Transcriptome Data Revealed the circRNA–miRNA–mRNA Regulatory Network during the Proliferation and Differentiation of Myoblasts in Shitou Goose
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Culture of Goose Myoblasts
2.2. RNA Isolation, Library Construction, and Sequencing
2.3. Read Mapping and Transcriptome Assembly
2.4. Identification of circRNAs
2.5. Differential Expression Analysis of circRNAs and mRNAs
2.6. Prediction of Targeting Relationship
2.7. Functional Enrichment Analysis
2.8. Establishment of the circRNA–miRNA–mRNA/hub Gene Network
2.9. Protein–Protein Interaction (PPI) Network Analysis
3. Results
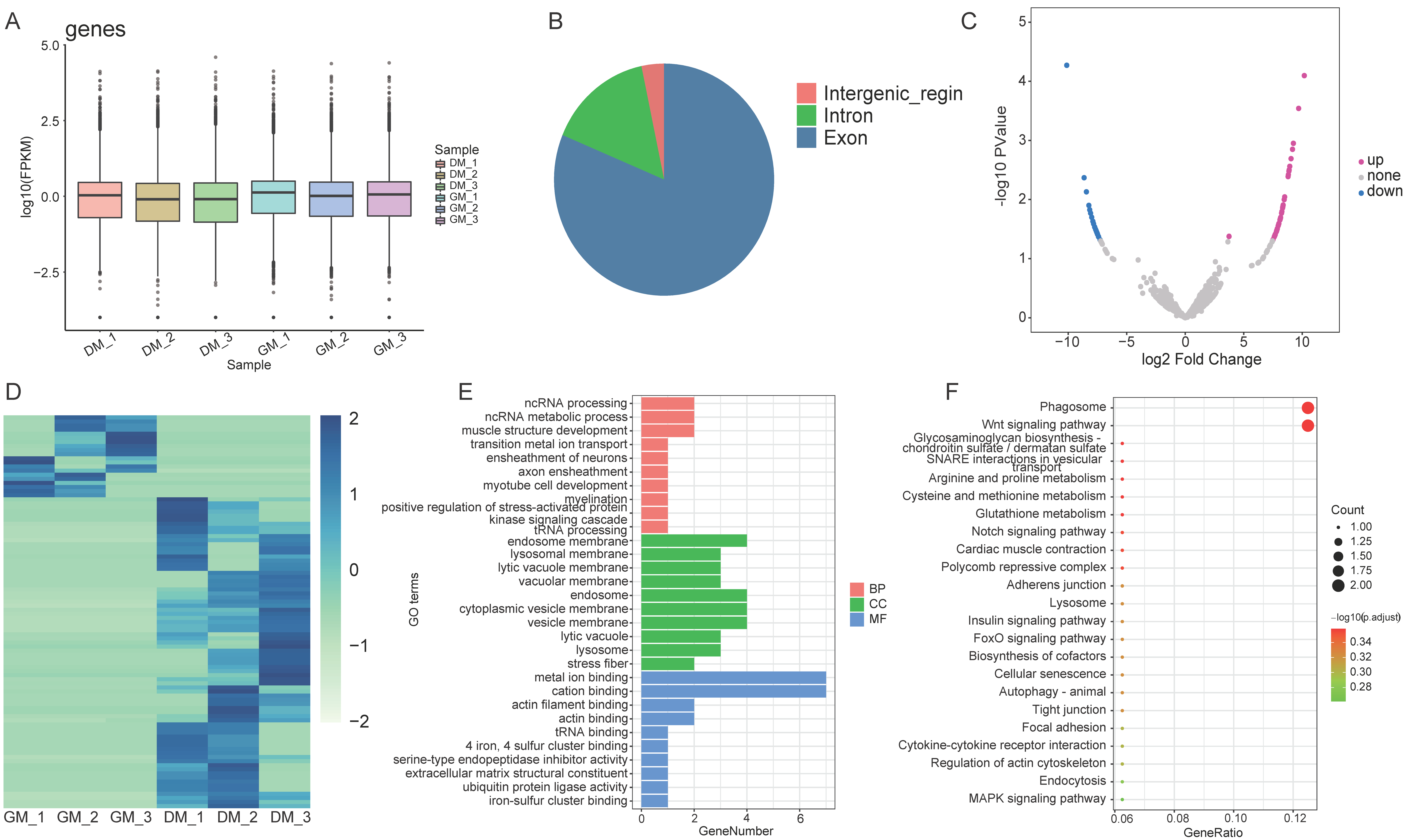
3.1. Analysis of DEcircRNAs in the Proliferation and Differentiation Stages of Myoblasts in Shitou Goose
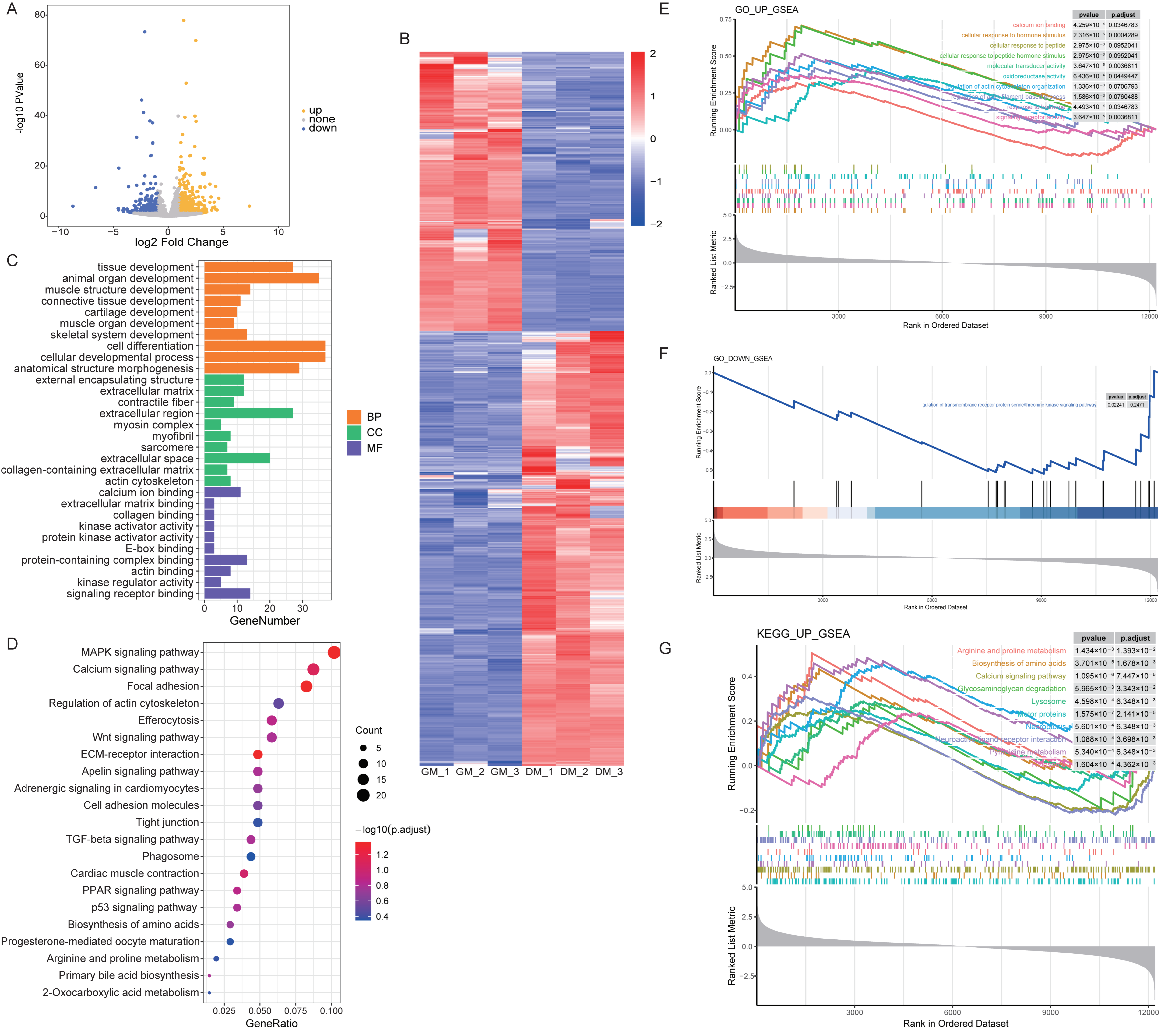
3.2. Analysis of DEmRNAs between Myoblasts and Myotubes in Shitou Goose
3.3. Construction of circRNA–miRNA Pairs and miRNA–mRNA Pairs
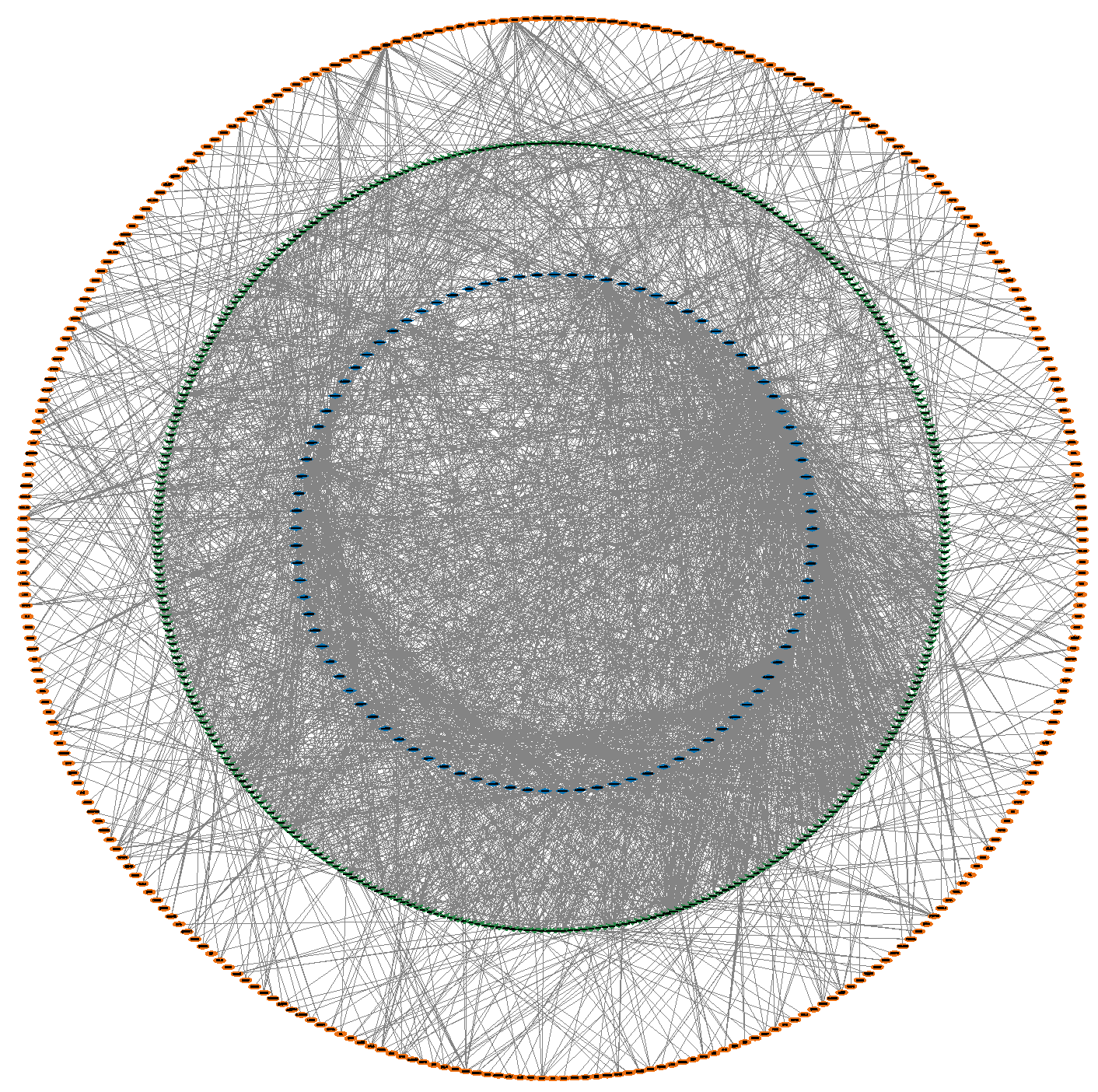
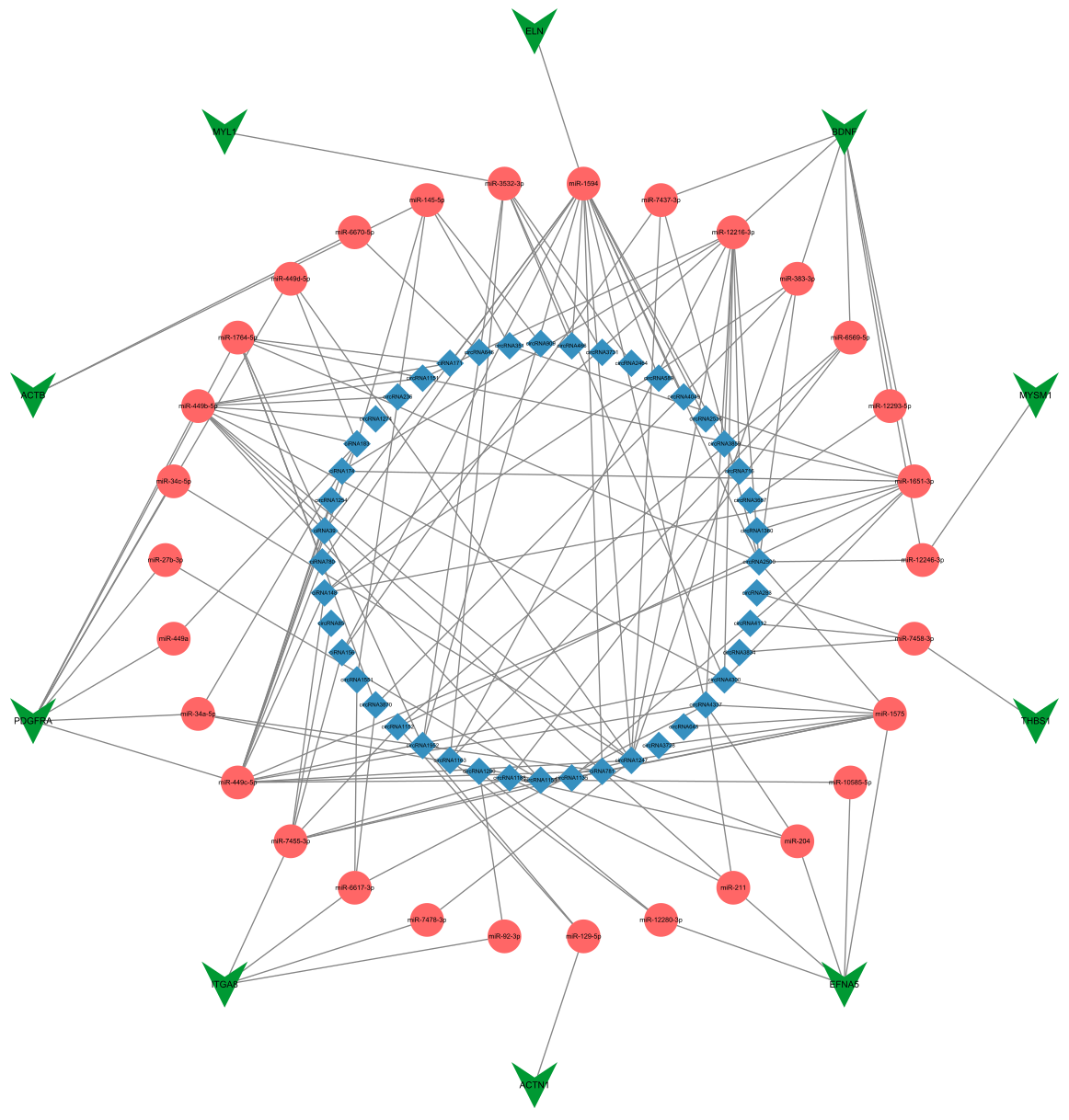
3.4. Construction of the circRNA–miRNA–mRNA Interaction Network
3.5. Establishment of the PPI Network and Identification of Hub Genes
3.6. Reconstructing the circRNA–miRNA–Hub Gene Interaction Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wereńska, M. Comparative study on the effects of sous-vide, microwave cooking, and stewing on functional properties and sensory quality of goose meat. Poult. Sci. 2023, 102, 103064. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shi, W.; Wang, Y.; Meng, X.; Dabbour, M.; Kumah Mintah, B.; Chen, X.; Chen, X.; He, R.; Ma, H. Mono-frequency ultrasonic-assisted thawing of frozen goose meat: Influence on thawing efficiency, product quality and microstructure. Ultrason. Sonochem. 2023, 98, 106489. [Google Scholar] [CrossRef]
- Hong, L.; Sun, Z.; Xu, D.; Li, W.; Cao, N.; Fu, X.; Huang, Y.; Tian, Y.; Li, B. Transcriptome and lipidome integration unveils mechanisms of fatty liver formation in Shitou geese. Poult. Sci. 2023, 103, 103280. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Ouyang, H.; Chen, X.; Jiang, D.; Tian, Y.; Huang, Y.; Shen, X. Comparative Transcriptome Analyses of Leg Muscle during Early Growth between Geese (Anser cygnoides) Breeds Differing in Body Size Characteristics. Genes 2023, 14, 1048. