune Function and Performance in Food-Producing Livestock: A Review. Microorganisms 2015, 3, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Shurson, G.C. Yeast and yeast derivatives in feed additives and ingredients: Sources, characteristics, animal responses, and quantification methods. Anim. Feed Sci. Technol. 2018, 235, 60–76. [Google Scholar] [CrossRef]
- Vohra, A.; Syal, P.; Madan, A. Probiotic yeasts in livestock sector. Anim. Feed Sci. Technol. 2016, 219, 31–47. [Google Scholar] [CrossRef]
- Kollár, R.; Reinhold, B.B.; Petráková, E.; Yeh, H.J.C.; Ashwell, G.; Drgonová, J.; Kapteyn, J.C.; Klis, F.M.; Cabib, E. Architecture of the Yeast cell wall: β(1→6)-glucan interconnects mannoprotein, β(1→3)-glucan, and Chitin. J. Biol. Chem. 1997, 272, 17762–17775. [Google Scholar] [CrossRef]
- Spring, P.; Wenk, C.; Connolly, A.; Kiers, A. A review of 733 published trials on Bio-Mos®, a mannan oligosaccharide, and Actigen®, a second generation mannose rich fraction, on farm and companion animals. J. Appl. Anim. Nutr. 2015, 3, e8. [Google Scholar] [CrossRef]
- Halas, V.; Nochta, I. Mannan Oligosaccharides in Nursery Pig Nutrition and Their Potential Mode of Action. Animals 2012, 2, 261–274. [Google Scholar] [CrossRef]
- Miguel, J.C.; Rodriguez-Zas, S.L.; Pettigrew, J.E. Efficacy of a mannan oligosaccharide (Bio-Mos®) for improving nursery pig performance. J. Swine Health Prod. 2004, 12, 296–307. [Google Scholar]
- Vetvicka, V.; Vannucci, L.; Sima, P. The Effects of β-Glucan on Pig Growth and Immunity. Open Biochem. J. 2014, 8, 89–93. [Google Scholar] [CrossRef]
- Volman, J.J.; Ramakers, J.D.; Plat, J. Dietary modulation of immune function by β-glucans. Physiol. Behav. 2008, 94, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Che, L.; Xu, Q.; Wu, C.; Luo, Y.; Huang, X.; Zhang, B.; Auclair, E.; Kiros, T.; Fang, Z.; Lin, Y.; et al. Effects of dietary live yeast supplementation on growth performance, diarrhoea severity, intestinal permeability and immunological parameters of weaned piglets challenged with enterotoxigenic Escherichia coli K88. Br. J. Nutr. 2017, 118, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Kiros, T.G.; Luise, D.; Derakhshani, H.; Petri, R.; Trevisi, P.; D’Inca, R.; Auclair, E.; Kessel, A.G. van Effect of live yeast Saccharomyces cerevisiae supplementation on the performance and cecum microbial profile of suckling piglets. PLoS ONE 2019, 14, e0219557. [Google Scholar] [CrossRef] [PubMed]
- Long, S.; He, T.; Kim, S.W.; Shang, Q.; Kiros, T.; Mahfuz, S.U.; Wang, C.; Piao, X. Live Yeast or Live Yeast Combined with Zinc Oxide Enhanced Growth Performance, Antioxidative Capacity, Immunoglobulins and Gut Health in Nursery Pigs. Animals 2021, 11, 1626. [Google Scholar] [CrossRef]
- National Research Council. Nutrient Requirements of Swine: Eleventh Revised Edition [Internet]; The National Academies Press: Washington, DC, USA, 2012; Available online: https://www.nap.edu/catalog/13298/nutrient-requirements-of-swine-eleventh-revised-edition (accessed on 1 September 2023).
- Knauer, M.; Baitinger, D.J. The Sow Body Condition Caliper. Appl. Eng. Agric. 2015, 31, 175–178. [Google Scholar]
- Gohl, D.M.; Vangay, P.; Garbe, J.; MacLean, A.; Hauge, A.; Becker, A.; Gould, T.J.; Clayton, J.B.; Johnson, T.J.; Hunter, R.; et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol. 2016, 34, 942–949. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- R Core Team R: The R Project for Statistical Computing [Internet]. Available online: https://www.r-project.org/ (accessed on 10 March 2023).
- McKnight, D.T.; Huerlimann, R.; Bower, D.S.; Schwarzkopf, L.; Alford, R.A.; Zenger, K.R. microDecon: A highly accurate read-subtraction tool for the post-sequencing removal of contamination in metabarcoding studies. Environ. DNA 2019, 1, 14–25. [Google Scholar] [CrossRef]
- Roberts, D.W. labdsv: Ordination and Multivariate Analysis for Ecology [Internet]. 2019. Available online: https://CRAN.R-project.org/package=labdsv (accessed on 18 January 2021).
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package [Internet]. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 18 January 2021).
