Characterization of the Complete Mitochondrial Genome of Schizothorax kozlovi (Cypriniformes, Cyprinidae, Schizothorax) and Insights into the Phylogenetic Relationships of Schizothorax
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Genome Sequencing and Assembly
2.3. Sequence Annotation and Analysis
2.4. Phylogenetic Analysis
3. Results
3.1. Genome Organization and Composition
3.2. Protein-Coding Genes and Codon Usage
3.3. Transfer RNAs, Ribosomal RNAs, and Noncoding Regions
3.4. Phylogenetic Relationships
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goodsell, D.S. Mitochondrion. Biochem. Mol. Biol. Educ. 2010, 38, 134–140. [Google Scholar] [CrossRef]
- Garesse, R.; Vallejo, C.G. Animal mitochondrial biogenesis and function: A regulatory cross-talk between two genomes. Gene 2001, 263, 1–16. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Hwang, D.S.; Lee, W.O.; Lee, J.S. Complete mitochondrial genome of the freshwater gudgeon, Pseudopungtungia nigra (Cypriniformes, Gobioninae). Mitochondrial DNA 2014, 25, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Park, J.; Xi, H.; Lee, G.S.; Kim, I.; Park, J. The complete mitochondrial genome of Ricania speculum (Walker, 1851) (Hemiptera: Ricaniidae): Investigation of intraspecific variations on mitochondrial genome. Mitochondrial DNA Part B 2020, 5, 3814–3816. [Google Scholar] [CrossRef]
- Galtier, N.; Nabholz, B.; Glémin, S.; Hurst, G.D. Mitochondrial DNA as a marker of molecular diversity: A reappraisal. Mol. Ecol. 2009, 18, 4541–4550. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.P.; Miya, M.; Mabuchi, K.; Nishida, M. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 2016, 17, 719. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Nielsen, R.; Hasegawa, M. Models of amino acid substitution and applications to mitochondrial protein evolution. Mol. Biol. Evol. 1998, 15, 1600–1611. [Google Scholar] [CrossRef]
- Zardoya, R.; Meyer, A. Cloning and characterization of a microsatellite in the mitochondrial control region of the African side-necked turtle, Pelomedusa subrufa. Gene 1998, 216, 149–153. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Caccone, A.; Gentile, G.; Burns, C.E.; Sezzi, E.; Bergman, W.; Ruelle, M.; Saltonstall, K.; Powell, J.R. Extreme difference in rate of mitochondrial and nuclear DNA evolution in a large ectotherm, Galápagos tortoises. Mol. Phylogenet. Evol. 2004, 31, 794–798. [Google Scholar] [CrossRef]