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Jin, J.; Xu, Z.; Zuo, B. Functions and Regulatory Mechanisms of lncRNAs in Skeletal Myogenesis, Muscle Disease and Meat Production. Cells 2019, 8, 1107. [Google Scholar] [CrossRef]
- Shen, L.; Liao, T.; Chen, Q.; Lei, Y.; Wang, L.; Gu, H.; Qiu, Y.; Zheng, T.; Yang, Y.; Wei, C.; et al. tRNA-derived small RNA, 5’tiRNA-Gly-CCC, promotes skeletal muscle regeneration through the inflammatory response. J. Cachexia Sarcopenia Muscle 2023, 14, 1033–1045. [Google Scholar] [CrossRef]
- Smith, J.A.B.; Murach, K.A.; Dyar, K.A.; Zierath, J.R. Exercise metabolism and adaptation in skeletal muscle. Nat. Rev. Mol. Cell Biol. 2023, 24, 607–632. [Google Scholar] [CrossRef]
- Sweeney, H.L.; Hammers, D.W. Muscle Contraction. Cold Spring Harb. Perspect. Biol. 2018, 10, a023200. [Google Scholar] [CrossRef]
- Huo, W.; Weng, K.; Li, Y.; Zhang, Y.; Zhang, Y.; Xu, Q.; Chen, G. Comparison of muscle fiber characteristics and glycolytic potential between slow- and fast-growing broilers. Poult. Sci. 2022, 101, 101649. [Google Scholar] [CrossRef]
- Yin, H.; Zhang, S.; Gilbert, E.R.; Siegel, P.B.; Zhu, Q.; Wong, E.A. Expression profiles of muscle genes in postnatal skeletal muscle in lines of chickens divergently selected for high and low body weight. Poult. Sci. 2014, 93, 147–154. [Google Scholar] [CrossRef]
- Wang, H.; He, K.; Zeng, X.; Zhou, X.; Yan, F.; Yang, S.; Zhao, A. Isolation and identification of goose skeletal muscle satellite cells and preliminary study on the function of C1q and tumor necrosis factor-related protein 3 gene. Anim. Biosci. 2021, 34, 1078–1087. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Shang, P.; Zhang, B.; Tian, X.; Nie, R.; Zhang, R.; Zhang, H. Function of the Porcine TRPC1 Gene in Myogenesis and Muscle Growth. Cells 2021, 10, 147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Y.; Zhu, C.; Li, F.; Liu, Z.; Li, X.; Shen, X.; Wu, Z.; Fu, M.; Xu, D.; et al. DNA Demethylation of Myogenic Genes May Contribute to Embryonic Leg Muscle Development Differences between Wuzong and Shitou Geese. Int. J. Mol. Sci. 2023, 24, 7188. [Google Scholar] [CrossRef]
