Whole-Genome Resequencing of Ujimqin Sheep Identifies Genes Associated with Vertebral Number
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Phenotypic Data
2.2. Sequencing
2.3. Quality Control and Read Mapping
2.4. Statistical Analysis
2.5. RNA-Seq Analysis
2.6. Genotyping Causal Variants of the ABCD4 Candidate Gene
3. Results
3.1. Investigation and Evaluation of Vertebral Number in Ujimqin Sheep
3.2. Whole-Genome Resequencing and SNP Identification
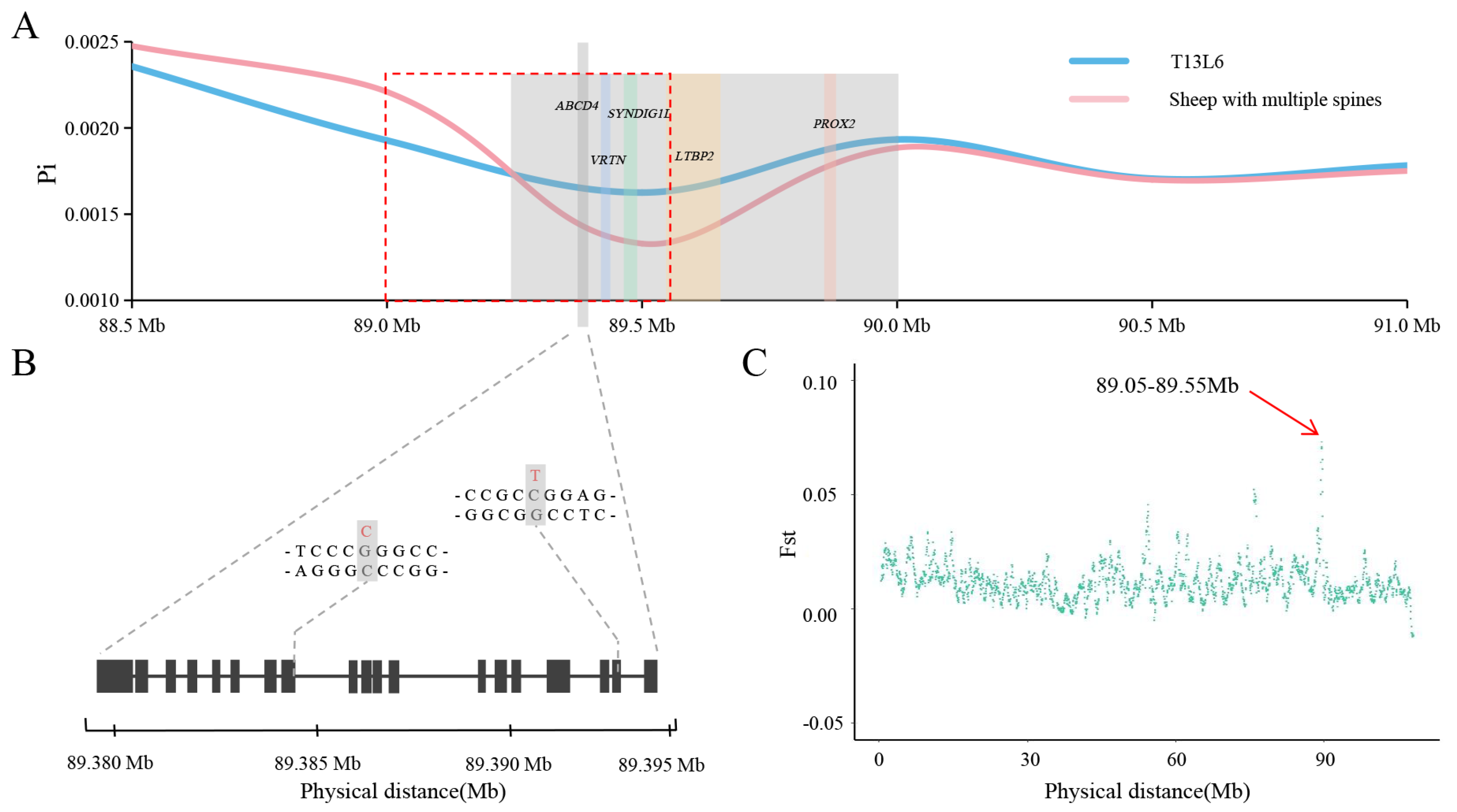
3.3. Selective-Sweep Analysis
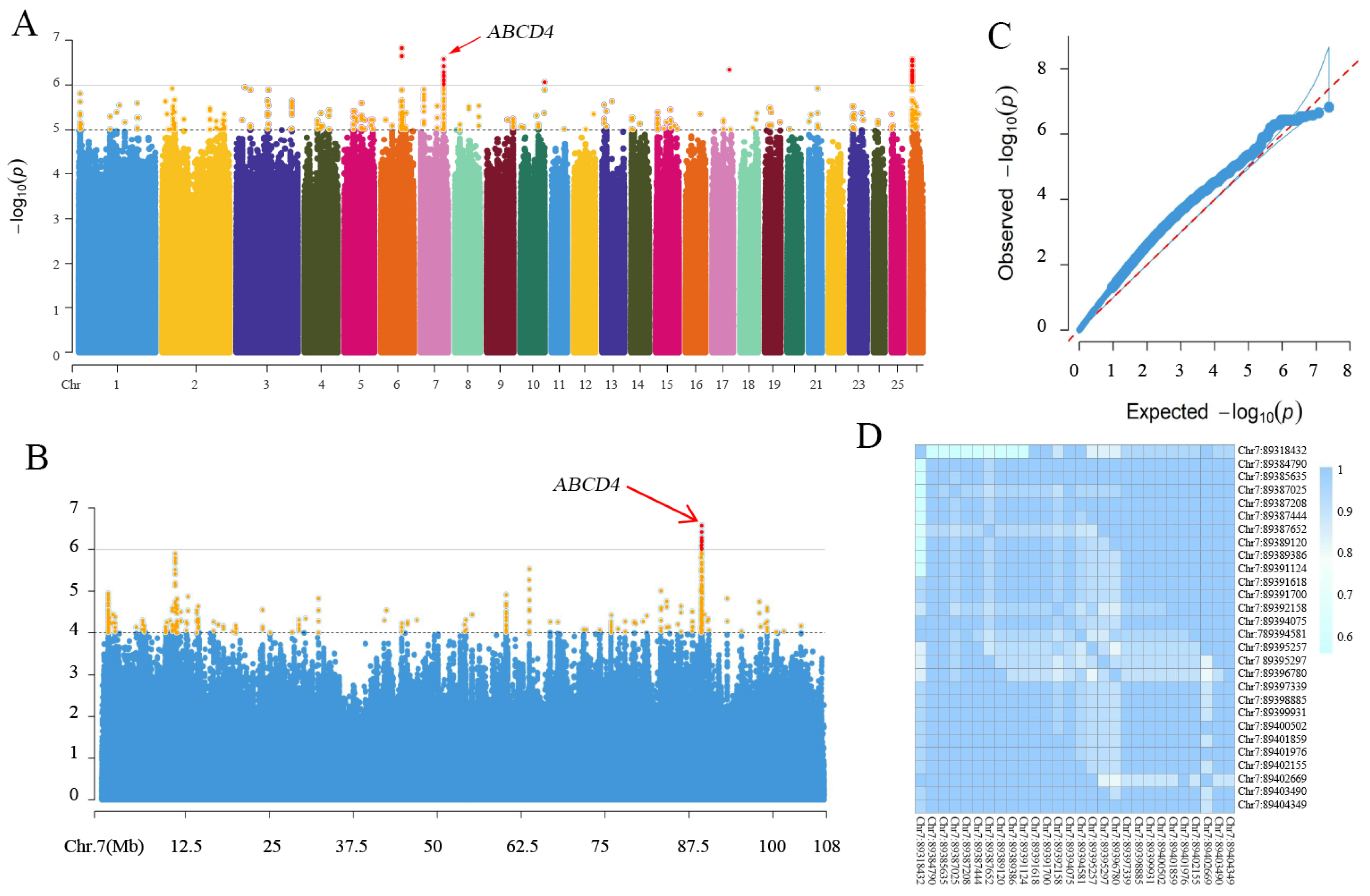
3.4. Genome-Wide Association Analysis
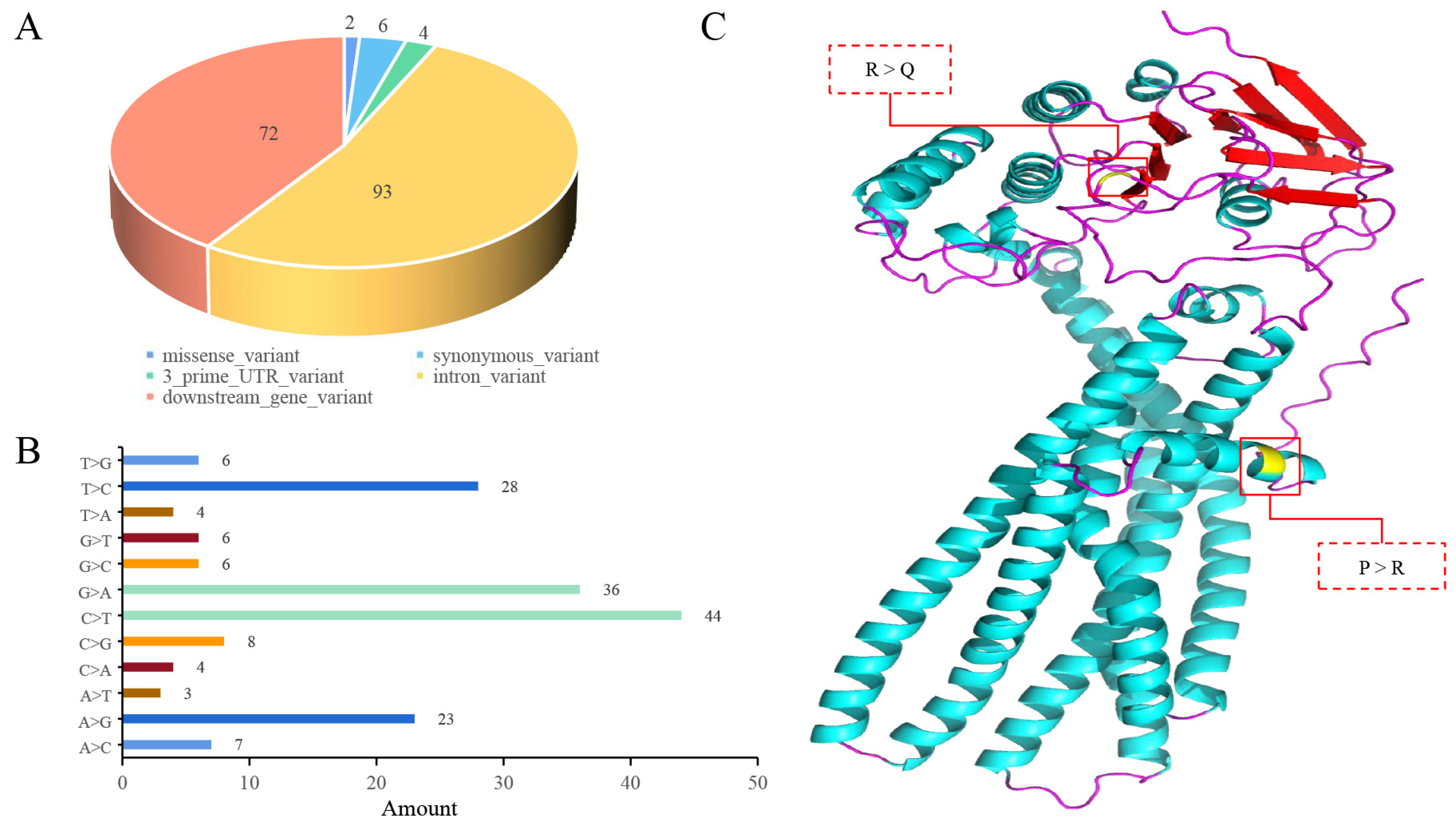
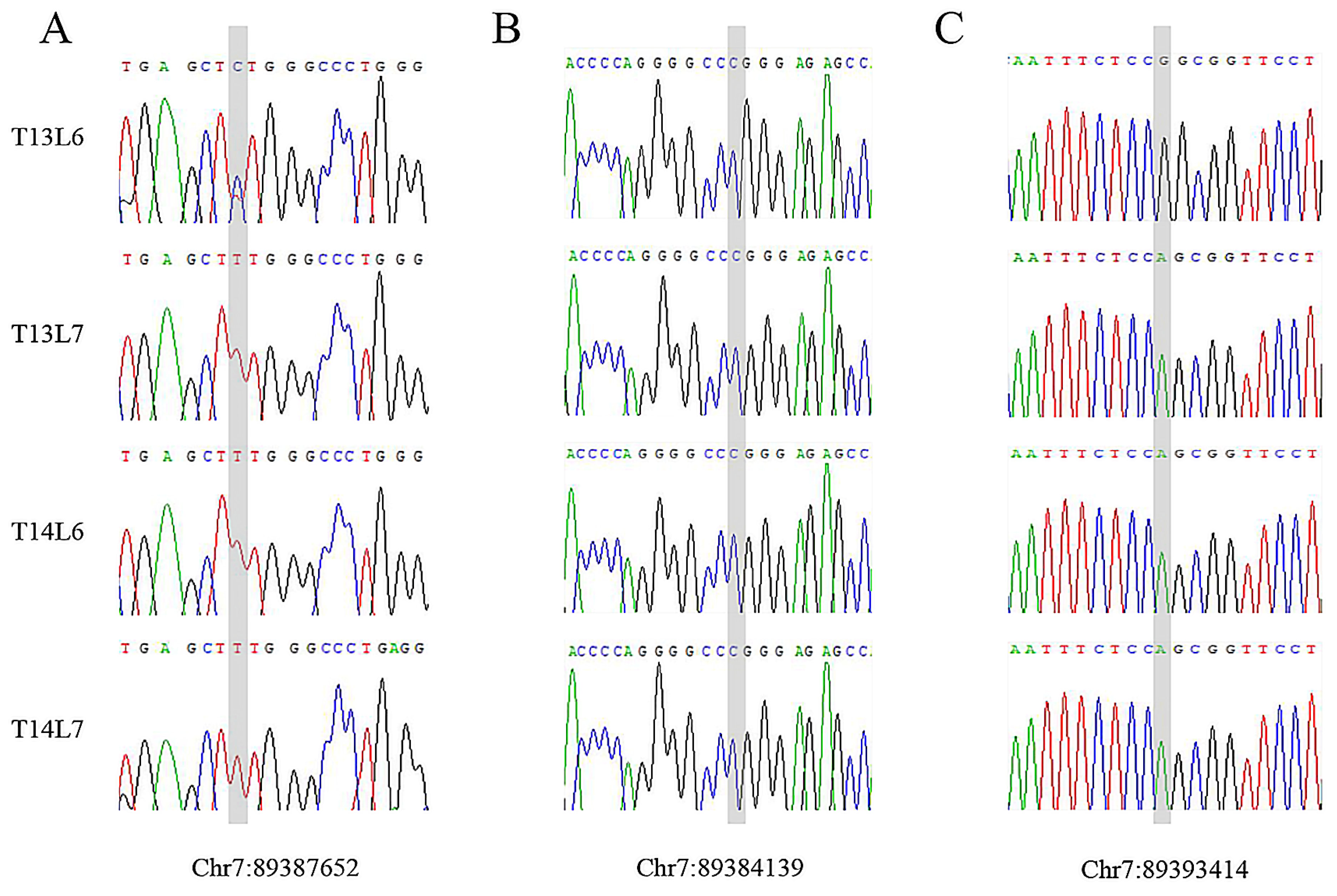
3.5. Mutation Analysis
3.6. SNP Screening and Sequencing Validation
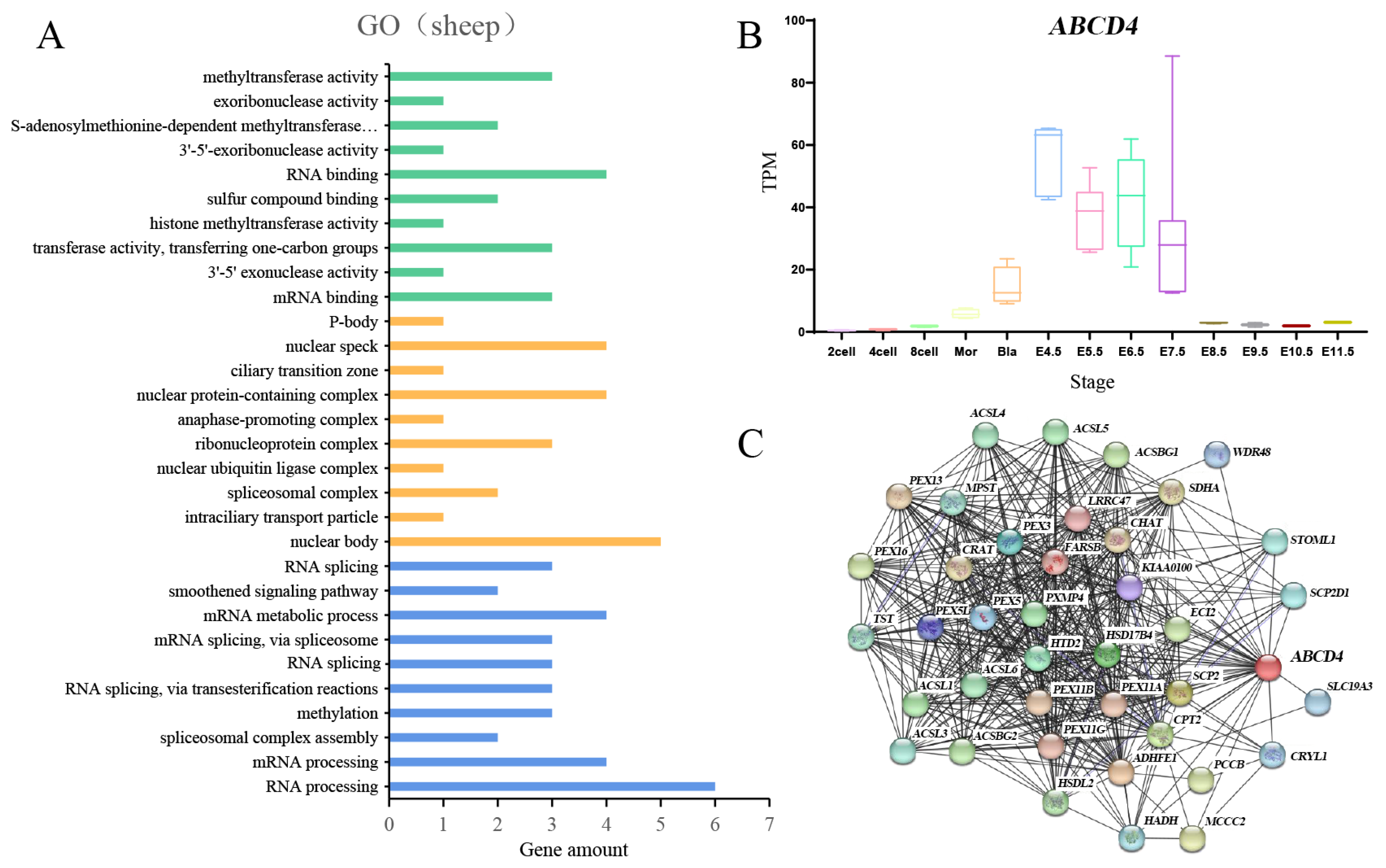
3.7. The Expression of ABCD4 and Related Gene Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| QTLs | quantitative trait loci |
| GWAS | genome-wide association study |
| C | cervical |
| T | thoracic |
| L | lumbar |
| S | sacral |
| Cd | caudal |
| QTN | quantitative trait nucleotide |
| SNP | single-nucleotide polymorphism |
| SSC7 | chromosome 7 |
| DR | digital radiography |
| GO | Gene Ontology |
| TPM | transcripts per kilobase million |