- Keith, P.; Lord, C.; Lorion, J.; Watanabe, S.; Tsukamoto, K.; Couloux, A.; Dettai, A. Phylogeny and biogeography of Sicydiinae (Teleostei: Gobiidae) inferred from mitochondrial and nuclear genes. Mar. Biol. 2011, 158, 311–326. [Google Scholar] [CrossRef]
- Shao, R.; Barker, S.C. Mitochondrial genomes of parasitic arthropods: Implications for studies of population genetics and evolution. Parasitology 2007, 134, 153–167. [Google Scholar] [CrossRef]
- Song, N.; Lin, A.; Zhao, X. Insight into higher-level phylogeny of Neuropterida: Evidence from secondary structures of mitochondrial rRNA genes and mitogenomic data. PLoS ONE 2018, 13, e0191826. [Google Scholar] [CrossRef]
- Ding, R.H. The Fishes of Sichuan, China; Sichuan Publishing House of Science and Technology: Chengdu, China, 1994. [Google Scholar]
- Lin, P.C.; Miao, Z.G.; Gao, X.; Liu, H.Z. Length-weight relationships of 11 fish species from the upper Jinsha River, China. J. Appl. Ichthyol. 2015, 31, 223–224. [Google Scholar] [CrossRef]
- Jiang, Z.; Jiang, J.; Wang, Y.; Zhang, E.; Zhang, Y. Red list of China’s vertebrates. Biodiv. Sci. 2016, 24, 500–551. [Google Scholar]
- Chen, Y.; Luo, Q. The fecundity of Schizothorax kozlovi from Wu River. Zool. Res. 1995, 16, 324–342. [Google Scholar]
- Zhang, X.; Dai, Y. Feeding habits and resources protection of Schizothorax kozlovi. J. Hydroecol. 2010, 32, 110–114. [Google Scholar] [CrossRef]
- He, Y.; Wu, X.; Zhu, Y.; Li, H.; Li, X.; Yang, D. Effect of rearing temperature on growth and thermal tolerance of Schizothorax (Racoma) kozlovi larvae and juveniles. J. Therm. Biol. 2014, 46, 24–30. [Google Scholar] [CrossRef]
- He, J.; He, Z.; Yang, D.; Ma, Z.; Chen, H.; Zhang, Q.; Deng, F.; Ye, L.; Pu, Y.; Zhang, M.; et al. Genetic variation in Schizothorax kozlovi Nikolsky in the upper reaches of the Chinese Yangtze River based on genotyping for simplified genome sequencing. Animals 2022, 12, 2181. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Gong, J.; Wu, X.; Zhu, Y.; Yang, D. Population structure of wild Schizothorax kozlovi in the upper Yangtze River based on mtDNA and stable isotopes, and their relationship with ambient temperature. Fishes 2022, 7, 292. [Google Scholar] [CrossRef]
- Wang, Y.; Shang, P.; Dai, Y.; Xu, D.; Dong, Y.; Huang, Z. The complete mitochondrial genome of a new species of the genus Schizothorax from Sichuan, China (Cypriniformes: Cyprinidae). Mitochondrial DNA Part B 2023, 8, 1356–1359. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.F.; Wu, C.Z. The Fishes of the Qinghai-Xizang Plateau; Sichuan Publishing House of Science and Technology: Chengdu, China, 1991. [Google Scholar]