- Chen, G.; Qi, L.; Zhang, S.; Peng, H.; Lin, Z.; Zhang, X.; Nie, Q.; Luo, W. Metabolomic, lipidomic, and proteomic profiles provide insights on meat quality differences between Shitou and Wuzong geese. Food Chem. 2024, 438, 137967. [Google Scholar] [CrossRef] [PubMed]
- Dey, B.K.; Pfeifer, K.; Dutta, A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014, 28, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Jiang, W.; Luo, H.; Tong, Q.; Niu, X.; Liu, X.; Miao, Y.; Wang, J.; Guo, Y.; Li, J.; et al. Long noncoding RNA lncMREF promotes myogenic differentiation and muscle regeneration by interacting with the Smarca5/p300 complex. Nucleic Acids Res. 2022, 50, 10733–10755. [Google Scholar] [CrossRef] [PubMed]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.; Xu, Z.; Zhang, D.; Xia, P.; Ling, J.; Tang, X.; Liu, X.; Xuan, R.; Zhang, M.; et al. Regulation of NcRNA-protein binding in diabetic foot. Biomed. Pharmacother. 2023, 160, 114361. [Google Scholar] [CrossRef]
- Beermann, J.; Piccoli, M.T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Liu, C.-X.; Chen, L.-L. Circular RNAs: Characterization, cellular roles, and applications. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.; Dinger, M. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, N.; Zhang, W.; Hei, W.; Cai, C.; Yang, Y.; Lu, C.; Gao, P.; Guo, X.; Cao, G.; et al. Comprehensive analysis of differentially expressed circRNAs and ceRNA regulatory network in porcine skeletal muscle. BMC Genom. 2021, 22, 320. [Google Scholar] [CrossRef]
- Yu, Z.; Xu, X.; Ai, N.; Wang, K.; Zhang, P.; Li, X.; LiuFu, S.; Liu, X.; Jiang, J.; Gu, J.; et al. Integrated analysis of circRNA, lncRNA, miRNA and mRNA to reveal the ceRNA regulatory network of postnatal skeletal muscle development in Ningxiang pig. Front. Cell Dev. Biol. 2023, 11, 1185823. [Google Scholar] [CrossRef]
- Lei, Q.; Hu, X.; Han, H.; Wang, J.; Liu, W.; Zhou, Y.; Cao, D.; Li, F.; Liu, J. Integrative analysis of circRNA, miRNA, and mRNA profiles to reveal ceRNA regulation in chicken muscle development from the embryonic to post-hatching periods. BMC Genom. 2022, 23, 342. [Google Scholar] [CrossRef]