| TMHs | transmembrane helices |
| Pro | proline |
| Fst | fixation index |
| mRNA | messenger RNA |
| snoRNA | small nucleolar RNA |
| miRNA | microRNA |
References
- Schimandle, J.H.; Boden, S.D. Spine Update. The Use of Animal Models to Study Spinal Fusion. Spine 1994, 19, 1998–2006. [Google Scholar] [CrossRef]
- Narita, Y.; Kuratani, S. Evolution of the Vertebral Formulae in Mammals: A Perspective on Developmental Constraints. J. Exp. Zoolog. B Mol. Dev. Evol. 2005, 304, 91–106. [Google Scholar] [CrossRef]
- Zhu, H.; Zhao, J.; Zhou, W.; Li, H.; Zhou, R.; Zhang, L.; Zhao, H.; Cao, J.; Zhu, X.; Hu, H.; et al. Ndrg2 Regulates Vertebral Specification in Differentiating Somites. Dev. Biol. 2012, 369, 308–318. [Google Scholar] [CrossRef][Green Version]
- King, J.W.B.; Roberts, R.C. Carcass Length in the Bacon Pig; Its Association with Vertebrae Numbers and Prediction from Radiographs of the Young Pig. Anim. Prod. 1960, 2, 59–65. [Google Scholar] [CrossRef]
- Donaldson, C.L.; Lambe, N.R.; Maltin, C.A.; Knott, S.; Bunger, L. Between- and within-Breed Variations of Spine Characteristics in Sheep. J. Anim. Sci. 2013, 91, 995–1004. [Google Scholar] [CrossRef]
