Advancements in Genetic Marker Exploration for Livestock Vertebral Traits with a Focus on China
Abstract
Simple Summary
Abstract
1. Introduction
2. Methodology and Criteria for Literature Search and Selection
3. Genetic Markers Associated with Number of Vertebrae in Pigs, Donkeys, and Sheep
3.1. Genetic Markers Associated with Number of Vertebrae in Pigs
3.2. Genetic Markers Associated with Number of Vertebrae in Donkeys
3.3. Genetic Markers Associated with Number of Vertebrae in Sheep
4. Comparative Analysis of Overlapping Genes Linked to Vertebral Traits in Pigs, Donkeys, and Sheep
4.1. Nuclear Receptor Subfamily 6, Group A, Member 1 (NR6A1)
4.2. Vertnin (VRTN)
4.3. Latent TGFβ Binding Protein-2 (LTBP2)
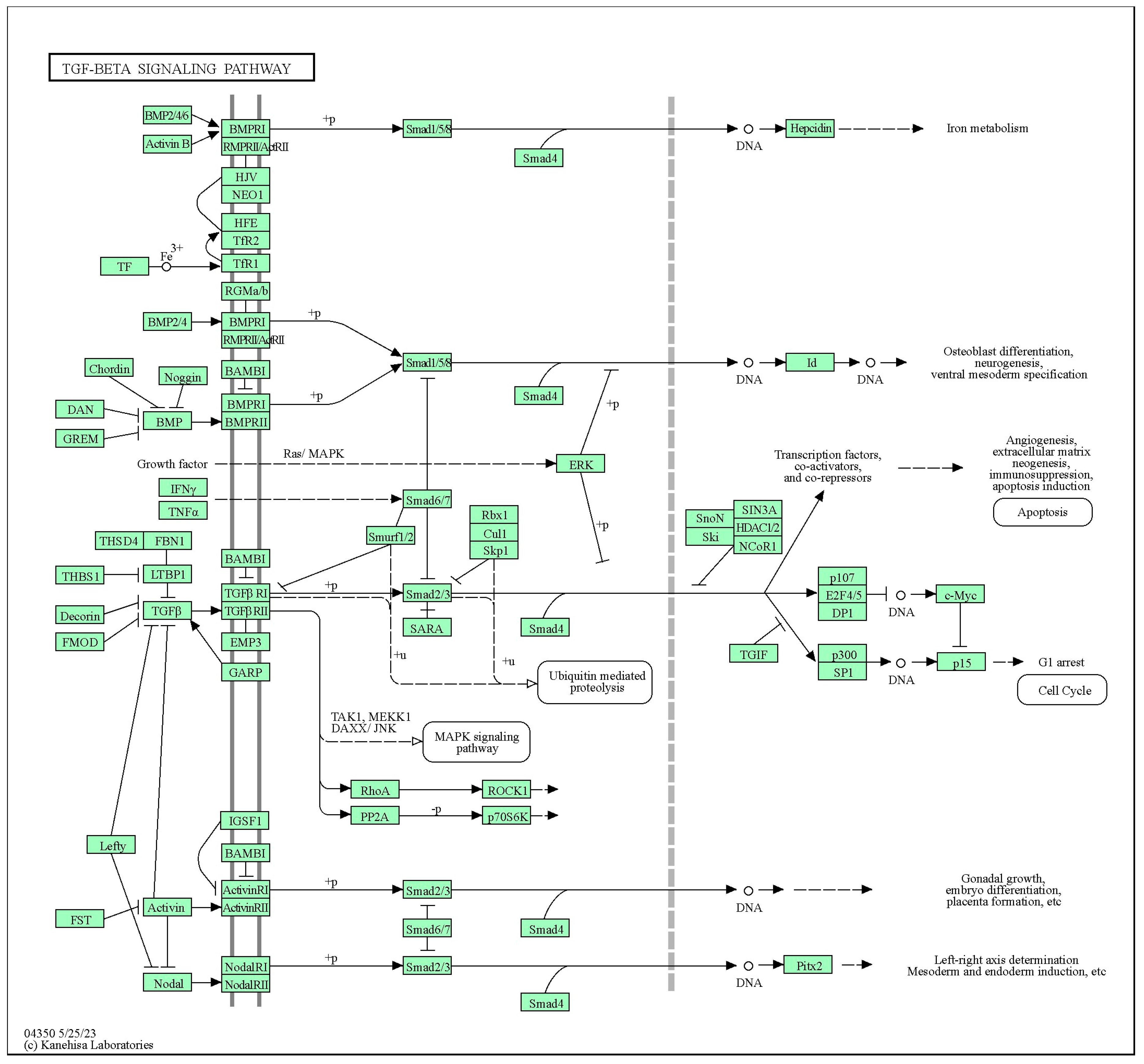
4.4. Bone Morphogenetic Proteins (BMPs)
4.5. Homeobox (Hox) Genes
5. Future Directions and Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Whitton, C.; Bogueva, D.; Marinova, D.; Phillips, C.J.C. Are We Approaching Peak Meat Consumption? Analysis of Meat Consumption from 2000 to 2019 in 35 Countries and Its Relationship to Gross Domestic Product. Animals 2021, 11, 3466. [Google Scholar] [CrossRef]
- Hötzel, M.J.; Vandresen, B. Brazilians’ attitudes toward meat consumption and production: Present and future challenges to the sustainability of the meat industry. Meat Sci. 2022, 192, 108893. [Google Scholar] [CrossRef]
- Chai, W.; Qu, H.; Ma, Q.; Zhu, M.; Li, M.; Zhan, Y.; Liu, Z.; Xu, J.; Yao, H.; Li, Z.; et al. RNA-seq analysis identifies differentially expressed genes in different types of donkey skeletal muscles. Anim. Biotechnol. 2023, 34, 1786–1795. [Google Scholar] [CrossRef]
- Ma, Q.; Kou, X.; Yang, Y.; Yue, Y.; Xing, W.; Feng, X.; Liu, G.; Wang, C.; Li, Y. Comparison of Lipids and Volatile Compounds in Dezhou Donkey Meat with High and Low Intramuscular Fat Content. Foods 2023, 12, 3269. [Google Scholar] [CrossRef]
- Chai, W.; Xu, J.; Qu, H.; Ma, Q.; Zhu, M.; Li, M.; Zhan, Y.; Wang, T.; Gao, J.; Yao, H.; et al. Differential proteomic analysis to identify potential biomarkers associated with quality traits of Dezhou donkey meat using a data-independent acquisition (DIA) strategy. LWT 2022, 166, 113792. [Google Scholar] [CrossRef]
- Dagevos, H.; Verbeke, W. Meat consumption and flexitarianism in the Low Countries. Meat Sci. 2022, 192, 108894. [Google Scholar] [CrossRef]
- Realini, C.E.; Ares, G.; Antúnez, L.; Brito, G.; Luzardo, S.; Del Campo, M.; Saunders, C.; Farouk, M.M.; Montossi, F.M. Meat insights: Uruguayan consumers’ mental associations and motives underlying consumption changes. Meat Sci. 2022, 192, 108901. [Google Scholar] [CrossRef]
- Wang, T.; Shi, X.; Liu, Z.; Ren, W.; Wang, X.; Huang, B.; Kou, X.; Liang, H.; Wang, C.; Chai, W. A novel A>G polymorphism in the intron 1 of LCORL gene is significantly associated with hide weight and body size in Dezhou donkey. Animals 2022, 12, 2581. [Google Scholar] [CrossRef]
- Gao, G.; Gao, N.; Li, S.; Kuang, W.; Zhu, L.; Jiang, W.; Yu, W.; Guo, J.; Li, Z.; Yang, C.; et al. Genome-wide association study of meat quality traits in a three-way crossbred commercial pig population. Front. Genet. 2021, 12, 614087. [Google Scholar] [CrossRef]
- Yan, S.U.; Li, Y.H.; Zhao, C.H.; Jun, T.E.; Wang, Y.H.; Wang, T.Q.; Shi, X.Y.; Liu, Z.W.; Li, H.J.; Wang, J.J.; et al. Genome-wide association study for numbers of vertebrae in Dezhou donkey population reveals new candidate genes. J. Integr. Agric. 2023, 22, 3159–3169. [Google Scholar]
- Wang, T.; Wang, X.; Liu, Z.; Shi, X.; Ren, W.; Huang, B.; Liang, H.; Wang, C.; Chai, W. Genotypes and haplotype combination of DCAF7 gene sequence variants are associated with the number of thoracolumbar vertebrae and carcass traits in Dezhou donkey. J. Appl. Anim. Res. 2023, 51, 31–39. [Google Scholar] [CrossRef]
- Wang, T.; Liu, Z.; Wang, X.; Li, Y.; Akhtar, F.; Li, M.; Zhang, Z.; Zhan, Y.; Shi, X.; Ren, W.; et al. Polymorphism Detection of PRKG2 Gene and Its Association with the Number of Thoracolumbar Vertebrae and Carcass Traits in Dezhou Donkey. BMC Genom. Data 2023, 24, 2. [Google Scholar] [CrossRef]