- Wu, Y.F. Systematic studies on the cyprinid fishes of the subfamily schizothoracinae from China. Acta Biologica Plateau Sinica 1984, 3, 119–140. [Google Scholar]
- He, D.; Chen, Y. Biogeography and molecular phylogeny of the genus Schizothorax (Teleostei: Cyprinidae) in China inferred from cytochrome b sequences. J. Biogeogr. 2006, 33, 1448–1460. [Google Scholar] [CrossRef]
- He, D.K.; Chen, Y.F.; Chen, Y.Y.; Chen, Z.M. Molecular phylogeny of the specialized schizothoracine fishes (Teleostei: Cyprinidae), with their implications for the uplift of the Qinghai-Tibetan Plateau. Chin. Sci. Bull. 2004, 49, 39–48. [Google Scholar] [CrossRef]
- Cao, W.X.; Chen, Y.Y.; Wu, Y.F. Origin and evolution of Schizothorax and their relationship with the uplift of the Qinghai-Xizang Plateau. In Ages, Amplitudes and Form Problems during the Uplift of the Qinghai-Xizang Plateau; The Qinghai-Tibet Plateau Comprehensive Scientific Expedition from Chinese Academy of Sciences, Ed.; Science Press: Beijing, China, 1981. [Google Scholar]
- Wu, Y.F.; Tan, Q.J. Characteristics of the fish-fauna of the characteristics of Qinghai-Xizang Plateau and its geological distribution and formation. Acta Zool. Sin. 1991, 37, 135–152. [Google Scholar]
- Chen, Y.F.; Cao, W.X. Schizothoracinae. In Fauna Sinica, Osteichthyes, Cypriniformes II; Yue, P.Q., Ed.; Science Press: Beijing, China, 2000. [Google Scholar]
- Heckel, J.J. Fische aus Caschmir; P.P. Mechitaristen: Wien, Austria, 1838. [Google Scholar]
- Chen, Y.Y. Fauna Sinica, Osteichthyes, Cypriniformes II; Science Press: Beijing, China, 1998. [Google Scholar]
- Bartlett, S.E.; Davidson, W.S. Identification of Thunnus tuna species by the polymerase chain reaction and direct sequence analysis of their mitochondrial cytochrome b genes. Can. J. Fish. Aquat. Sci. 1991, 48, 309–317. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High throughput Sequence Data; Babraham Institute: Cambridge, UK, 2013. [Google Scholar]