- Yang, G.; Ren, J.; Zhang, Z.; Huang, L. Genetic Evidence for the Introgression of Western NR6A1 Haplotype into Chinese Licha Breed Associated with Increased Vertebral Number. Anim. Genet. 2009, 40, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Burgos, C.; Latorre, P.; Altarriba, J.; Carrodeguas, J.A.; Varona, L.; López-Buesa, P. Allelic Frequencies of NR6A1 and VRTN, Two Genes That Affect Vertebrae Number in Diverse Pig Breeds: A Study of the Effects of the VRTN Insertion on Phenotypic Traits of a Duroc × Landrace-Large White Cross. Meat Sci. 2015, 100, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Huang, L.; Yang, M.; Fan, Y.; Li, L.; Fang, S.; Deng, W.; Cui, L.; Zhang, Z.; Ai, H.; et al. Possible Introgression of the VRTN Mutation Increasing Vertebral Number, Carcass Length and Teat Number from Chinese Pigs into European Pigs. Sci. Rep. 2016, 6, 19240. [Google Scholar] [CrossRef] [PubMed]
- Vasiliou, V.; Pappa, A.; Petersen, D.R. Role of Aldehyde Dehydrogenases in Endogenous and Xenobiotic Metabolism. Chem. Biol. Interact. 2000, 129, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Walkley, S.U.; Suzuki, K. Consequences of NPC1 and NPC2 Loss of Function in Mammalian Neurons. Biochim. Biophys. Acta 2004, 1685, 48–62. [Google Scholar] [CrossRef]