- Jiang, N.; Liu, C.; Lan, T.; Zhang, Q.; Cao, Y.; Pu, G.; Niu, P.; Zhang, Z.; Li, Q.; Zhou, J.; et al. Polymorphism of VRTN Gene g. 20311_20312ins291 Was Associated with the Number of Ribs, Carcass Diagonal Length and Cannon Bone Circumference in Suhuai Pigs. Animals 2020, 10, 484. [Google Scholar] [CrossRef]
- Fogel, J.L.; Lakeland, D.L.; Mah, I.K.; Mariani, F.V. A minimally sufficient model for rib proximal-distal patterning based on genetic analysis and agent-based simulations. eLife 2017, 6, e29144. [Google Scholar] [CrossRef]
- Borchers, N.; Reinsch, N.; Kalm, E. The number of ribs and vertebrae in a Piétrain cross: Variation, heritability and effects on performance traits. J. Anim. Breed. Genet. 2004, 121, 392–403. [Google Scholar] [CrossRef]
- Burgos, C.; Latorre, P.; Altarriba, J.; Carrodeguas, J.; Varona, L.; López Buesa, P. Allelic frequencies of NR6A1 and VRTN, two genes that affect vertebrae number in diverse pig breeds: A study of the effects of the VRTN insertion on phenotypic traits of a Duroc×Landrace–Large White cross. Meat Sci. 2015, 100, 150–155. [Google Scholar] [CrossRef]
- Donaldson, C.L.; Lambe, N.R.; Maltin, C.A.; Knott, S.; Bunger, L. Between-and within-breed variations of spine characteristics in sheep. J. Anim. Sci. 2013, 91, 995–1004. [Google Scholar] [CrossRef]
- Liu, Z.; Gao, Q.; Wang, T.; Chai, W.; Zhan, Y.; Akhtar, F.; Zhang, Z.; Li, Y.; Shi, X.; Wang, C. Multi-Thoracolumbar Variations and NR6A1 Gene Polymorphisms Potentially Associated with Body Size and Carcass Traits of Dezhou Donkey. Animals 2022, 12, 1349. [Google Scholar] [CrossRef]
- Yang, J.; Wen, Y.; Feng, Z.; Ke, M.; Gao, X.; An, D. Correlation analysis for beef performance and multi-vertebra properties of Jinchuan yak. J. Domest. Anim. Ecol. 2015, 36, 26–30. [Google Scholar]
- Palombo, V.; D’Andrea, M.; Licastro, D.; Dal Monego, S.; Sgorlon, S.; Sandri, M.; Stefanon, B. Single-step genome-wide association study identifies QTL signals for untrimmed and trimmed thigh weight in Italian crossbred pigs for dry-cured ham production. Animals 2021, 11, 1612. [Google Scholar] [CrossRef]
- Yang, W.; Liu, Z.; Zhao, Q.; Du, H.; Yu, J.; Wang, H.; Liu, X.; Liu, H.; Jing, X.; Yang, H.; et al. Population Genetic Structure and Selection Signature Analysis of Beijing Black Pig. Front. Genet. 2022, 13, 860669. [Google Scholar] [CrossRef]
- Wang, C.; Li, H.; Guo, Y.; Huang, J.; Sun, Y.; Min, J.; Wang, J.; Fang, X.; Zhao, Z.; Wang, S.; et al. Donkey genomes provide new insights into domestication and selection for coat color. Nat. Commun. 2020, 1, 6014. [Google Scholar] [CrossRef]
- Chen, W.; Fang, G.F.; Wang, S.D.; Wang, H.; Zeng, Y.-Q. Longissimus lumborum muscle transcriptome analysis of Laiwu and Yorkshire pigs differing in intramuscular fat content. Genes Genom. 2017, 39, 759–766. [Google Scholar] [CrossRef]
- Takasuga, A. PLAG1 and NCAPG-LCORL in livestock. Anim. Sci. J. 2016, 87, 159–167. [Google Scholar] [CrossRef]
- Liu, J.; Sun, S.; Han, L.; Li, X.; Sun, Z. Association between single nucleotide polymorphism of ActRIIB gene and vertebra number variation in small tail Han sheep. Acta Vet. Et. Zootech. Sin. 2010, 41, 951–954. [Google Scholar]
- Yue, J.; Guo, H.; Zhou, W.; Liu, X.; Wang, L.; Gao, H.; Hou, X.; Zhang, Y.; Yan, H.; Wei, X. Polymorphism Sites of TGFβ3 Gene and Its Association Analysis with Vertebral Number of Porcine. Chin. Anim. Husb. Vet. Med. 2018, 45, 738–744. [Google Scholar]
- Niu, N.; Liu, Q.; Hou, X.; Liu, X.; Wang, L.; Zhao, F.; Gao, H.; Shi, L.; Wang, L.; Zhang, L. Genome-wide association study revealed ABCD4 on SSC7 and GREB1L and MIB1 on SSC6 as crucial candidate genes for rib number in Beijing Black pigs. Anim. Genet. 2022, 53, 690–695. [Google Scholar] [CrossRef]