- Dong, X.; Xing, J.; Liu, Q.; Ye, M.; Zhou, Z.; Li, Y.; Huang, R.; Li, Z.; Nie, Q. CircPLXNA2 Affects the Proliferation and Apoptosis of Myoblast through circPLXNA2/gga-miR-12207-5P/MDM4 Axis. Int. J. Mol. Sci. 2023, 24, 5459. [Google Scholar] [CrossRef]
- Wang, J.; Ren, Q.; Hua, L.; Chen, J.; Zhang, J.; Bai, H.; Li, H.; Xu, B.; Shi, Z.; Cao, H.; et al. Comprehensive Analysis of Differentially Expressed mRNA, lncRNA and circRNA and Their ceRNA Networks in the Longissimus Dorsi Muscle of Two Different Pig Breeds. Int. J. Mol. Sci. 2019, 20, 1107. [Google Scholar] [CrossRef]
- Tang, R.; Zhu, L.; Zhu, P.; Yin, R.; Zheng, C. The Effect of Blood Clots on the Quality of RNA Extracted from PAXgene Blood RNA Tubes. Biopreserv. Biobank. 2023, ahead of print. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–460. [Google Scholar] [CrossRef]
- Kim, D.; Salzberg, S.L. TopHat-Fusion: An algorithm for discovery of novel fusion transcripts. Genome Biol. 2011, 12, R72. [Google Scholar] [CrossRef]
- Zhang, X.O.; Dong, R.; Zhang, Y.; Zhang, J.L.; Luo, Z.; Zhang, J.; Chen, L.L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef]
- Wei, Y.; Tian, Y.; Li, X.; Amevor, F.K.; Shen, X.; Zhao, J.; Zhao, X.; Zhang, X.; Huang, W.; Hu, J.; et al. Circular RNA circFNDC3AL Upregulates BCL9 Expression to Promote Chicken Skeletal Muscle Satellite Cells Proliferation and Differentiation by Binding to miR-204. Front. Cell Dev. Biol. 2021, 9, 736749. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Du, J.; Shen, L.; Tan, Z.; Jiang, D.; Jiang, A.; Li, Q.; Tang, G.; Jiang, Y.; Wang, J.; et al. MiR-204-5p regulates C2C12 myoblast differentiation by targeting MEF2C and ERRγ. Biomed. Pharmacother. 2018, 101, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Ravenscroft, G.; Zaharieva, I.T.; Bortolotti, C.A.; Lambrughi, M.; Pignataro, M.; Borsari, M.; Sewry, C.A.; Phadke, R.; Haliloglu, G.; Ong, R.; et al. Bi-allelic mutations in MYL1 cause a severe congenital myopathy. Hum. Mol. Genet. 2018, 27, 4263–4272. [Google Scholar] [CrossRef] [PubMed]