- Lin, H.; Peddada, S.D. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020, 11, 3514. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Zanello, G.; Meurens, F.; Serreau, D.; Chevaleyre, C.; Melo, S.; Berri, M.; D’Inca, R.; Auclair, E.; Salmon, H. Effects of dietary yeast strains on immunoglobulin in colostrum and milk of sows. Vet. Immunol. Immunopathol. 2013, 152, 20–27. [Google Scholar] [CrossRef]
- Jang, Y.D.; Kang, K.W.; Piao, L.G.; Jeong, T.S.; Auclair, E.; Jonvel, S.; D’Inca, R.; Kim, Y.Y. Effects of live yeast supplementation to gestation and lactation diets on reproductive performance, immunological parameters and milk composition in sows. Livest. Sci. 2013, 152, 167–173. [Google Scholar] [CrossRef]
- Peng, X.; Yan, C.; Hu, L.; Huang, Y.; Fang, Z.; Lin, Y.; Xu, S.; Feng, B.; Li, J.; Zhuo, Y.; et al. Live yeast supplementation during late gestation and lactation affects reproductive performance, colostrum and milk composition, blood biochemical and immunological parameters of sows. Anim. Nutr. 2020, 6, 288–292. [Google Scholar] [CrossRef]
- van Heugten, E.; Funderburke, D.W.; Dorton, K.L. Growth performance, nutrient digestibility, and fecal microflora in weanling pigs fed live yeast1. J. Anim. Sci. 2003, 81, 1004–1012. [Google Scholar] [CrossRef]
- Kiros, T.G.; Derakhshani, H.; Pinloche, E.; D’Inca, R.; Marshall, J.; Auclair, E.; Khafipour, E.; Van Kessel, A. Effect of live yeast Saccharomyces cerevisiae (Actisaf Sc 47) supplementation on the performance and hindgut microbiota composition of weanling pigs. Sci. Rep. 2018, 8, 5315. [Google Scholar] [CrossRef]
- Willems, A.; Collins, M.D. Pseudoramibacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2015; pp. 1–8. [Google Scholar] [CrossRef]
- Feng, K.; Wang, Q.; Li, H.; Du, X.; Zhang, Y. Microbial mechanism of enhancing methane production from anaerobic digestion of food waste via phase separation and pH control. J. Environ. Manag. 2021, 288, 112460. [Google Scholar] [CrossRef]
- Wang, C.; Wei, S.; Xu, B.; Hao, L.; Su, W.; Jin, M.; Wang, Y. Bacillus subtilis and Enterococcus faecium co-fermented feed regulates lactating sow’s performance, immune status and gut microbiota. Microb. Biotechnol. 2021, 14, 614–627. [Google Scholar] [CrossRef]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef]
- Ramayo-Caldas, Y.; Mach, N.; Lepage, P.; Levenez, F.; Denis, C.; Lemonnier, G.; Leplat, J.-J.; Billon, Y.; Berri, M.; Doré, J.; et al. Phylogenetic network analysis applied to pig gut microbiota identifies an ecosystem structure linked with growth traits. ISME J. 2016, 10, 2973–2977. [Google Scholar] [CrossRef]
- Dou, S.; Gadonna-Widehem, P.; Rome, V.; Hamoudi, D.; Rhazi, L.; Lakhal, L.; Larcher, T.; Bahi-Jaber, N.; Pinon-Quintana, A.; Guyonvarch, A.; et al. Characterisation of Early-Life Fecal Microbiota in Susceptible and Healthy Pigs to Post-Weaning Diarrhoea. PLoS ONE 2017, 12, e0169851. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, J.; Dong, Z.; Li, G.; Wang, J.; Li, Y.; Wan, D.; Yang, H.; Yin, Y. Effect of Dietary Copper on Intestinal Microbiota and Antimicrobial Resistance Profiles of Escherichia coli in Weaned Piglets. Front. Microbiol. 2019, 10, 484922. [Google Scholar] [CrossRef]
- De Rodas, B.; Youmans, B.P.; Danzeisen, J.L.; Tran, H.; Johnson, T.J. Microbiome profiling of commercial pigs from farrow to finish. J. Anim. Sci. 2018, 96, 1778–1794. [Google Scholar] [CrossRef]
- Wang, X.; Tsai, T.; Deng, F.; Wei, X.; Chai, J.; Knapp, J.; Apple, J.; Maxwell, C.V.; Lee, J.A.; Li, Y.; et al. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome 2019, 7, 109. [Google Scholar] [CrossRef]
- Vangroenweghe, F.A.C.J.; Boone, M. Vaccination with an Escherichia coli F4/F18 Vaccine Improves Piglet Performance Combined with a Reduction in Antimicrobial Use and Secondary Infections Due to Streptococcus suis. Animals 2022, 12, 2231. [Google Scholar] [CrossRef] [PubMed]
- Correa-Fiz, F.; Neila-Ibáñez, C.; López-Soria, S.; Napp, S.; Martinez, B.; Sobrevia, L.; Tibble, S.; Aragon, V.; Migura-Garcia, L. Feed additives for the control of post-weaning Streptococcus suis disease and the effect on the faecal and nasal microbiota. Sci. Rep. 2020, 10, 20354. [Google Scholar] [CrossRef] [PubMed]
- McAllister, J.S.; Kurtz, H.J.; Short, E.C., Jr. Changes in the Intestinal Flora of Young Pigs with Postweaning Diarrhea or Edema Disease. J. Anim. Sci. 1979, 49, 868–879. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Nie, K.; Xu, Y.; Zhang, H.; Xie, F.; Xu, L.; Zhang, Z.; Ding, Y.; Yin, Z.; Zhang, X. Fecal Microbial Structure and Metabolic Profile in Post-Weaning Diarrheic Piglets. Genes 2023, 14, 1166. [Google Scholar] [CrossRef]
- Rhouma, M.; Braley, C.; Thériault, W.; Thibodeau, A.; Quessy, S.; Fravalo, P. Evolution of pig fecal microbiota composition and diversity in response to enterotoxigenic Escherichia coli infection and colistin treatment in weaned piglets. Microorganisms 2021, 9, 1459. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).