- Yan, G.; Qiao, R.; Zhang, F.; Xin, W.; Xiao, S.; Huang, T.; Zhang, Z.; Huang, L. Imputation-Based Whole-Genome Sequence Association Study Rediscovered the Missing QTL for Lumbar Number in Sutai Pigs. Sci. Rep. 2017, 7, 615. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhang, H.; Zhang, Z.; Gao, J.; Yang, J.; Wu, Z.; Fan, Y.; Xing, Y.; Li, L.; Xiao, S.; et al. VRTN Is Required for the Development of Thoracic Vertebrae in Mammals. Int. J. Biol. Sci. 2018, 14, 667–681. [Google Scholar] [CrossRef]
- Fan, Y.; Xing, Y.; Zhang, Z.; Ai, H.; Ouyang, Z.; Ouyang, J.; Yang, M.; Li, P.; Chen, Y.; Gao, J.; et al. A Further Look at Porcine Chromosome 7 Reveals VRTN Variants Associated with Vertebral Number in Chinese and Western Pigs. PLoS ONE 2013, 8, e62534. [Google Scholar] [CrossRef]
- Li, C.; Li, M.; Li, X.; Ni, W.; Xu, Y.; Yao, R.; Wei, B.; Zhang, M.; Li, H.; Zhao, Y.; et al. Whole-Genome Resequencing Reveals Loci Associated with Thoracic Vertebrae Number in Sheep. Front. Genet. 2019, 10, 674. [Google Scholar] [CrossRef]
- Zhang, Z.; Sun, Y.; Du, W.; He, S.; Liu, M.; Tian, C. Effects of Vertebral Number Variations on Carcass Traits and Genotyping of Vertnin Candidate Gene in Kazakh Sheep. Asian-Australas. J. Anim. Sci. 2017, 30, 1234–1238. [Google Scholar] [CrossRef] [PubMed]