- Duan, X.; Dong, X.; Li, J.; Lü, J.; Guo, B.; Xu, K.; Ye, Y. The complete mitochondrial genome of Pilumnopeus Makianus (Brachyura: Pilumnidae), novel gene rearrangements, and phylogenetic relationships of Brachyura. Genes 2022, 13, 1943. [Google Scholar] [CrossRef]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; dePamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De Novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016, 45, e18. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Yang, T.; Xu, G.; Gu, B.; Shi, Y.; Mzuka, H.L.; Shen, H. The complete mitochondrial genome sequences of the Philomycus bilineatus (Stylommatophora: Philomycidae) and phylogenetic analysis. Genes 2019, 10, 198. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Xiang, C.-Y.; Gao, F.; Jakovlić, I.; Lei, H.-P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.-T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Wang, I.C.; Lin, H.-D.; Liang, C.-M.; Huang, C.-C.; Wang, R.-D.; Yang, J.-Q.; Wang, W.-K. Complete mitochondrial genome of the freshwater fish Onychostoma lepturum (Teleostei, Cyprinidae): Genome characterization and phylogenetic analysis. ZooKeys 2020, 1005, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Mar-Silva, A.F.; Arroyave, J.; Díaz-Jaimes, P. The complete mitochondrial genome of the Mexican-endemic cavefish Ophisternon infernale (Synbranchiformes, Synbranchidae): Insights on patterns of selection and implications for synbranchiform phylogenetics. ZooKeys 2022, 1089, 1–23. [Google Scholar] [CrossRef]
- Mao, L.; Zeng, Y.; Li, J.; Zhang, F.; Liu, Y.; Gong, J. The complete mitochondrial genome sequence and phylogenetic analysis of Gnathopogon herzensteini (Cypriniformes, Cyprinidae, Gobioninae). Biologia 2021, 76, 1087–1094. [Google Scholar] [CrossRef]
- Li, W.; Liu, Y.; Xu, Q. Complete mitochondrial genome of Schizothorax gongshanensis (Cypriniformes: Cyprinidae). Mitochondrial DNA Part B 2016, 1, 528–529. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, S.; Kumazawa, Y.; Araki, T.; Himeno, H.; Miura, K.-I.; Watanabe, K. Strand-specific nucleotide composition bias in echinoderm and vertebrate mitochondrial genomes. J. Mol. Evol. 1991, 32, 511–520. [Google Scholar] [CrossRef]
- Cui, L.; Cao, R.; Dong, Y.; Gao, X.; Cen, J.; Lu, S. The first complete mitochondrial genome of the flathead Cociella crocodilus (Scorpaeniformes: Platycephalidae) and the phylogenetic relationships within Scorpaeniformes based on whole mitogenomes. Genes 2019, 10, 533. [Google Scholar] [CrossRef]
- Rustam, D.; Yuan, X.; Zhang, Q.; Han, J. Study on the phylogeny of Schizothoracids based on complete mitochondrial genome. J. Fish. Sci. China 2022, 29, 781–791. [Google Scholar] [CrossRef]