- Shi, X.; Li, Y.; Wang, T.; Ren, W.; Huang, B.; Wang, X.; Liu, Z.; Liang, H.; Kou, X.; Chen, Y.; et al. Association of HOXC8 Genetic Polymorphisms with Multi-Vertebral Number and Carcass Weight in Dezhou Donkey. Genes 2022, 13, 2175. [Google Scholar] [CrossRef]
- Xu, J.; Jiang, A.; Zhang, C.; Zheng, Y.; Zhang, T.; Zhou, L. Potential of eight mutations for marker-assisted breeding in Chinese Lulai black pigs. Can. J. Anim. Sci. 2022, 102, 431–439. [Google Scholar] [CrossRef]
- Liu, C.; Hou, L.; Zhao, Q.; Zhou, W.; Liu, K.; Liu, Q.; Zhou, T.; Xu, B.; Li, P.; Huang, R. The selected genes NR6A1, RSAD2-CMPK2, and COL3A1 contribute to body size variation in Meishan pigs through different patterns. J. Anim. Sci. 2023, 101, skad304. [Google Scholar] [CrossRef]
- Xie, L.; Qin, J.; Yao, T.; Tang, X.; Cui, D.; Chen, L.; Rao, L.; Xiao, S.; Zhang, Z.; Huang, L. Genetic dissection of 26 meat cut, meat quality and carcass traits in four pig populations. Genet. Sel. Evol. 2023, 55, 43. [Google Scholar] [CrossRef]
- Xie, L.; Qin, J.; Rao, L.; Cui, D.; Tang, X.; Chen, L.; Xiao, S.; Zhang, Z.; Huang, L. Genetic dissection and genomic prediction for pork cuts and carcass morphology traits in pig. J. Anim. Sci. Biotechnol. 2023, 14, 116. [Google Scholar] [CrossRef]
- Zhou, F.; Quan, J.; Ruan, D.; Qiu, Y.; Ding, R.; Xu, C.; Ye, Y.; Cai, G.; Liu, L.; Zhang, Z.; et al. Identification of Candidate Genes for Economically Important Carcass Cutting in Commercial Pigs through GWAS. Animals 2023, 13, 3243. [Google Scholar] [CrossRef]
- Wang, X.; Ran, X.; Niu, X.; Huang, S.; Li, S.; Wang, J. Whole-genome sequence analysis reveals selection signatures for important economic traits in Xiang pigs. Sci. Rep. 2022, 12, 11823. [Google Scholar] [CrossRef]
- Niu, N.; Wang, H.; Shi, G.; Liu, X.; Liu, H.; Liu, Q.; Yang, M.; Wang, L.; Zhang, L. Genome scanning reveals novel candidate genes for vertebral and teat number in the Beijing Black Pig. Anim. Genet. 2021, 52, 734–738. [Google Scholar] [CrossRef]
- Li, L.Y.; Xiao, S.J.; Tu, J.M.; Zhang, Z.K.; Zheng, H.; Huang, L.B.; Huang, Z.Y.; Yan, M.; Liu, X.D.; Guo, Y.M. A further survey of the quantitative trait loci affecting swine body size and carcass traits in five related pig populations. Anim. Genet. 2021, 52, 621–632. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Yu, D.; Hao, J.; Zhang, L.; Adeola, A.C.; Mao, B.; Gao, Y.; Wu, S.; Zhu, C.; et al. Single-cell RNA sequencing reveals thoracolumbar vertebra heterogeneity and rib-genesis in pigs. Genom. Proteom. Bioinform. 2021, 19, 423–436. [Google Scholar] [CrossRef]
- Zhang, H.; Zhuang, Z.; Yang, M.; Ding, R.; Quan, J.; Zhou, S.; Gu, T.; Xu, Z.; Zheng, E.; Cai, G.; et al. Genome-wide detection of genetic loci and candidate genes for body conformation traits in Duroc× Landrace× Yorkshire crossbred pigs. Front. Genet. 2021, 12, 664343. [Google Scholar] [CrossRef]
- Liu, Q.; Yue, J.; Niu, N.; Liu, X.; Yan, H.; Zhao, F.; Hou, X.; Gao, H.; Shi, L.; Wang, L.; et al. Genome-wide association analysis identified BMPR1A as a novel candidate gene affecting the number of thoracic vertebrae in a Large White× Minzhu intercross pig population. Animals 2020, 10, 2186. [Google Scholar] [CrossRef]
- Zheng, Y.; Cai, C.H.; Wei, C.H.; Wang, X.Y.; Wei, W.A.; Bo, G.A.; Wimmers, K.; Mao, J.D.; Song, C.Y. Two new SINE insertion polymorphisms in pig Vertnin (VRTN) gene revealed by comparative genomic alignment. J. Integr. Agric. 2020, 19, 2514–2522. [Google Scholar] [CrossRef]
- Duan, Y.; Zhang, H.; Zhang, Z.; Gao, J.; Yang, J.; Wu, Z.; Fan, Y.; Xing, Y.; Li, L.; Xiao, S.; et al. VRTN is required for the development of thoracic vertebrae in mammals. Int. J. Biol. Sci. 2018, 14, 667. [Google Scholar] [CrossRef]
- Van Son, M.; Lopes, M.S.; Martell, H.J.; Derks, M.F.; Gangsei, L.E.; Kongsro, J.; Wass, M.N.; Grindflek, E.H.; Harlizius, B. A QTL for the number of teats shows breed-specific effects on the number of vertebrae in pigs: Bridging the gap between molecular and quantitative genetics. Front. Genet. 2019, 10, 272. [Google Scholar] [CrossRef]