- Charité, J.; de Graaff, W.; Deschamps, J. Specification of Multiple Vertebral Identities by Ectopically Expressed Hoxb-8. Dev. Dyn. Off. Public Am. Assoc. Anat. 1995, 204, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.-J.; Yang, Y.; Wang, X.-Y.; Di, R.; Chu, M.-X.; Liu, Q.-Y. Expression Analysis and Single-Nucleotide Polymorphisms of SYNDIG1L and UNC13C Genes Associated with Thoracic Vertebral Numbers in Sheep (Ovis aries). Arch. Anim. Breed. 2021, 64, 131–138. [Google Scholar] [CrossRef]
- Li, C.; Liu, K.; Dai, J.; Li, X.; Liu, X.; Ni, W.; Li, H.; Wang, D.; Qiao, J.; Wang, Y.; et al. Whole-Genome Resequencing to Investigate the Determinants of the Multi-Lumbar Vertebrae Trait in Sheep. Gene 2022, 809, 146020. [Google Scholar] [CrossRef]
- Sun, Y.; Li, Y.; Zhao, C.; Teng, J.; Wang, Y.; Wang, T.; Shi, X.; Liu, Z.; Li, H.; Wang, J.; et al. Genome-wide association study for numbers of vertebrae in Dezhou donkey population reveals new candidate genes. J. Integr. Agric. 2023, 22, 3159–3169. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup the Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Yin, L. CMplot: Circle Manhattan Plot. Available online: https://github.com/YinLiLin/CMplot (accessed on 2 March 2021).