- Ling, F.; Fang, W.; Chen, Y.; Li, J.; Liu, X.; Wang, L.; Zhang, H.; Chen, S.; Mei, Y.; Du, H.; et al. Identification of novel transcripts from the porcine MYL1 gene and initial characterization of its promoters. Mol. Cell. Biochem. 2010, 343, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Martin, S.; Rochat, A.; Mademtzoglou, D.; Morais, J.; de Reyniès, A.; Auradé, F.; Chang, T.H.; Zammit, P.S.; Relaix, F. Gene Expression Profiling of Muscle Stem Cells Identifies Novel Regulators of Postnatal Myogenesis. Front. Cell Dev. Biol. 2016, 4, 58. [Google Scholar] [CrossRef] [PubMed]
- Swartz, M.E.; Eberhart, J.; Pasquale, E.B.; Krull, C.E. EphA4/ephrin-A5 interactions in muscle precursor cell migration in the avian forelimb. Development 2001, 128, 4669–4680. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Zhang, Y.; Wu, P.; Li, D.; Lu, Y.; Shen, P.; Yang, T.; Shi, G.; Chen, Q.; Yuan, H.; et al. CircSTX6 promotes pancreatic ductal adenocarcinoma progression by sponging miR-449b-5p and interacting with CUL2. Mol. Cancer 2022, 21, 121. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Zhao, C.; Qian, S.; Tai, Y.; Guo, Y.; Tang, C.; Huang, Z.; Gao, J. Proangiogenic role of circRNA-007371 in liver fibrosis. Cell Prolif. 2023, 56, e13432. [Google Scholar] [CrossRef]
- Wang, K.; Long, B.; Liu, F.; Wang, J.X.; Liu, C.Y.; Zhao, B.; Zhou, L.Y.; Sun, T.; Wang, M.; Yu, T.; et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016, 37, 2602–2611. [Google Scholar] [CrossRef]
- Yao, Z.; Xu, R.; Yuan, L.; Xu, M.; Zhuang, H.; Li, Y.; Zhang, Y.; Lin, N. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019, 10, 945. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, H.; Li, Z.; Li, F.; Liang, L.; Zou, Y.; Shen, H.; Li, J.; Xia, Y.; Cheng, Z.; et al. Circular RNA ACTN4 promotes intrahepatic cholangiocarcinoma progression by recruiting YBX1 to initiate FZD7 transcription. J. Hepatol. 2022, 76, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.; Wang, J.; Song, C.; Wu, J.; Cao, X.; Huang, Y.; Lan, X.; Lei, C.; Huang, B.; Chen, H. Biogenesis and ceRNA role of circular RNAs in skeletal muscle myogenesis. Int. J. Biochem. Cell Biol. 2019, 117, 105621. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Cui, C.; Tang, S.; Han, S.; Zhang, Y.; Xia, L.; Tan, B.; Ma, M.; Kang, H.; Yu, J.; et al. MyoG-enhanced circGPD2 regulates chicken skeletal muscle development by targeting miR-203a. Int. J. Biol. Macromol. 2022, 222 Pt B, 2212–2224. [Google Scholar] [CrossRef]