- Chen, Y.; Cheng, Q.; Qiao, H.; Zhu, Y.; Chen, W.; Ren, G. The complete mitochondrial genome sequence of Schizothorax wangchiachii (Cypriniformes: Cyprinidae). Mitochondrial DNA 2013, 24, 353–355. [Google Scholar] [CrossRef]
- Zhang, H.; Fang, W.; Zhao, X.; Jiang, X.; Stroiński, A.; Qin, D. Comparative analysis of the complete mitochondrial genomes of five species of Ricaniidae (Hemiptera: Fulgoromorpha) and phylogenetic implications. Biology 2022, 11, 92. [Google Scholar] [CrossRef]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Ben Slimen, H.; Awadi, A.; Tolesa, Z.G.; Knauer, F.; Alves, P.C.; Makni, M.; Suchentrunk, F. Positive selection on the mitochondrial ATP synthase 6 and the NADH dehydrogenase 2 genes across 22 hare species (genus Lepus). J. Zool. Syst. Evol. Res. 2018, 56, 428–443. [Google Scholar] [CrossRef]
- Pons, J.; Bauzà-Ribot, M.M.; Jaume, D.; Juan, C. Next-generation sequencing, phylogenetic signal and comparative mitogenomic analyses in Metacrangonyctidae (Amphipoda: Crustacea). BMC Genom. 2014, 15, 566. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Brown, W.M.; Boore, J.L. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc. Natl. Acad. Sci. USA 2000, 97, 13738–13742. [Google Scholar] [CrossRef]
- Li, T.; Yang, J.; Li, Y.; Cui, Y.; Xie, Q.; Bu, W.; Hillis, D.M. A mitochondrial genome of Rhyparochromidae (Hemiptera: Heteroptera) and a comparative analysis of related mitochondrial genomes. Sci. Rep. 2016, 6, 35175. [Google Scholar] [CrossRef] [PubMed]
- Varani, G.; McClain, W.H. The G·U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000, 1, 18–23. [Google Scholar] [CrossRef]
- Mao, M.; Gu, J.; Liu, R.; Chen, Y.; Wu, L.; Jiang, Z.; Jiang, J. Analysis of complete mitochondrial genome sequences of Gadus macrocephalus. Acta Hydrobiol. Sin. 2019, 43, 17–26. [Google Scholar] [CrossRef]
- Yu, X.; Tan, W.; Zhang, H.; Jiang, W.; Gao, H.; Wang, W.; Liu, Y.; Wang, Y.; Tian, X. Characterization of the complete mitochondrial genome of Harpalus sinicus and its implications for phylogenetic analyses. Genes 2019, 10, 724. [Google Scholar] [CrossRef]
- Shao, R.; Barker, S.C.; Mitani, H.; Aoki, Y.; Fukunaga, M. Evolution of duplicate control regions in the mitochondrial genomes of Metazoa: A case study with Australasian Ixodes Ticks. Mol. Biol. Evol. 2004, 22, 620–629. [Google Scholar] [CrossRef]