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Xu, D.; Feng, Z.; Hou, W.-T.; Jiang, Y.-L.; Wang, L.; Sun, L.; Zhou, C.-Z.; Chen, Y. Cryo-EM Structure of Human Lysosomal Cobalamin Exporter ABCD4. Cell Res. 2019, 29, 1039–1041. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An Enhanced Web Server for Large-Scale Expression Profiling and Interactive Analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein–Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Bovine HapMap Consortium; Gibbs, R.A.; Taylor, J.F.; Van Tassell, C.P.; Barendse, W.; Eversole, K.A.; Gill, C.A.; Green, R.D.; Hamernik, D.L.; Kappes, S.M.; et al. Genome-Wide Survey of SNP Variation Uncovers the Genetic Structure of Cattle Breeds. Science 2009, 324, 528–532. [Google Scholar] [CrossRef]
- Brito, L.F.; McEwan, J.C.; Miller, S.; Bain, W.; Lee, M.; Dodds, K.; Newman, S.-A.; Pickering, N.; Schenkel, F.S.; Clarke, S. Genetic Parameters for Various Growth, Carcass and Meat Quality Traits in a New Zealand Sheep Population. Small Rumin. Res. 2017, 154, 81–91. [Google Scholar] [CrossRef]
- Li, C.; Zhang, X.; Cao, Y.; Wei, J.; You, S.; Jiang, Y.; Cai, K.; Wumaier, W.; Guo, D.; Qi, J.; et al. Multi-Vertebrae Variation Potentially Contribute to Carcass Length and Weight of Kazakh Sheep. Small Rumin. Res. 2017, 150, 8–10. [Google Scholar] [CrossRef]
- Pan, Z.; Li, S.; Liu, Q.; Wang, Z.; Zhou, Z.; Di, R.; Miao, B.; Hu, W.; Wang, X.; Hu, X.; et al. Whole-Genome Sequences of 89 Chinese Sheep Suggest Role of RXFP2 in the Development of Unique Horn Phenotype as Response to Semi-Feralization. GigaScience 2018, 7, giy019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-C.; Yue, J.-W.; Pu, L.; Wang, L.-G.; Liu, X.; Liang, J.; Yan, H.; Zhao, K.-B.; Li, N.; Shi, H.-B.; et al. Genome-Wide Study Refines the Quantitative Trait Locus for Number of Ribs in a Large White × Minzhu Intercross Pig Population and Reveals a New Candidate Gene. Mol. Genet. Genom. 2016, 291, 1885–1890. [Google Scholar] [CrossRef] [PubMed]
- Coelho, D.; Kim, J.C.; Miousse, I.R.; Fung, S.; du Moulin, M.; Buers, I.; Suormala, T.; Burda, P.; Frapolli, M.; Stucki, M.; et al. Mutations in ABCD4 Cause a New Inborn Error of Vitamin B12 Metabolism. Nat. Genet. 2012, 44, 1152–1155. [Google Scholar] [CrossRef] [PubMed]
- Van Son, M.; Lopes, M.S.; Martell, H.J.; Derks, M.F.L.; Gangsei, L.E.; Kongsro, J.; Wass, M.N.; Grindflek, E.H.; Harlizius, B. A QTL for Number of Teats Shows Breed Specific Effects on Number of Vertebrae in Pigs: Bridging the Gap Between Molecular and Quantitative Genetics. Front. Genet. 2019, 10, 272. [Google Scholar] [CrossRef] [PubMed]
