Polymorphism of the CSN3 3’UTR in Dairy Cows Causes Changes in bta-miR-708 Binding Ability and κ-Casein Expression
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Animals and Samples
2.3. RNA Extraction and cDNA Synthesis
2.4. Full-Length CSN3 3’UTR Amplification and Sequencing
2.5. Quantitative Real-Time PCR (qPCR) of CSN1S1, CSN1S2, CSN2, and CSN3
2.6. Western Blot Analysis of CSN3
2.7. Bioinformatics Analysis of CSN3 3’UTR Polymorphisms
2.8. Dual Fluorescence Assay for miRNA Binding
2.9. Assessment of Dairy Herd Improvement (DHI) Data
2.10. Genomic DNA Sequencing of the CSN3 3’UTR
2.11. Statistical Analysis
3. Results
3.1. CSN3 3’UTR Polymorphisms
3.2. The Effect of CSN3 3’UTR Polymorphisms on the Gene Expression of CSN1S1, CSN1S2, CSN2, and CSN3
3.3. The Effect of CSN3 3’UTR Polymorphisms on the CSN3 Protein
3.4. The Effect of CSN3 3’UTR Polymorphisms on Its Secondary Structure and Possibly Binding miRNAs
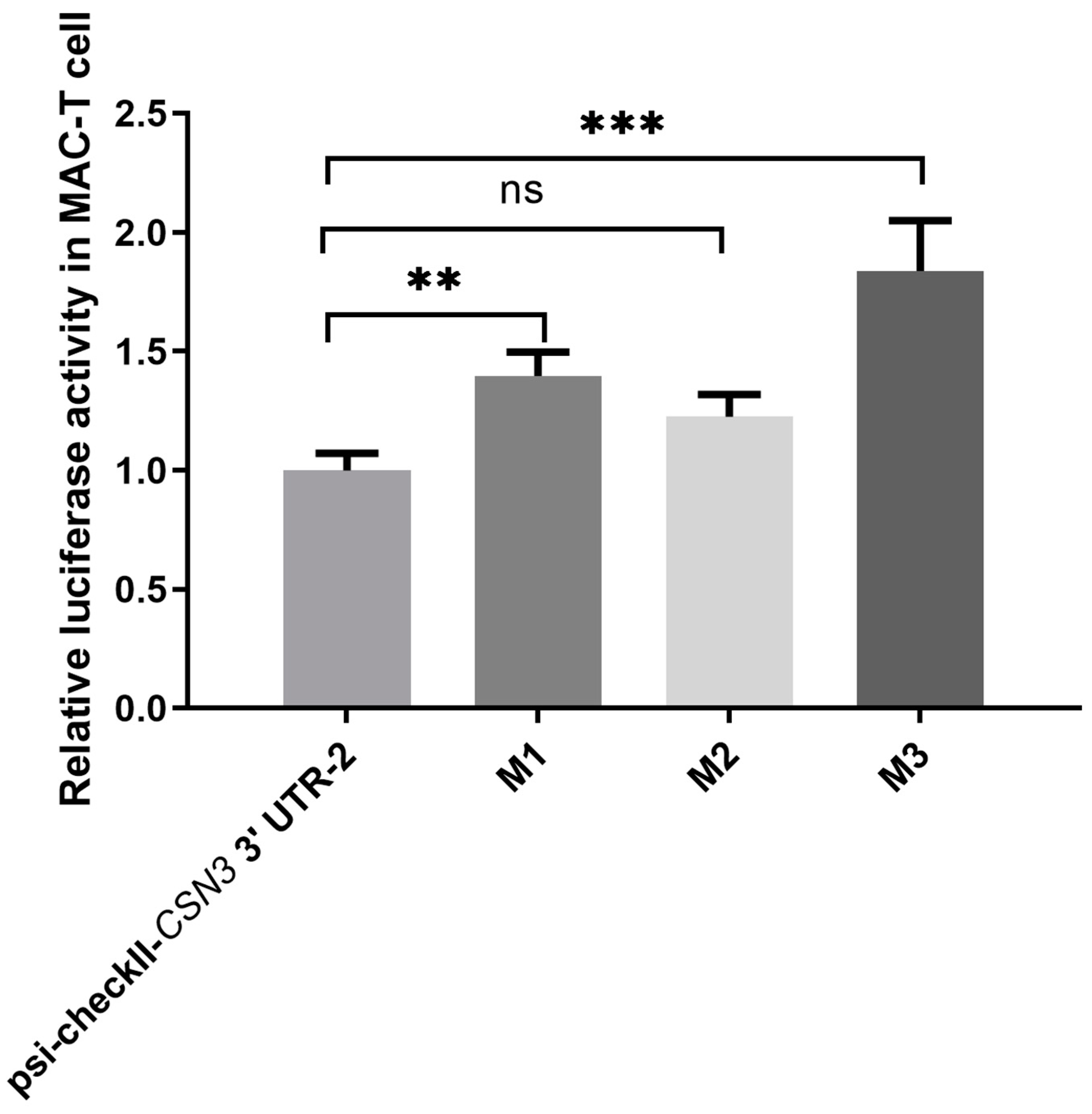
3.5. The Effect of CSN3 3’UTR Polymorphisms on miRNA Binding
3.6. The Effect of CSN3 3’UTR Polymorphisms on DHI Data in Dairy Cows
3.7. The Corresponding Genomic DNA Sequence of the CSN3 3’UTR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Inoue, R.; Tsukahara, T. Composition and physiological functions of the porcine colostrum. Anim. Sci. J. 2021, 92, e13618. [Google Scholar] [CrossRef]
- Paul, A.; Martin, F.; Simard, B.; Scher, J.; Gaiani, C.; le Floch-Fouere, C.; Jeantet, R.; Burgain, J. Deciphering the segregation of proteins in high-protein dairy powders after spray-drying. J. Dairy. Sci. 2023, 106, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhang, Z.; Qiu, L.; Zhang, Y.; Miao, Y. Polymorphisms of the kappa casein (CSN3) gene and inference of its variants in water buffalo (Bubalus bubalis). Arch. Anim. Breed. 2019, 62, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Madende, M.; Osthoff, G. Comparative genomics of casein genes. J. Dairy. Res. 2019, 86, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Day, L.; Williams, R.P.; Otter, D.; Augustin, M.A. Casein polymorphism heterogeneity influences casein micelle size in milk of individual cows. J. Dairy Sci. 2015, 98, 3633–3644. [Google Scholar] [CrossRef]
- Dadousis, C.; Biffani, S.; Cipolat-Gotet, C.; Nicolazzi, E.L.; Rosa, G.J.M.; Gianola, D.; Rossoni, A.; Santus, E.; Bittante, G.; Cecchinato, A. Genome-wide association study for cheese yield and curd nutrient recovery in dairy cows. J. Dairy. Sci. 2017, 100, 1259–1271. [Google Scholar] [CrossRef]