- Chen, M.M.; Zhao, Y.P.; Zhao, Y.; Deng, S.L.; Yu, K. Regulation of Myostatin on the Growth and Development of Skeletal Muscle. Front. Cell Dev. Biol. 2021, 9, 785712. [Google Scholar] [CrossRef]
- Park, J.W.; Lee, J.H.; Han, J.S.; Shin, S.P.; Park, T.S. Muscle differentiation induced by p53 signaling pathway-related genes in myostatin-knockout quail myoblasts. Mol. Biol. Rep. 2020, 47, 9531–9540. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Franco, D.; Kalra, R.; Draper, I.; Pacak, C.A.; Asakura, A.; Kang, P.B. The Notch signaling pathway in skeletal muscle health and disease. Muscle Nerve 2022, 66, 530–544. [Google Scholar] [CrossRef]
- Ilha, J.; do Espírito-Santo, C.C.; de Freitas, G.R. mTOR Signaling Pathway and Protein Synthesis: From Training to Aging and Muscle Autophagy. Adv. Exp. Med. Biol. 2018, 1088, 139–151. [Google Scholar] [CrossRef]
- Keren, A.; Tamir, Y.; Bengal, E. The p38 MAPK signaling pathway: A major regulator of skeletal muscle development. Mol. Cell. Endocrinol. 2006, 252, 224–230. [Google Scholar] [CrossRef]
- Tao, H.; Yang, J.J.; Shi, K.H.; Li, J. Wnt signaling pathway in cardiac fibrosis: New insights and directions. Metabolism 2016, 65, 30–40. [Google Scholar] [CrossRef]
- Rudnicki, M.A.; Williams, B.O. Wnt signaling in bone and muscle. Bone 2015, 80, 60–66. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, J.; Luo, H.; Meng, X.; Chen, M.; Zhu, D. Wnt signaling pathway in cancer immunotherapy. Cancer Lett. 2022, 525, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Chen, N.; Fu, Z.; Zhang, Q. Progress of Wnt Signaling Pathway in Osteoporosis. Biomolecules 2023, 13, 483. [Google Scholar] [CrossRef] [PubMed]
- van Amerongen, R.; Berns, A. Knockout mouse models to study Wnt signal transduction. Trends Genet. 2006, 22, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Yin, L.; Lin, Z.; Yu, C.; Li, J.; Ren, P.; Yang, C.; Qiu, M.; Liu, Y. miR-136-5p/FZD4 axis is critical for Wnt signaling-mediated myogenesis and skeletal muscle regeneration. J. Cell. Physiol. 2023, ahead of print. [Google Scholar] [CrossRef]