- Zheng, C.; Nie, L.; Wang, J.; Zhou, H.; Hou, H.; Wang, H.; Liu, J. Recombination and evolution of duplicate control regions in the mitochondrial genome of the Asian big-headed turtle, Platysternon megacephalum. PLoS ONE 2013, 8, e82854. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Briolay, J.; Galtier, N.; Brito, R.M.; Bouvet, Y. Molecular phylogeny of Cyprinidae inferred from cytochrome b DNA sequences. Mol. Phylogenet. Evol. 1998, 9, 100–108. [Google Scholar] [CrossRef] [PubMed]
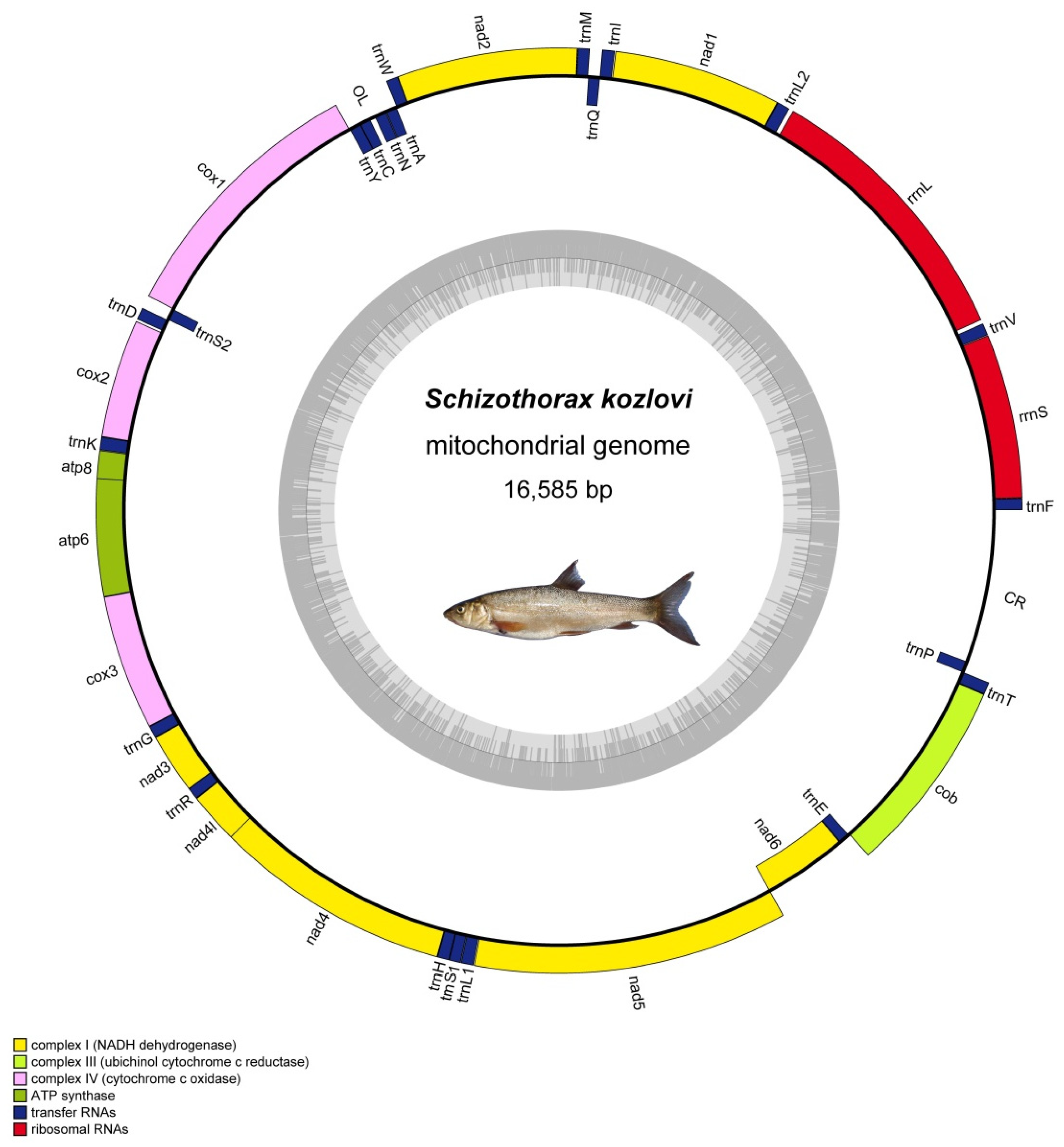
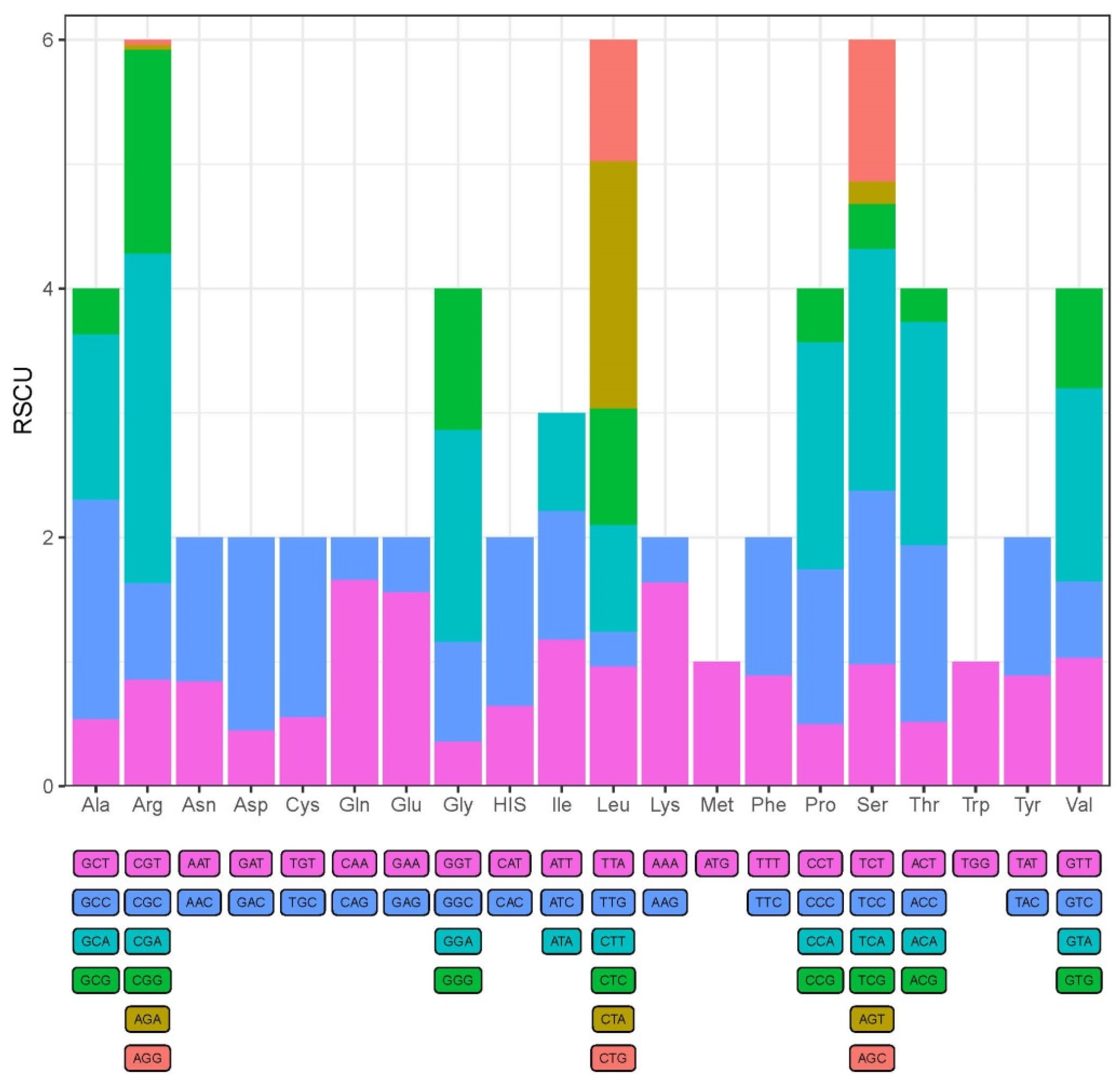
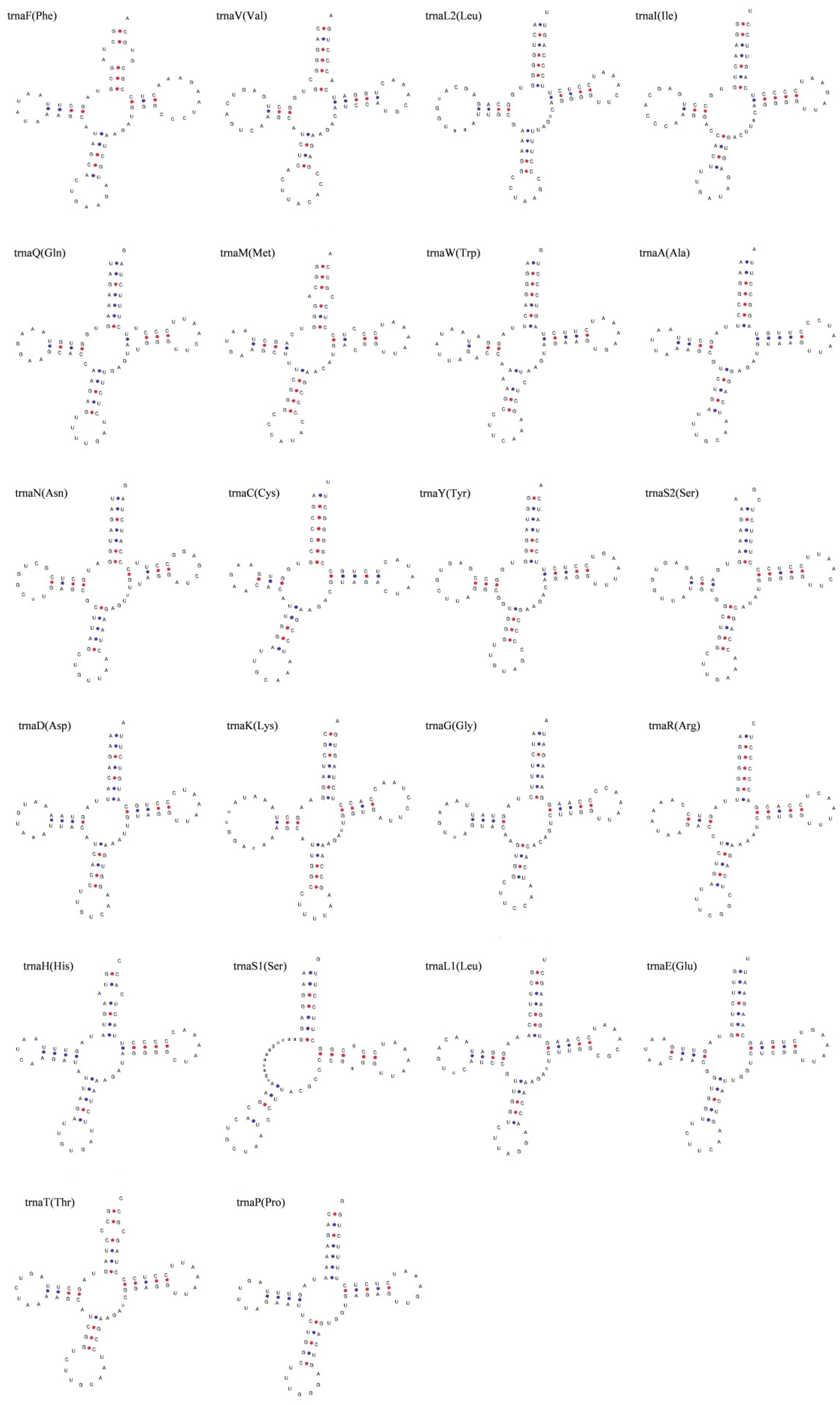
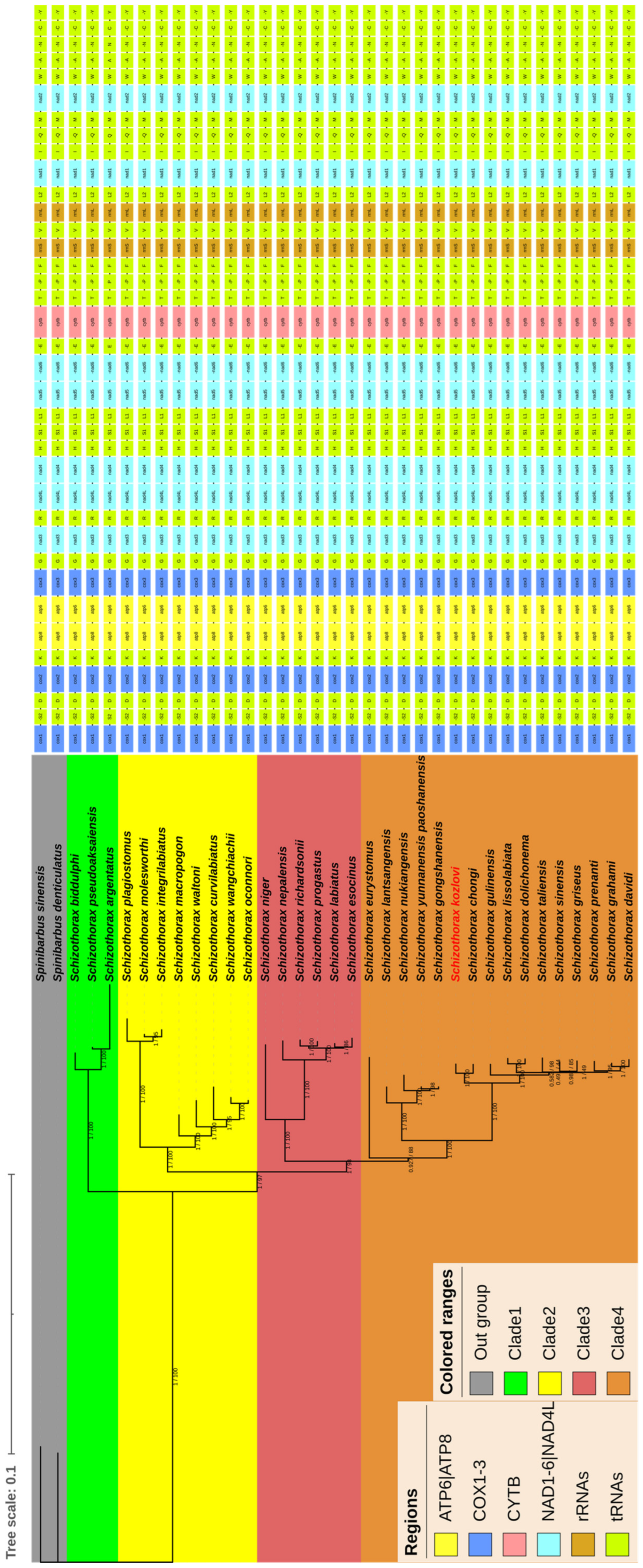
| Species | Size (bp) | Accession Number |
|---|---|---|
| Schizothorax argentatus | 16,587 | NC_061395.1 |
| Schizothorax biddulphi | 16,588 | OR812523.1 |
| Schizothorax chongi | 16,584 | NC_024621.1 |
| Schizothorax curvilabiatus | 16,578 | NC_035994.1 |
| Schizothorax davidi | 16,576 | NC_026205.1 |
| Schizothorax dolichonema | 16,583 | NC_023979.1 |
| Schizothorax esocinus | 16,591 | KT210882.1 |
| Schizothorax eurystomus | 16,590 | ON920824.1 |
| Schizothorax gongshanensis | 16,591 | NC_031803.1 |
| Schizothorax grahami | 16,584 | NC_029708.1 |
| Schizothorax griseus | 16,586 | NC_046462.1 |
| Schizothorax gulinensis | 16,587 | NC_079827.1 |
| Schizothorax integrilabiatus | 16,588 | NC_036746.1 |
| Schizothorax kozlovi | 16,585 | OR416862.1 |
| Schizothorax labiatus | 16,590 | KT944287.1 |
| Schizothorax lantsangensis | 16,580 | NC_026294.1 |
| Schizothorax lissolabiata | 16,583 | NC_027162.1 |
| Schizothorax macropogon | 16,588 | NC_020339.1 |
| Schizothorax molesworthi | 16,585 | MG171194.1 |
| Schizothorax nepalensis | 16,589 | AP011207.1 |
| Schizothorax niger | 16,585 | NC_022866.1 |
| Schizothorax nukiangensis | 16,585 | NC_027940.1 |
| Schizothorax oconnori | 16,616 | KT833107.1 |
| Schizothorax plagiostomus | 16,576 | NC_023531.1 |
| Schizothorax prenanti | 16,587 | NC_023829.1 |
| Schizothorax progastus | 16,575 | NC_023366.1 |
| Schizothorax pseudoaksaiensis | 16,586 | KM243919.1 |
| Schizothorax richardsonii | 16,592 | NC_021448.1 |
| Schizothorax sinensis | 16,571 | NC_056907.1 |
| Schizothorax taliensis | 16,578 | NC_037516.1 |
| Schizothorax waltoni | 16,589 | NC_020606.1 |
| Schizothorax wangchiachii | 16,593 | NC_020360.1 |
| Schizothorax yunnanensis paoshanensis | 16,585 | KP892531.1 |
| Spinibarbus denticulatus | 16,589 | AP013335.1 |
| Spinibarbus sinensis | 16,591 | NC_022465.1 |
| Gene | Location | Size (bp) | Intergenic Nucleotide | Start Codon | Stop Codon | Anticodon | Strand |