- Dadousis, C.; Biffani, S.; Cipolat-Gotet, C.; Nicolazzi, E.; Rossoni, A.; Santus, E.; Bittante, G.; Cecchinato, A. Genome-wide association of coagulation properties, curd firmness modeling, protein percentage, and acidity in milk from Brown Swiss cows. J. Dairy Sci. 2016, 99, 3654–3666. [Google Scholar] [CrossRef]
- Hobor, S.; Kunej, T.; Dovc, P. Polymorphisms in the kappa casein (CSN3) gene in horse and comparative analysis of its promoter and coding region. Anim. Genet. 2008, 39, 520–530. [Google Scholar] [CrossRef]
- Caroli, A.M.; Chessa, S.; Erhardt, G.J. Invited review: Milk protein polymorphisms in cattle: Effect on animal breeding and human nutrition. J. Dairy Sci. 2009, 92, 5335–5352. [Google Scholar] [CrossRef]
- Djedovic, R.; Bogdanovic, V.; Perisic, P.; Stanojevic, D.; Popovic, J.; Brka, M. Relationship between genetic polymorphism of κ-casein and quantitative milk yield traits in cattle breeds and crossbreds in Serbia. Genetika 2015, 47, 23–32. [Google Scholar] [CrossRef]
- Ibeagha-Awemu, E.M.; Kgwatalala, P.; Zhao, X. A critical analysis of production-associated DNA polymorphisms in the genes of cattle, goat, sheep, and pig. Mamm. Genome 2008, 19, 591–617. [Google Scholar] [CrossRef] [PubMed]
- Mohr, U.; Koczan, D.; Linder, D.; Hobom, G.; Erhardt, G. A single point mutation results in A allele-specific exon skipping in the bovine αS1-casein mRNA. Gene 1994, 143, 187–192. [Google Scholar] [CrossRef]
- Rando, A.; Di Gregorio, P.; Ramunno, L.; Mariani, P.; Fiorella, A.; Senese, C.; Marletta, D.; Masina, P. Characterization of the CSN1AG allele of the bovine αs1-casein locus by the insertion of a relict of a long interspersed element. J. Dairy Sci. 1998, 81, 1735–1742. [Google Scholar] [CrossRef]
- Prinzenberg, E.M.; Weimann, C.; Brandt, H.; Bennewitz, J.; Kalm, E.; Schwerin, M.; Erhardt, G. Polymorphism of the bovine CSN1S1 promoter: Linkage mapping, intragenic haplotypes, and effect on milk production traits. J. Dairy Sci. 2003, 86, 2696–2705. [Google Scholar] [CrossRef]
- Damiani, G.; Caroli, A.; Leone, P.; Budelli, E.; Florio, S. Effect of Bov-A2 Sine elements on quantitative traits. In Proceedings of the XIV National Congress ASPA, Florence, Italy, 12–15 June 2001; University of Florence Press: Florence, Italy, 2001; p. 49. [Google Scholar]
- Cosenza, G.; Mauriello, R.; Garro, G.; Auzino, B.; Iannaccone, M.; Costanzo, A.; Chianese, L.; Pauciullo, A. Casein composition and differential translational efficiency of casein transcripts in donkey’s milk. J. Dairy Res. 2019, 86, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Li, W.Q.; Li, W.L.; Wang, X.Y.; Zhang, H.L.; Wang, L.F.; Gao, T.Y. Comparison of miRNA profiles in milk-derived extracellular vesicles and bovine mammary glands. Int. Dairy J. 2022, 134, 105444. [Google Scholar] [CrossRef]