- Chen, M.; Wei, X.; Song, M.; Jiang, R.; Huang, K.; Deng, Y.; Liu, Q.; Shi, D.; Li, H. Circular RNA circMYBPC1 promotes skeletal muscle differentiation by targeting MyHC. Mol. Ther. Nucleic Acids 2021, 24, 352–368. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Yu, J.; Guo, L.; Byers, M.S.; Wang, Z.; Chen, X.; Xu, H.; Nie, Q. Circular RNA circHIPK3 Promotes the Proliferation and Differentiation of Chicken Myoblast Cells by Sponging miR-30a-3p. Cells 2019, 8, 177. [Google Scholar] [CrossRef] [PubMed]
- Geng, T.; Liu, Y.; Xu, Y.; Jiang, Y.; Zhang, N.; Wang, Z.; Carmichael, G.G.; Taylor, H.S.; Li, D.; Huang, Y. H19 lncRNA Promotes Skeletal Muscle Insulin Sensitivity in Part by Targeting AMPK. Diabetes 2018, 67, 2183–2198. [Google Scholar] [CrossRef]
- Cuvertino, S.; Stuart, H.M.; Chandler, K.E.; Roberts, N.A.; Armstrong, R.; Bernardini, L.; Bhaskar, S.; Callewaert, B.; Clayton-Smith, J.; Davalillo, C.H.; et al. ACTB Loss-of-Function Mutations Result in a Pleiotropic Developmental Disorder. Am. J. Hum. Genet. 2017, 101, 1021–1033. [Google Scholar] [CrossRef]
- Yang, X.; Pang, Y.; Zhang, J.; Shi, J.; Zhang, X.; Zhang, G.; Yang, S.; Wang, J.; Hu, K.; Wang, J.; et al. High Expression Levels of ACTN1 and ACTN3 Indicate Unfavorable Prognosis in Acute Myeloid Leukemia. J. Cancer 2019, 10, 4286–4292. [Google Scholar] [CrossRef]
- Matsumoto, M.; Tsuru, H.; Suginobe, H.; Narita, J.; Ishii, R.; Hirose, M.; Hashimoto, K.; Wang, R.; Yoshihara, C.; Ueyama, A.; et al. Atomic force microscopy identifies the alteration of rheological properties of the cardiac fibroblasts in idiopathic restrictive cardiomyopathy. PLoS ONE 2022, 17, e0275296. [Google Scholar] [CrossRef]
- Muñoz, L.M.; Zayachkivsky, A.; Kunz, R.B.; Hunt, J.M.; Wang, G.; Scott, S.A. Ephrin-A5 inhibits growth of embryonic sensory neurons. Dev. Biol. 2005, 283, 397–408. [Google Scholar] [CrossRef]
- Zmojdzian, M.; Jagla, K. The relationship between muscle stem cells and motor neurons. Cell. Mol. Life Sci. 2021, 78, 5043–5049. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Shi, H.; Li, Z.; Kang, J.; Li, M.; Liu, M.; Liu, Y.; Zhao, J.; Dou, T.; Jia, J.; et al. Identification of New Genes and Genetic Variant Loci Associated with Breast Muscle Development in the Mini-Cobb F2 Chicken Population Using a Genome-Wide Association Study. Genes 2022, 13, 2153. [Google Scholar] [CrossRef] [PubMed]