- Amundson, L.A.; Hernandez, L.L.; Crenshaw, T.D. Gene expression of matrix metalloproteinase 9 (MMP9), matrix metalloproteinase 13 (MMP13), vascular endothelial growth factor (VEGF), and fibroblast growth factor 23 (FGF23) in femur and vertebra tissues of the hypovitaminosis D kyphotic pig model. Br. J. Nutr. 2018, 120, 404–414. [Google Scholar] [CrossRef]
- Zhang, L.C.; Xin, L.I.; Liang, J.; Hua, Y.A.; Zhao, K.B.; Na, L.I.; Lei, P.U.; Shi, H.B.; Zhang, Y.B.; Wang, L.G.; et al. Quantitative trait loci for the number of vertebrae on Sus scrofa chromosomes 1 and 7 independently influence the numbers of thoracic and lumbar vertebrae in pigs. J. Integr. Agric. 2015, 14, 2027–2033. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, T.; Shi, X.; Wang, X.; Ren, W.; Huang, B.; Wang, C. Identification of LTBP2 gene polymorphisms and their association with thoracolumbar vertebrae number, body size, and carcass traits in Dezhou donkeys. Front. Genet. 2022, 13, 969959. [Google Scholar] [CrossRef]
- Huang, B.; Khan, M.Z.; Chai, W.; Ullah, Q.; Wang, C. Exploring Genetic Markers: Mitochondrial DNA and Genomic Screening for Biodiversity and Production Traits in Donkeys. Animals 2023, 13, 2725. [Google Scholar] [CrossRef]
- Koltes, J.E.; Mishra, B.P.; Kumar, D.; Kataria, R.S.; Totir, L.R.; Fernando, R.L.; Cobbold, R.; Steffen, D.; Coppieters, W.; Georges, M.; et al. A nonsense mutation in cGMP-dependent type II protein kinase (PRKG2) causes dwarfism in American Angus cattle. Proc. Natl. Acad. Sci. USA 2009, 106, 19250–19255. [Google Scholar] [CrossRef]
- Soranzo, N.; Rivadeneira, F.; Chinappen-Horsley, U.; Malkina, I.; Richards, J.B.; Hammond, N.; Stolk, L.; Nica, A.; Inouye, M.; Hofman, A.; et al. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009, 5, e1000445. [Google Scholar] [CrossRef]
- Tsuchida, A.; Yokoi, N.; Namae, M.; Fuse, M.; Masuyama, T.; Sasaki, M.; Kawazu, S.; Komeda, K. Phenotypic Characterization of the Komeda Miniature Rat Ishikawa, an Animal Model of Dwarfism Caused by a Mutation in Prkg2. Comp. Med. 2008, 58, 560–567. [Google Scholar]
- Garces, G.R.; Turba, M.E.; Muracchini, M.; Diana, A.; Jagannathan, V.; Gentilini, F.; Leeb, T. PRKG2 Splice Site Variant in Dogo Argentino Dogs with Disproportionate Dwarfism. Genes 2021, 12, 1489. [Google Scholar] [CrossRef]
- Yi, X.; Wu, P.; Liu, J.; He, S.; Gong, Y.; Xiong, J.; Xu, X.; Li, W. Candidate kinases for adipogenesis and osteoblastogenesis from human bone marrow mesenchymal stem cells. Mol. Omics 2021, 17, 790–795. [Google Scholar] [CrossRef]
- Zhong, Y.J.; Yang, Y.; Wang, X.Y.; Di, R.; Chu, M.X.; Liu, Q.Y. Expression analysis and single-nucleotide polymorphisms of SYNDIG1L and UNC13C genes associated with thoracic vertebral numbers in sheep (Ovis aries). Arch. Anim. Breed. 2021, 64, 131–138. [Google Scholar] [CrossRef]
- Li, S.; Luo, R.; Lai, D.; Ma, M.; Hao, F.; Qi, X.; Liu, X. Whole-genome resequencing of Ujumqin sheep to investigate the determinants of the multi-vertebral trait. Genome 2018, 61, 653–661. [Google Scholar] [CrossRef]
- Li, C.; Zhang, X.; Cao, Y.; Wei, J.; You, S.; Jiang, Y.; Chen, C. Multivertebrae variation potentially contributes to carcass length and weight of Kazakh sheep. Small Rumin. Res. 2017, 150, 8–10. [Google Scholar] [CrossRef]
- Zhang, Z.; Sun, Y.; Du, W.; He, S.; Liu, M.; Tian, C. Effects of vertebral number variations on carcass traits and genotyping of Vertnin candidate gene in Kazakh sheep. Asian-Australas. J. Anim. Sci. 2017, 30, 123. [Google Scholar] [CrossRef]
- Mi, T.; Liu, K.; Guo, T.; Li, L.; Wang, Y.; Li, C.; Cui, Y.; Dai, J.; Zhang, Y.; Hu, S. Analysis of the eighth intron polymorphism of NR6A1 gene in sheep and its correlation with lumbar spine number. Anim. Biotechnol. 2023, 34, 218–224. [Google Scholar] [CrossRef]
- Li, C.; Liu, K.; Dai, J.; Li, X.; Liu, X.; Ni, W.; Li, H.; Wang, D.; Qiao, J.; Wang, Y.; et al. Whole-genome resequencing to investigate the determinants of the multi-lumbar vertebrae trait in sheep. Gene 2022, 809, 146020. [Google Scholar] [CrossRef]