- Cui, X.; Hou, Y.; Yang, S.; Xie, Y.; Zhang, S.; Zhang, Y.; Zhang, Q.; Lu, X.; Liu, G.; Sun, D. Transcriptional profiling of mammary gland in Holstein cows with extremely different milk protein and fat percentage using RNA sequencing. BMC Genom. 2014, 15, 226. [Google Scholar] [CrossRef] [PubMed]
- Krappmann, K.; Weikard, R.; Kühn, C. Evaluation of a replacement method for mammary gland biopsies by comparing gene expression in udder tissue and mammary epithelial cells isolated from milk. Res. Vet. Sci. 2012, 93, 970–974. [Google Scholar] [CrossRef] [PubMed]
- Cánovas, A.; Rincón, G.; Bevilacqua, C.; Islas-Trejo, A.; Brenaut, P.; Hovey, R.C.; Boutinaud, M.; Morgenthaler, C.; VanKlompenberg, M.K.; Martin, P.; et al. Comparison of five different RNA sources to examine the lactating bovine mammary gland transcriptome using RNA-Sequencing. Sci. Rep. 2014, 4, 5297. [Google Scholar] [CrossRef]
- Suárez-Vega, A.; Gutiérrez-Gil, B.; Klopp, C.; Robert-Granie, C.; Tosser-Klopp, G.; Arranz, J.J. Characterization and Comparative Analysis of the Milk Transcriptome in Two Dairy Sheep Breeds using RNA Sequencing. Sci. Rep. 2015, 5, 18399. [Google Scholar] [CrossRef]
- Michailidou, S.; Gelasakis, A.; Banos, G.; Arsenos, G.; Argiriou, A. Comparative Transcriptome Analysis of Milk Somatic Cells During Lactation Between Two Intensively Reared Dairy Sheep Breeds. Front. Genet. 2021, 12, 700489. [Google Scholar] [CrossRef] [PubMed]
- Shock, D.A.; LeBlanc, S.J.; Leslie, K.E.; Hand, K.; Godkin, M.A.; Coe, J.B.; Kelton, D.F. Exploring the Characteristics and Dynamics of Ontario Dairy Herds Experiencing Increases in Bulk Milk Somatic Cell Count During the Summer. J. Dairy Sci. 2015, 98, 3741–3753. [Google Scholar] [CrossRef] [PubMed]
- Haltia, L.; Honkanen-Buzalski, T.; Spiridonova, I.; Olkonen, A.; Myllys, V. A study of bovine mastitis, milking procedures and management practices on 25 Estonian dairy herds. Acta Vet. Scand. 2006, 48, 22. [Google Scholar] [CrossRef][Green Version]
- Xu, H.; Shao, J.; Fang, J.; Yin, B.; Zhang, L.; Zhang, J.; Xia, G. miR-381 Targets KCTD15 to Regulate Bovine Preadipocyte Differentiation In Vitro. Horm. Metab. Res. 2021, 53, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Langschied, F.; Leisegang, M.S.; Brandes, R.P.; Ebersberger, I. ncOrtho: Efficient and reliable identification of miRNA orthologs. Nucleic Acids Res. 2023, 51, e71. [Google Scholar] [CrossRef]