- Marek, I.; Lichtneger, T.; Cordasic, N.; Hilgers, K.F.; Volkert, G.; Fahlbusch, F.; Rascher, W.; Hartner, A.; Menendez-Castro, C. Alpha8 Integrin (Itga8) Signalling Attenuates Chronic Renal Interstitial Fibrosis by Reducing Fibroblast Activation, Not by Interfering with Regulation of Cell Turnover. PLoS ONE 2016, 11, e0150471. [Google Scholar] [CrossRef] [PubMed]

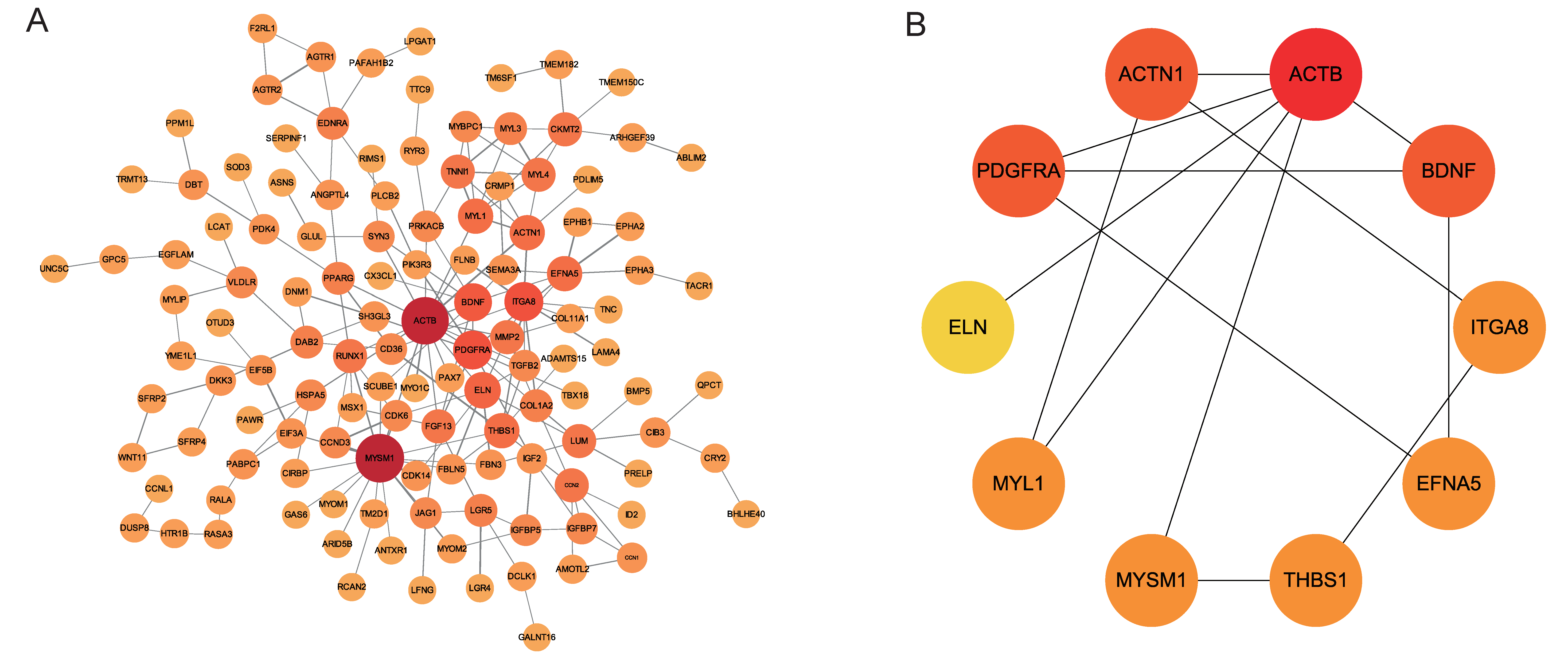
| Name | Score | Gene Description |
|---|---|---|
| ACTB | 6 | actin beta |
| ACTN1 | 3 | actinin alpha 1 |
| BDNF | 3 | brain-derived neurotrophic factor |
| PDGFRA | 3 | platelet-derived growth factor receptor alpha |
| MYL1 | 2 | myosin light chain 1 |
| EFNA5 | 2 | ephrin-A5 |
| MYSM1 | 2 | Myb-like, SWIRM, and MPN domains 1 |
| THBS1 | 2 | thrombospondin 1 |
| ITGA8 | 2 | integrin subunit alpha 8 |
| ELN | 1 | elastin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, R.; Chen, J.; Dong, X.; Zhang, X.; Luo, W. Transcriptome Data Revealed the circRNA–miRNA–mRNA Regulatory Network during the Proliferation and Differentiation of Myoblasts in Shitou Goose. Animals 2024, 14, 576. https://doi.org/10.3390/ani14040576
Huang R, Chen J, Dong X, Zhang X, Luo W. Transcriptome Data Revealed the circRNA–miRNA–mRNA Regulatory Network during the Proliferation and Differentiation of Myoblasts in Shitou Goose. Animals. 2024; 14(4):576. https://doi.org/10.3390/ani14040576
Chicago/Turabian StyleHuang, Rongqin, Jiahui Chen, Xu Dong, Xiquan Zhang, and Wen Luo. 2024. "Transcriptome Data Revealed the circRNA–miRNA–mRNA Regulatory Network during the Proliferation and Differentiation of Myoblasts in Shitou Goose" Animals 14, no. 4: 576. https://doi.org/10.3390/ani14040576
APA StyleHuang, R., Chen, J., Dong, X., Zhang, X., & Luo, W. (2024). Transcriptome Data Revealed the circRNA–miRNA–mRNA Regulatory Network during the Proliferation and Differentiation of Myoblasts in Shitou Goose. Animals, 14(4), 576. https://doi.org/10.3390/ani14040576