- Kalds, P.; Luo, Q.; Sun, K.; Zhou, S.; Chen, Y.; Wang, X. Trends towards revealing the genetic architecture of sheep tail patterning: Promising genes and investigatory pathways. Anim. Genet. 2021, 52, 799–812. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, X.; Li, F.; Liu, T.; Hu, Z.; Gao, N.; Yuan, L.; Li, X.; Zhao, Y.; Zhao, L.; et al. Whole-genome resequencing identified candidate genes associated with the number of ribs in Hu sheep. Genomics 2021, 113, 2077–2084. [Google Scholar] [CrossRef]
- Zhao, F.; Deng, T.; Shi, L.; Wang, W.; Zhang, Q.; Du, L.; Wang, L. Genomic scan for selection signature reveals fat deposition in Chinese indigenous sheep with extreme tail types. Animals 2020, 10, 773. [Google Scholar] [CrossRef]
- Li, C.; Li, M.; Li, X.; Ni, W.; Xu, Y.; Yao, R.; Wei, B.; Zhang, M.; Li, H.; Zhao, Y.; et al. Whole-genome resequencing reveals loci associated with thoracic vertebrae number in sheep. Front. Genet. 2019, 10, 674. [Google Scholar] [CrossRef]
- Zhang, X.; Li, C.; Li, X.; Liu, Z.; Ni, W.; Hazi, W.; Cao, Y.; Yao, Y.; Wang, D.; Hou, X.; et al. Expression profiles of MicroRNAs from multiple lumbar spine in sheep. Gene 2018, 678, 105–114. [Google Scholar] [CrossRef]
- Moradi, M.H.; Mahmodi, R.; Farahani, A.H.; Karimi, M.O. Genome-Wide Evaluation of Copy Gain and Loss Variations in Three Afghan Sheep Breeds. Sci. Rep. 2022, 12, 14286. [Google Scholar] [CrossRef]
- Ahbara, A.; Bahbahani, H.; Almathen, F.; Al Abri, M.; Agoub, M.O.; Abeba, A.; Kebede, A.; Musa, H.H.; Mastrangelo, S.; Pilla, F.; et al. Genome-Wide Variation, Candidate Regions, and Genes Associated With Fat Deposition and Tail Morphology in Ethiopian Indigenous Sheep. Front. Genet. 2019, 9, 699. [Google Scholar] [CrossRef]
- Chang, Y.C.; Manent, J.; Schroeder, J.; Wong, S.F.; Hauswirth, G.M.; Shylo, N.A.; Moore, E.L.; Achilleos, A.; Garside, V.; Polo, J.M.; et al. Nr6a1 controls Hox expression dynamics and is a master regulator of vertebrate trunk development. Nat. Commun. 2022, 13, 7766. [Google Scholar] [CrossRef]
- Wang, Y.; Wan, X.; Hao, Y.; Zhao, Y.; Du, L.; Huang, Y.; Liu, Z.; Wang, Y.; Wang, N.; Zhang, P. NR6A1 regulates lipid metabolism through mammalian target of rapamycin complex 1 in HepG2 cells. Cell Commun. Signal. 2019, 17, 77. [Google Scholar] [CrossRef]
- Zhang, X.; Li, C.; Li, X.; Liu, Z.; Ni, W.; Cao, Y.; Yao, Y.; Islamov, E.; Wei, J.; Hou, X.; et al. Association analysis of polymorphism in the NR6A1 gene with the lumbar vertebrae number traits in sheep. Genes Genom. 2019, 41, 1165–1171. [Google Scholar] [CrossRef]
- Yan, G.; Qiao, R.; Zhang, F.; Xin, W.; Xiao, S.; Huang, T.; Zhang, Z.; Huang, L. Imputation-based whole-genome sequence association study rediscovered the missing QTL for lumbar number in Sutai pigs. Sci. Rep. 2017, 7, 615. [Google Scholar] [CrossRef]
- Rubin, C.J.; Megens, H.J.; Martinez, B.A.; Maqbool, K.; Andersson, L. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. USA 2012, 109, 19529–19536. [Google Scholar] [CrossRef]
- Fang, X.; Lai, Z.; Liu, J.; Zhang, C.; Li, S.; Wu, F.; Zhou, Z.; Lei, C.; Dang, R. A Novel 13 bp Deletion within the NR6A1 Gene Is Significantly Associated with Growth Traits in Donkeys. Animals 2019, 9, 681. [Google Scholar] [CrossRef]
- Mikawa, S.; Morozumi, T.; Shimanuki, S.I.; Hayashi, T.; Uenishi, H.; Domukai, M.; Okumura, N.; Awata, T. Fine mapping of a swine quantitative trait locus for number of vertebrae and analysis of an orphan nuclear receptor, germ cell nuclear factor (NR6A1). Genome Res. 2007, 17, 586–593. [Google Scholar] [CrossRef]
- Yang, G.; Ren, J.; Zhang, Z.; Huang, L. Genetic evidence for the introgression of Western NR6A1 haplotype into Chinese Licha breed associated with increased vertebral number. Anim. Genet. 2009, 40, 247–250. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, M.; Ye, R.; Ma, Y.; Lei, C. Effects of increased vertebral number on carcass weight in PIC pigs. Anim. Sci. J. 2017, 88, 2057–2062. [Google Scholar] [CrossRef]