- Vanvanhossou, S.F.U.; Giambra, I.J.; Yin, T.; Brügemann, K.; Dossa, L.H.; König, S. First DNA Sequencing in Beninese Indigenous Cattle Breeds Captures New Milk Protein Variants. Genes 2021, 12, 1702. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, Y.; Song, F.; Zheng, H.; Hu, L.; Lu, H.; Liu, P.; Hao, X.; Zhang, W.; Chen, K. Functional SNP in the microRNA-367 binding site in the 3’UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc. Natl. Acad. Sci. USA 2011, 108, 13653–13658. [Google Scholar] [CrossRef]



| Allele Position in 3’UTR and SNP ID | 26 A>G rs450438851 | 27 C>T (No SNP ID) | 36 T>C (No SNP ID) | 37 T>C rs209024052 | 48 T>C rs209359743 | 49 G>A rs135528518 | 80 T>C (No SNP ID) | 92 A>G rs135796226 | 104 A>T (No SNP ID) | 122 C>T rs134221650 | 129 T>C (No SNP ID) | 131 T>C rs136843602 | 141 T>C (No SNP ID) | 154 T>C (No SNP ID) | 170 G>A rs134516686 | 178 A>G (No SNP ID) | 183 T>C (No SNP ID) | 187 T>C (No SNP ID) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype/ Number of Mutant Bases (Sample Number) | |||||||||||||||||||
| Haplotype 1/No mutation point (No. 4,6,8,14,16,21,22, 25,28,29,32,36,38,40,42,43) | |||||||||||||||||||
| Haplotype 2/One mutation point (No. 5) | C | ||||||||||||||||||
| Haplotype 3/One mutation point (No. 12) | T | ||||||||||||||||||
| Haplotype 4/One mutation point (No. 48) | T | ||||||||||||||||||
| Haplotype 5/Two mutation points (No. 2) | T | C | |||||||||||||||||
| Haplotype 6/Two mutation points (No. 35) | C | A | |||||||||||||||||
| Haplotype 7/Four mutation points (No. 50) | T | C | C | A | |||||||||||||||
| Haplotype 8/Six mutation points (No. 13,46) | C | C | A | G | T | C | |||||||||||||
| Haplotype 9/Six mutation points (No. 1,17) | C | C | A | G | C | A | |||||||||||||
| Haplotype 10/Seven mutation points (No. 3,7,9,10,11,15,18, 19,24,26,31,34,39,41,44,45, 47) | C | C | A | G | T | C | A | ||||||||||||
| Haplotype 11/Seven mutation points (No. 33,37) | C | C | A | G | T | C | A | ||||||||||||
| Haplotype 12/Eight mutation points (No. 20) | C | C | A | G | T | C | C | A | |||||||||||
| Haplotype 13/Eight mutation points (No. 23) | C | C | A | G | T | C | A | G | |||||||||||
| Haplotype 14/Eight mutation points (No. 27) | C | C | A | C | G | T | C | A | |||||||||||
| Haplotype 15/Eight mutation points (No. 49) | C | C | A | G | T | C | A | C | |||||||||||
| Haplotype 16/Nine mutation points (No. 30) | G | C | C | A | G | T | C | C | A | ||||||||||
| Base mutation frequency | 2% | 2% | 2% | 52% | 56% | 56% | 2% | 56% | 2% | 56% | 2% | 60% | 2% | 2% | 56% | 2% | 2% | 4% | |
| Gene | CSN1S1 | CSN1S2 | CSN2 | CSN3 | |