|---|---|---|---|---|---|---|---|
| trnF | 1–69 | 69 | 0 | --- | --- | GAA | H |
| rrnS | 70–1023 | 954 | 2 | --- | --- | --- | H |
| trnV | 1026–1097 | 72 | 22 | --- | --- | TAC | H |
| rrnL | 1120–2748 | 1629 | 24 | --- | --- | --- | H |
| trnL2 | 2773–2848 | 76 | 0 | --- | --- | TAA | H |
| nad1 | 2849–3823 | 975 | 4 | ATG | TAA | --- | H |
| trnI | 3828–3899 | 72 | −2 | --- | --- | GAT | H |
| trnQ | 3898–3968 | 71 | 2 | --- | --- | TTG | L |
| trnM | 3971–4039 | 69 | 0 | --- | --- | CAT | H |
| nad2 | 4040–5086 | 1047 | −2 | ATG | TAG | --- | H |
| trnW | 5085–5155 | 71 | 2 | --- | --- | TCA | H |
| trnA | 5158–5226 | 69 | 1 | --- | --- | TGC | L |
| trnN | 5228–5300 | 73 | 2 | --- | --- | GTT | L |
| OL | 5303–5334 | 32 | −1 | --- | --- | --- | H |
| trnC | 5334–5400 | 67 | −1 | --- | --- | GCA | L |
| trnY | 5400–5470 | 71 | 1 | --- | --- | GTA | L |
| cox1 | 5472–7022 | 1551 | 0 | GTG | TAA | --- | H |
| trnS2 | 7023–7093 | 71 | 3 | --- | --- | TGA | L |
| trnD | 7097–7168 | 72 | 12 | --- | --- | GTC | H |
| cox2 | 7181–7871 | 691 | 0 | ATG | T-- | --- | H |
| trnK | 7872–7947 | 76 | 1 | --- | --- | TTT | H |
| atp8 | 7949–8113 | 165 | −7 | ATG | TAG | --- | H |
| atp6 | 8107–8790 | 684 | −1 | ATG | TAA | --- | H |
| cox3 | 8790–9575 | 786 | −1 | ATG | TAA | --- | H |
| trnG | 9575–9646 | 72 | 0 | --- | --- | TCC | H |
| nad3 | 9647–9997 | 351 | −2 | ATG | TAG | --- | H |
| trnR | 9996–10,065 | 70 | 0 | --- | --- | TCG | H |
| nad4l | 10,066–10,362 | 297 | −7 | ATG | TAA | --- | H |
| nad4 | 10,356–11,736 | 1381 | 0 | ATG | T-- | --- | H |
| trnH | 11,737–11,805 | 69 | 0 | --- | --- | GTG | H |
| trnS1 | 11,806–11,873 | 68 | 1 | --- | --- | GCT | H |
| trnL1 | 11,875–11,947 | 73 | 3 | --- | --- | TAG | H |
| nad5 | 11,951–13,774 | 1824 | −4 | ATG | TAA | --- | H |
| nad6 | 13,771–14,292 | 522 | 0 | ATG | TAA | --- | L |
| trnE | 14,293–14,361 | 69 | 4 | --- | --- | TTC | L |
| cob | 14,366–15,506 | 1141 | 0 | ATG | T-- | --- | H |
| trnT | 15,507–15,578 | 72 | −1 | --- | --- | TGT | H |
| trnP | 15,578–15,647 | 70 | 15 | --- | --- | TGG | L |
| CR | 15,663–16,483 | 821 | 102 | --- | --- | --- | H |
| Regions | Size (bp) | A (%) | T (%) | G (%) | C (%) | A + T (%) | AT-Skew | GC-Skew |
|---|---|---|---|---|---|---|---|---|
| cox1 | 1551 | 26.24 | 29.08 | 18.63 | 26.05 | 55.32 | −0.05 | −0.17 |
| cox2 | 691 | 29.96 | 27.21 | 16.93 | 25.90 | 57.17 | 0.05 | −0.21 |