- Green, H.E.; Oliveira, H.R.; Alvarenga, A.B.; Scramlin-Zuelly, S.; Grossi, D.; Schinckel, A.P.; Brito, L.F. Genomic background of biotypes related to growth, carcass and meat quality traits in Duroc pigs based on principal component analysis. J. Anim. Breed. Genet. 2023, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Le, T.H.; Christensen, O.F.; Nielsen, B.; Sahana, G. Genome-wide association study for conformation traits in three Danish pig breeds. Genet. Sel. Evol. 2017, 49, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.C.; Yue, J.W.; Pu, L.; Wang, L.G.; Liu, X.; Liang, J.; Yan, H.; Zhao, K.B.; Li, N.; Shi, H.B.; et al. Genome-wide study refines the quantitative trait locus for number of ribs in a Large White× Minzhu intercross pig population and reveals a new candidate gene. Mol. Genet. Genom. 2016, 291, 1885–1890. [Google Scholar] [CrossRef]
- Mikawa, S.; Sato, S.; Nii, M.; Morozumi, T.; Yoshioka, G.; Imaeda, N.; Yamaguchi, T.; Hayashi, T.; Awata, T. Identification of a second gene associated with variation in vertebral number in domestic pigs. BMC Genet. 2011, 12, 5. [Google Scholar] [CrossRef]
- Nakano, H.; Sato, S.; Uemoto, Y.; Kikuchi, T.; Shibata, T.; Kadowaki, H.; Kobayashi, E.; Suzuki, K. Effect of VRTN gene polymorphisms on D uroc pig production and carcass traits, and their genetic relationships. Anim. Sci. J. 2015, 86, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Huang, L.; Yang, M.; Fan, Y.; Li, L.; Fang, S.; Deng, W.; Cui, L.; Zhang, Z.; Ai, H.; et al. Possible introgression of the VRTN mutation increasing vertebral number, carcass length and teat number from Chinese pigs into European pigs. Sci. Rep. 2016, 6, 19240. [Google Scholar] [CrossRef]
- Rohrer, G.A.; Nonneman, D.J.; Wiedmann, R.T.; Schneider, J.F. A study of vertebra number in pigs confirms the association of vertnin and reveals additional QTL. BMC Genet. 2015, 16, 129. [Google Scholar] [CrossRef]
- Hirose, K.; Mikawa, S.; Okumura, N.; Noguchi, G.; Fukawa, K.; Kanaya, N.; Mikawa, A.; Arakawa, A.; Ito, T.; Hayashi, Y.; et al. Association of swine vertnin (VRTN) gene with production traits in D uroc pigs improved using a closed nucleus breeding system. Anim. Sci. J. 2013, 84, 213–221. [Google Scholar] [CrossRef]
- Ren, D.R.; Ren, J.; Ruan, G.F.; Guo, Y.M.; Wu, L.H.; Zhou, L.H.; Li, L.; Zhang, Z.Y.; Huang, L.S. Mapping and fine mapping of quantitative trait loci for the number of vertebrae in a W hite D uroc× C hinese E rhualian intercross resource population. Anim. Genet. 2012, 43, 545–551. [Google Scholar] [CrossRef]
- Bodmer, N.K.; Knutsen, R.H.; Roth, R.A.; Castile, R.M.; Brodt, M.D.; Gierasch, C.M.; Broekelmann, T.J.; Gibson, M.A.; Haspel, J.A.; Lake, S.P.; et al. Multi-organ phenotypes in mice lacking latent TGFβ binding protein 2 (LTBP2). Dev. Dyn. 2023, 253, 233–254. [Google Scholar] [CrossRef]
- Dabovic, B.; Chen, Y.; Colarossi, C.; Zambuto, L.; Obata, H. BEYOND CARRIER PROTEINS Bone defects in latent TGF-β binding protein (Ltbp)-3 null mice; a role for Ltbp in TGF-β presentation. J. Endocrinol. 2002, 175, 129–141. [Google Scholar] [CrossRef]
- Du, X.; Cai, L.; Xie, J.; Zhou, X. The role of TGF-beta3 in cartilage development and osteoarthritis. Bone Res. 2023, 11, 2. [Google Scholar] [CrossRef]
- Nistala, H.; Lee-Arteaga, S.; Smaldone, S.; Siciliano, G.; Carta, L.; Ono, R.N.; Sengle, G.; Arteaga-Solis, E.; Levasseur, R.; Ducy, P.; et al. Fibrillin-1 and-2 differentially modulate endogenous TGF-β and BMP bioavailability during bone formation. J. Cell Biol. 2010, 190, 1107–1121. [Google Scholar] [CrossRef]
- Park, H.B.; Han, S.H.; Lee, J.B.; Cho, I.C. Rapid Communication: High-resolution quantitative trait loci analysis identifies LTBP2 encoding latent transforming growth factor beta binding protein 2 associated with thoracic vertebrae number in a large F2 intercross between Landrace and Korean native pigs. J. Anim. Sci. 2017, 95, 1957–1962. [Google Scholar]