|---|---|---|---|---|---|
| Group | |||||
| Haplotype 1 group (n = 12) | 1 ± 0.41 | 1 ± 0.41 | 1 ± 0.30 | 1 ± 0.24 | |
| Haplotype 10 group (n = 12) | 0.49 ± 0.22 | 0.78 ± 0.32 | 1.40 ± 0.81 | 0.04 ± 0.02 | |
| p | 0.339 | 0.341 | 0.622 | 0.001 | |
| Index | 305-d Milk Production | Fat-Corrected Milk | Fat (%) | Protein (%) | Fat/Protein Ratio | |
|---|---|---|---|---|---|---|
| Group | ||||||
| Haplotype 1 group (n = 16) | 7899.93 ± 456.12 | 29.81 ± 3.08 | 3.51 ± 0.15 | 3.41 ± 0.09 | 0.95 ± 0.03 | |
| Haplotype 10 group (n = 17) | 7825.38 ± 662.29 | 31.58 ± 2.86 | 3.56 ± 0.17 | 3.58 ± 0.10 | 0.96 ± 0.04 | |
| p | 0.928 | 0.678 | 0.828 | 0.241 | 0.822 | |
| Index | 305-d Milk Production | Fat-Corrected Milk | Fat (%) | Protein (%) | Fat/Protein Ratio | |
|---|---|---|---|---|---|---|
| Group | ||||||
| Haplotype 1 (n = 7) | 9057.62 ± 303.89 | 33.08 ± 3.30 | 3.18 ± 0.39 | 3.54 ± 0.10 | 0.90 ± 0.10 | |
| Haplotype 10 (n = 5) | 9071.40 ± 1278.61 | 27.89 ± 5.35 | 3.96 ± 0.38 | 3.86 ± 0.24 | 1.04 ± 0.10 | |
| p | 0.990 | 0.404 | 0.197 | 0.207 | 0.360 | |
| Month | March | April | May | June | July | August | |
|---|---|---|---|---|---|---|---|
| Group | |||||||
| Haplotype 1 | 3.41 ± 0.09 | 3.17 ± 0.06 | 3.23 ± 0.07 | 3.52 ± 0.09 | 3.50 ± 0.10 | 3.54 ± 0.10 | |
| Haplotype 10 | 3.58 ± 0.10 | 3.36 ± 0.11 | 3.33 ± 0.11 | 3.67 ± 0.14 | 3.61 ± 0.16 | 3.86 ± 0.24 | |
| p | 0.241 | 0.117 | 0.459 | 0.362 | 0.576 | 0.207 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Wang, X.; Wu, P.; Xu, X.; Liu, W.; Zhang, G.; Zhang, L.; Fu, T.; Gao, T. Polymorphism of the CSN3 3’UTR in Dairy Cows Causes Changes in bta-miR-708 Binding Ability and κ-Casein Expression. Animals 2024, 14, 3462. https://doi.org/10.3390/ani14233462
Li W, Wang X, Wu P, Xu X, Liu W, Zhang G, Zhang L, Fu T, Gao T. Polymorphism of the CSN3 3’UTR in Dairy Cows Causes Changes in bta-miR-708 Binding Ability and κ-Casein Expression. Animals. 2024; 14(23):3462. https://doi.org/10.3390/ani14233462
Chicago/Turabian StyleLi, Wenqing, Xiaoyang Wang, Pinhui Wu, Xiuyang Xu, Wei Liu, Guozhi Zhang, Liyang Zhang, Tong Fu, and Tengyun Gao. 2024. "Polymorphism of the CSN3 3’UTR in Dairy Cows Causes Changes in bta-miR-708 Binding Ability and κ-Casein Expression" Animals 14, no. 23: 3462. https://doi.org/10.3390/ani14233462
APA StyleLi, W., Wang, X., Wu, P., Xu, X., Liu, W., Zhang, G., Zhang, L., Fu, T., & Gao, T. (2024). Polymorphism of the CSN3 3’UTR in Dairy Cows Causes Changes in bta-miR-708 Binding Ability and κ-Casein Expression. Animals, 14(23), 3462. https://doi.org/10.3390/ani14233462