| atp8 | 165 | 33.33 | 29.70 | 12.73 | 24.24 | 63.03 | 0.06 | −0.31 |
| atp6 | 684 | 29.24 | 28.80 | 15.35 | 26.61 | 58.04 | 0.01 | −0.27 |
| cox3 | 786 | 27.61 | 26.84 | 17.56 | 27.99 | 54.45 | 0.01 | −0.23 |
| nad3 | 351 | 27.92 | 28.77 | 15.38 | 27.92 | 56.69 | −0.02 | −0.29 |
| nad1 | 975 | 24.10 | 25.13 | 20.41 | 30.36 | 49.23 | −0.02 | −0.20 |
| nad5 | 1824 | 29.22 | 25.16 | 16.01 | 29.61 | 54.38 | 0.07 | −0.30 |
| nad4 | 1381 | 28.39 | 26.36 | 16.58 | 28.67 | 54.75 | 0.04 | −0.27 |
| nad4l | 297 | 24.92 | 28.62 | 16.84 | 29.63 | 53.54 | −0.07 | −0.28 |
| nad6 | 522 | 13.22 | 38.12 | 32.18 | 16.48 | 51.34 | −0.49 | 0.32 |
| cob | 1141 | 26.47 | 28.66 | 17.09 | 27.78 | 55.13 | −0.04 | −0.24 |
| nad2 | 1047 | 28.37 | 22.25 | 17.57 | 31.81 | 50.62 | 0.12 | −0.29 |
| PCGs | 11,415 | 27.03 | 27.24 | 17.88 | 27.85 | 54.27 | −0.003 | −0.22 |
| tRNAs | 1562 | 28.10 | 26.70 | 23.75 | 21.45 | 54.80 | 0.03 | 0.05 |
| rRNAs | 2583 | 34.34 | 20.36 | 21.25 | 24.04 | 54.70 | 0.26 | −0.06 |
| OL | 32 | 15.63 | 18.75 | 31.25 | 34.38 | 34.38 | −0.09 | −0.05 |
| CR | 821 | 33.13 | 33.74 | 14.01 | 19.12 | 66.87 | −0.01 | −0.15 |
| genome | 16,585 | 29.59 | 25.42 | 17.94 | 27.05 | 55.01 | 0.08 | −0.20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, Q.; Chen, L.; Zhang, F.; Xu, J.; Zeng, Y. Characterization of the Complete Mitochondrial Genome of Schizothorax kozlovi (Cypriniformes, Cyprinidae, Schizothorax) and Insights into the Phylogenetic Relationships of Schizothorax. Animals 2024, 14, 721. https://doi.org/10.3390/ani14050721
Qin Q, Chen L, Zhang F, Xu J, Zeng Y. Characterization of the Complete Mitochondrial Genome of Schizothorax kozlovi (Cypriniformes, Cyprinidae, Schizothorax) and Insights into the Phylogenetic Relationships of Schizothorax. Animals. 2024; 14(5):721. https://doi.org/10.3390/ani14050721
Chicago/Turabian StyleQin, Qiang, Lin Chen, Fubin Zhang, Jianghaoyue Xu, and Yu Zeng. 2024. "Characterization of the Complete Mitochondrial Genome of Schizothorax kozlovi (Cypriniformes, Cyprinidae, Schizothorax) and Insights into the Phylogenetic Relationships of Schizothorax" Animals 14, no. 5: 721. https://doi.org/10.3390/ani14050721
APA StyleQin, Q., Chen, L., Zhang, F., Xu, J., & Zeng, Y. (2024). Characterization of the Complete Mitochondrial Genome of Schizothorax kozlovi (Cypriniformes, Cyprinidae, Schizothorax) and Insights into the Phylogenetic Relationships of Schizothorax. Animals, 14(5), 721. https://doi.org/10.3390/ani14050721