- Sun, X.; Essalmani, R.; Susan-Resiga, D.; Prat, A.; Seidah, N.G. Latent transforming growth factor β-binding proteins-2 and-3 inhibit the proprotein convertase 5/6A. J. Biol. Chem. 2011, 286, 29063–29073. [Google Scholar] [CrossRef]
- Ma, Q.; Yang, Y.; Mao, F.; Zhou, Q.; Wang, L.; Chen, G. Genome-wide identification, phylogeny and expression analysis of the bmp gene family associated with development and skeleton deformity in cobia (Rachycentron canadum). Aquac. Rep. 2023, 31, 101644. [Google Scholar] [CrossRef]
- Li, X.; Cao, X. BMP signaling and skeletogenesis. Ann. N. Y. Acad. Sci. 2006, 1068, 26–40. [Google Scholar] [CrossRef]
- Cao, X.; Chen, D. The BMP signaling and in vivo bone formation. Gene 2005, 357, 1–8. [Google Scholar] [CrossRef]
- Pregizer, S.; Mortlock, D.P. Control of BMP gene expression by long-range regulatory elements. Cytokine Growth Factor. Rev. 2009, 20, 509–515. [Google Scholar] [CrossRef][Green Version]
- Peskin, B.; Norman, J.; Bagwell, J.; Lin, A.; Adhyapok, P.; Di Talia, S.; Bagnat, M. Dynamic BMP signaling mediates notochord segmentation in zebrafish. Curr. Biol. 2023, 33, P2574–P2581.E3. [Google Scholar] [CrossRef]
- Pogoda, H.M.; Riedl-Quinkertz, I.; Hammerschmidt, M. Direct BMP signaling to chordoblasts is required for the initiation of segmented notochord sheath mineralization in zebrafish vertebral column development. Front. Endocrinol. 2023, 14, 1107339. [Google Scholar] [CrossRef]
- Costantini, A.; Guasto, A.; Cormier-Daire, V. TGF-β and BMP Signaling Pathways in Skeletal Dysplasia with Short and Tall Stature. Annu. Rev. Genom. Hum. Genet. 2023, 24, 225–253. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.; Tsuji, K.; Cox, K.; Harfe, B.D.; Rosen, V.; Tabin, C.J. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006, 2, e216. [Google Scholar] [CrossRef]
- Wan, M.; Cao, X. BMP signaling in skeletal development. Biochem. Biophys. Res. Commun. 2005, 328, 651–657. [Google Scholar] [CrossRef]
- Chen, Y.; Shan, X.; Jiang, H.; Sun, L.; Guo, Z. Regulation of litter size in sheep (Ovis aries) by the GDF9 and BMP15 genes. Ann. Agric. Sci. 2023, 68, 148–158. [Google Scholar] [CrossRef]
- Lu, X.; Li, L.; Wu, N.; Chen, W.; Hong, S.; Xu, M.; Ding, Y.; Gao, Y. BMP9 functions as a negative regulator in the myogenic differentiation of primary mouse myoblasts. Biosci. Biotechnol. Biochem. 2023, 87, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, G.; Yang, L.; Sun, M.; Zhang, Z.; Xu, Z.; Gao, Y.; Jiang, X.; Su, Z.; Li, X.; et al. BMP7 expression in mammalian cortical radial glial cells increases the length of the neurogenic period. Protein Cell 2023, 15, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Bottasso-Arias, N.; Burra, K.; Sinner, D.; Riede, T. Disruption of BMP4 signaling is associated with laryngeal birth defects in a mouse model. Dev. Biol. 2023, 500, 10–21. [Google Scholar] [CrossRef]
- Bottasso-Arias, N.; Leesman, L.; Burra, K.; Snowball, J.; Shah, R.; Mohanakrishnan, M.; Xu, Y.; Sinner, D. BMP4 and Wnt signaling interact to promote mouse tracheal mesenchyme morphogenesis. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2022, 322, L224–L242. [Google Scholar] [CrossRef]
- Mallo, M. Reassessing the role of Hox genes during vertebrate development and evolution. Trends Genet. 2018, 34, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Iimura, T.; Denans, N.; Pourquié, O. Establishment of Hox vertebral identities in the embryonic spine precursors. Curr. Top. Dev. Biol. 2009, 88, 201–234. [Google Scholar] [PubMed]
- Lappin, T.R.; Grier, D.G.; Thompson, A.; Halliday, H.L. HOX genes: Seductive science, mysterious mechanisms. Ulst. Med. J. 2006, 75, 23. [Google Scholar]
- Carpenter, E.M. Hox genes and spinal cord development. Dev. Neurosci. 2002, 24, 24–34. [Google Scholar] [CrossRef]