- Niu, N.; Liu, Q.; Hou, X.; Liu, X.; Wang, L.; Zhao, F.; Gao, H.; Shi, L.; Wang, L.; Zhang, L. Genome-Wide Association Study Revealed ABCD4 on SSC7 and GREB1L and MIB1 on SSC6 as Crucial Candidate Genes for Rib Number in Beijing Black Pigs. Anim. Genet. 2022, 53, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Tseng, W.-C.; Johnson Escauriza, A.J.; Tsai-Morris, C.-H.; Feldman, B.; Dale, R.K.; Wassif, C.A.; Porter, F.D. The Role of Niemann–Pick Type C2 in Zebrafish Embryonic Development. Development 2021, 148, dev194258. [Google Scholar] [CrossRef]
- Jo, B.-S.; Choi, S.S. Introns: The Functional Benefits of Introns in Genomes. Genom. Inform. 2015, 13, 112–118. [Google Scholar] [CrossRef]
- Morgan, J.T.; Fink, G.R.; Bartel, D.P. Excised Linear Introns Regulate Growth in Yeast. Nature 2019, 565, 606–611. [Google Scholar] [CrossRef]
- Parenteau, J.; Maignon, L.; Berthoumieux, M.; Catala, M.; Gagnon, V.; Abou Elela, S. Introns Are Mediators of Cell Response to Starvation. Nature 2019, 565, 612–617. [Google Scholar] [CrossRef]
- Vaes, B.L.T.; Lute, C.; Blom, H.J.; Bravenboer, N.; de Vries, T.J.; Everts, V.; Dhonukshe-Rutten, R.A.; Müller, M.; de Groot, L.C.P.G.M.; Steegenga, W.T. Vitamin B12 Deficiency Stimulates Osteoclastogenesis via Increased Homocysteine and Methylmalonic Acid. Calcif. Tissue Int. 2009, 84, 413–422. [Google Scholar] [CrossRef]
- Liu, Z.; Choi, S.-W.; Crott, J.W.; Keyes, M.K.; Jang, H.; Smith, D.E.; Kim, M.; Laird, P.W.; Bronson, R.; Mason, J.B. Mild Depletion of Dietary Folate Combined with Other B Vitamins Alters Multiple Components of the Wnt Pathway in Mouse Colon. J. Nutr. 2007, 137, 2701–2708. [Google Scholar] [CrossRef][Green Version]
- Ciappio, E.D.; Liu, Z.; Brooks, R.S.; Mason, J.B.; Bronson, R.T.; Crott, J.W. Maternal B Vitamin Supplementation from Preconception through Weaning Suppresses Intestinal Tumorigenesis in Apc1638N Mouse Offspring. Gut 2011, 60, 1695–1702. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-González, J.G.; Santillán, M.; Fowler, A.C.; Mackey, M.C. The Segmentation Clock in Mice: Interaction between the Wnt and Notch Signalling Pathways. J. Theor. Biol. 2007, 248, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka-Kobayashi, K.; Matsumiya, M.; Niino, Y.; Isomura, A.; Kori, H.; Miyawaki, A.; Kageyama, R. Coupling Delay Controls Synchronized Oscillation in the Segmentation Clock. Nature 2020, 580, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Fan, L.; Zhao, L.; Su, Y. The Interaction of Notch and Wnt Signaling Pathways in Vertebrate Regeneration. Cell Regen. Lond. Engl. 2021, 10, 11. [Google Scholar] [CrossRef] [PubMed]