- Hauswirth, G.M.; Garside, V.C.; Wong, L.S.; Bildsoe, H.; Manent, J.; Chang, Y.C.; Nefzger, C.M.; Firas, J.; Chen, J.; Rossello, F.J.; et al. Breaking constraint of mammalian axial formulae. Nat. Commun. 2022, 13, 243. [Google Scholar] [CrossRef] [PubMed]
- Van Den Akker, E.; Fromental-Ramain, C.; de Graaff, W.; Le Mouellic, H.; Brûlet, P.; Chambon, P.; Deschamps, J. Axial skeletal patterning in mice lacking all paralogous group 8 Hox genes. Development 2001, 128, 1911–1921. [Google Scholar] [CrossRef]
- Tang, J.; Liu, Y.; Xie, S.; Ma, S.; Jiang, M. Advances in Candidate Genes on Vertebral Number Trait of Pig. Chin. J. Anim. Sci. 2022, 58, 92–98. [Google Scholar]
- Zhao, J.; Zhang, L.; Qi, C.; Batu. Relationship between methylation of Hoxc8 gene and the numbers of thoracic vertebrae in Mongolia sheep. Heilongjiang Anim. Sci. Vet. 2011, 5, 5. [Google Scholar]
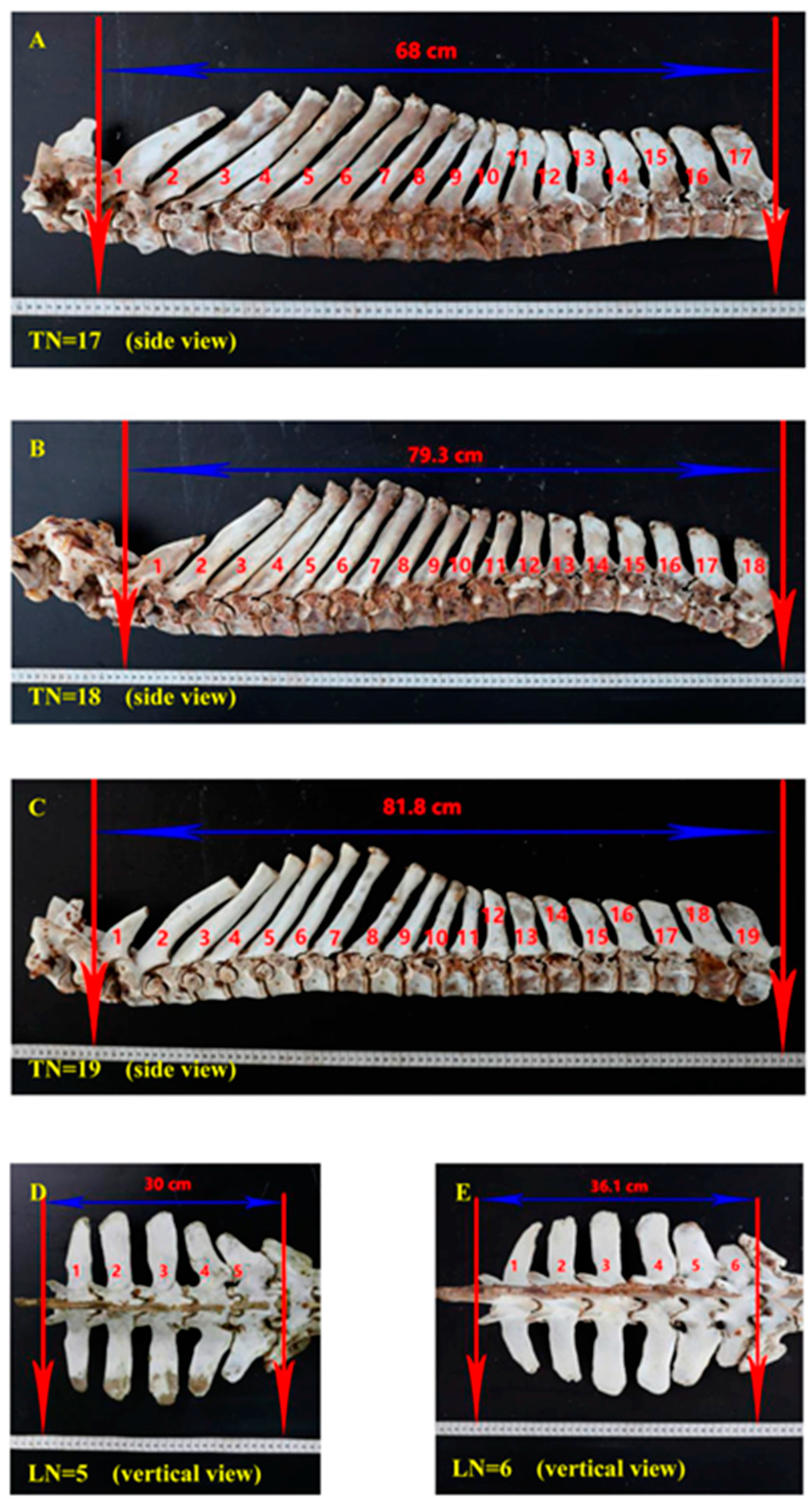
| Genes | Associated Traits | Breeds | Country | Reference |
|---|---|---|---|---|
| RSAD2-CMPK2, COL3A1 |
| Meishan pigs | China | [30] |
| HMGA1, VRTN, BMP2 |
| Duroc × Landrace × Yorkshire crossbred pigs | [31,32] | |
| TIMP2, EML1, SMN1 |
| Pigs | [33] | |
| NR6A1, LTBP2 |
| Xiang pigs | [34] | |
| GREB1L, ABCD4, VRTN, MIB1 |
| Beijing Black pigs | [27] | |
| ABCD4 Hox family genes (HOXB 1–7, 9, and 13), NTRK2 |
| Beijing Black pigs | [35] | |
| NR6A1, VRTN PLAG1, BMP2 MC4R |
| Shanxia Black pigs | [36] | |
| HOXA10 |
| Pigs | [37] | |
| BMP2 |
| Duroc × (Landrace × Yorkshire) hybrid pigs | [38] | |
| VRTN, LTBP2, BMPR1A, FOS |
| White × Minzhu crossbred pigs | [39] | |
| VRTN |
| Sujiang, Meishan, Bama, Erhualian, and Tibetan pigs | [40] | |
| VRTN |
| Suhuai pigs | [13] | |
| VRTN |
| Pigs | [41] | |
| VRTN |
| Duroc, Landrace, and Large White pigs | Norway | [42] |
| MMP9, VEGF |
| Pigs | USA | [43] |
| NR6A1 |
| Large White × Minzhu pigs | China | [44] |
| VRTN, FOS, PROX2, TGFB3 |
|
| Genetic Markers | Biological Effect | Breed | Country | Reference |
|---|---|---|---|---|
| DCAF7 |
| Dezhou donkeys | China | [11] |
| PRKG2 |
| [12] | ||
| NR6A1 |
| [18] | ||
| LTBP2 |
| [46] | ||
| HOXC8 |
| [28] | ||
| NLGN1, DCC, FBXO4 SLC26A7, TOX, LRP5 WNT7A, LOC123286078, LOC123280142, GABBR2, LOC123277146, LOC123277359, BMP7, B3GAT1, EML2 |
| [10] |
| Genes | Associated Traits | Breeds | Country | Reference |
|---|---|---|---|---|
| NR6A1 |
| Xinjiang Kazakh sheep | China | [56] |
| SFRP4 |
| Duolang sheep | [57] | |
| SYNDIG1L, UNC13C |
| Han sheep and Sunite sheep | [52] | |
| TBXT |
| Sheep | [58] | |
| MGAT4A, KCNH1 CPOX, CPQ |
| Hu sheep | [59] | |
| LTBP2, SYNDIG1L |
| Large fat-tailed sheep, Altay sheep, Tibetan sheep | [60] | |
| VRTN, HoxA |
| Xinjiang Kazakh sheep | [61] | |
| NDRG2 |
| Kazakh sheep | [62] | |
| VRTN |
| China Kazakh sheep | [55] | |
| NID2, ACAN |
| Afghani sheep | Iran | [63] |
| ALX4, HOXB13, BMP4 EYA2, SULF2 |
| Ethiopian indigenous sheep | Ethiopia | [64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.Z.; Chen, W.; Huang, B.; Liu, X.; Wang, X.; Liu, Y.; Chai, W.; Wang, C. Advancements in Genetic Marker Exploration for Livestock Vertebral Traits with a Focus on China. Animals 2024, 14, 594. https://doi.org/10.3390/ani14040594
Khan MZ, Chen W, Huang B, Liu X, Wang X, Liu Y, Chai W, Wang C. Advancements in Genetic Marker Exploration for Livestock Vertebral Traits with a Focus on China. Animals. 2024; 14(4):594. https://doi.org/10.3390/ani14040594
Chicago/Turabian StyleKhan, Muhammad Zahoor, Wenting Chen, Bingjian Huang, Xiaotong Liu, Xinrui Wang, Yihong Liu, Wenqiong Chai, and Changfa Wang. 2024. "Advancements in Genetic Marker Exploration for Livestock Vertebral Traits with a Focus on China" Animals 14, no. 4: 594. https://doi.org/10.3390/ani14040594
APA StyleKhan, M. Z., Chen, W., Huang, B., Liu, X., Wang, X., Liu, Y., Chai, W., & Wang, C. (2024). Advancements in Genetic Marker Exploration for Livestock Vertebral Traits with a Focus on China. Animals, 14(4), 594. https://doi.org/10.3390/ani14040594