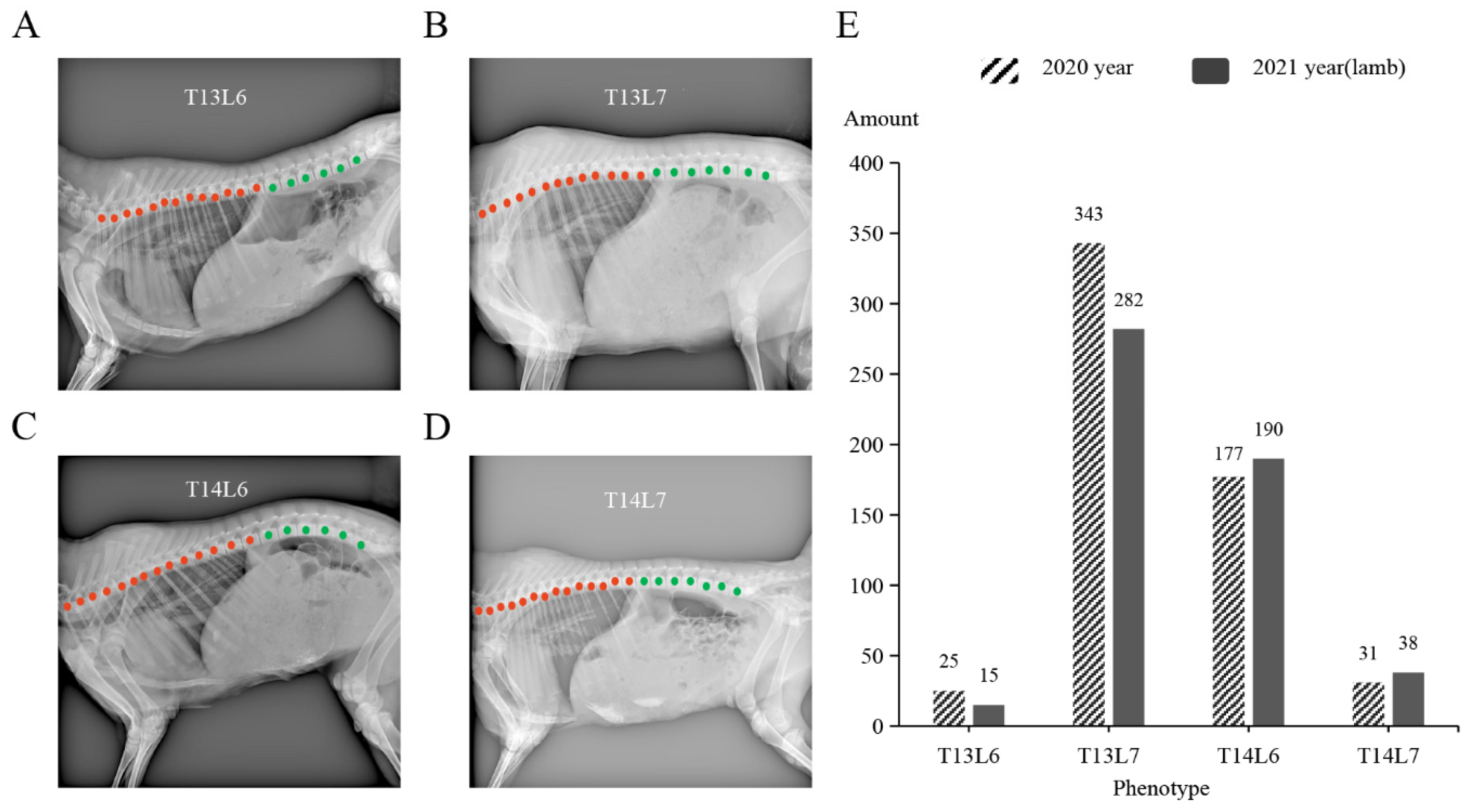
| Year | Number | T13L6 | T13L7 | T14L6 | T14L7 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Number | Proportion | Number | Proportion | Number | Proportion | Number | Proportion | ||
| 2020 | 576 | 25 | 4.3 | 343 | 59.6 | 177 | 30.7 | 31 | 5.4 |
| 2021 | 525 | 15 | 2.9 | 282 | 53.7 | 190 | 36.2 | 38 | 7.2 |
| Total | 1101 | 40 | 3.6 | 625 | 56.8 | 367 | 33.3 | 69 | 6.3 |
| Paternal Phenotype | Maternal Phenotype (Number) | Offspring (T13L6) | Offspring (T13L7) | Offspring (T14L6) | Offspring (T14L7) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Number | Proportion | Number | Proportion | Number | Proportion | Number | Proportion | ||
| T14L7 | T13L6 (11) | 2 | 18.2 | 8 | 72.7 | 1 | 9.1 | - | - |
| T14L7 | T13L7 (148) | 1 | 0.7 | 91 | 61.5 | 49 | 33.1 | 7 | 4.7 |
| T14L7 | T14L6 (94) | 1 | 1.1 | 25 | 26.6 | 56 | 56.9 | 12 | 12.7 |
| T14L7 | T14L7 (12) | 5 | 41.7 | 5 | 41.7 | 2 | 16.6 | - | - |
| Chromosome | Bin_Start | Bin_End | Fst |
|---|---|---|---|
| 7 | 89,050,001 | 89,550,000 | 0.0729947 |
| 7 | 89,100,001 | 89,600,000 | 0.0707678 |
| 7 | 89,150,001 | 89,650,000 | 0.0701816 |
| 7 | 89,000,001 | 89,500,000 | 0.0642738 |
| 7 | 89,200,001 | 89,700,000 | 0.0653491 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, C.; Zhang, Y.; Ma, T.; Wu, D.; Yang, Y.; Wang, D.; Li, X.; Guo, S.; Yang, S.; Song, Y.; et al. Whole-Genome Resequencing of Ujimqin Sheep Identifies Genes Associated with Vertebral Number. Animals 2024, 14, 677. https://doi.org/10.3390/ani14050677
Zhou C, Zhang Y, Ma T, Wu D, Yang Y, Wang D, Li X, Guo S, Yang S, Song Y, et al. Whole-Genome Resequencing of Ujimqin Sheep Identifies Genes Associated with Vertebral Number. Animals. 2024; 14(5):677. https://doi.org/10.3390/ani14050677
Chicago/Turabian StyleZhou, Chuanqing, Yue Zhang, Teng Ma, Dabala Wu, Yanyan Yang, Daqing Wang, Xiunan Li, Shuchun Guo, Siqi Yang, Yongli Song, and et al. 2024. "Whole-Genome Resequencing of Ujimqin Sheep Identifies Genes Associated with Vertebral Number" Animals 14, no. 5: 677. https://doi.org/10.3390/ani14050677
APA StyleZhou, C., Zhang, Y., Ma, T., Wu, D., Yang, Y., Wang, D., Li, X., Guo, S., Yang, S., Song, Y., Zhang, Y., Zuo, Y., & Cao, G. (2024). Whole-Genome Resequencing of Ujimqin Sheep Identifies Genes Associated with Vertebral Number. Animals, 14(5), 677. https://doi.org/10.3390/ani14050677






