Enteric Methane Emission in Livestock Sector: Bibliometric Research from 1986 to 2024 with Text Mining and Topic Analysis Approach by Machine Learning Algorithms
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
2.2. Text Mining
- Convert text to lowercase;
- Remove strange symbols and punctuations (“@”, “/”, “*”);
- Remove numbers and extra white spaces;
- Remove common English language words such as articles, prepositions, and conjunctions (e.g., “the,” “a,” “and,” “on,” “at,” etc.) as they provide little information about the contents of the corpus;
- Remove stop words: “emission,” “enteric,” “methane,” “buffalo,” “cow,” “sheep,” “goat,” “ruminants,” “cattle,” “additive,” “microbiome,” “microbiota”.
2.3. Topic Analysis
3. Results
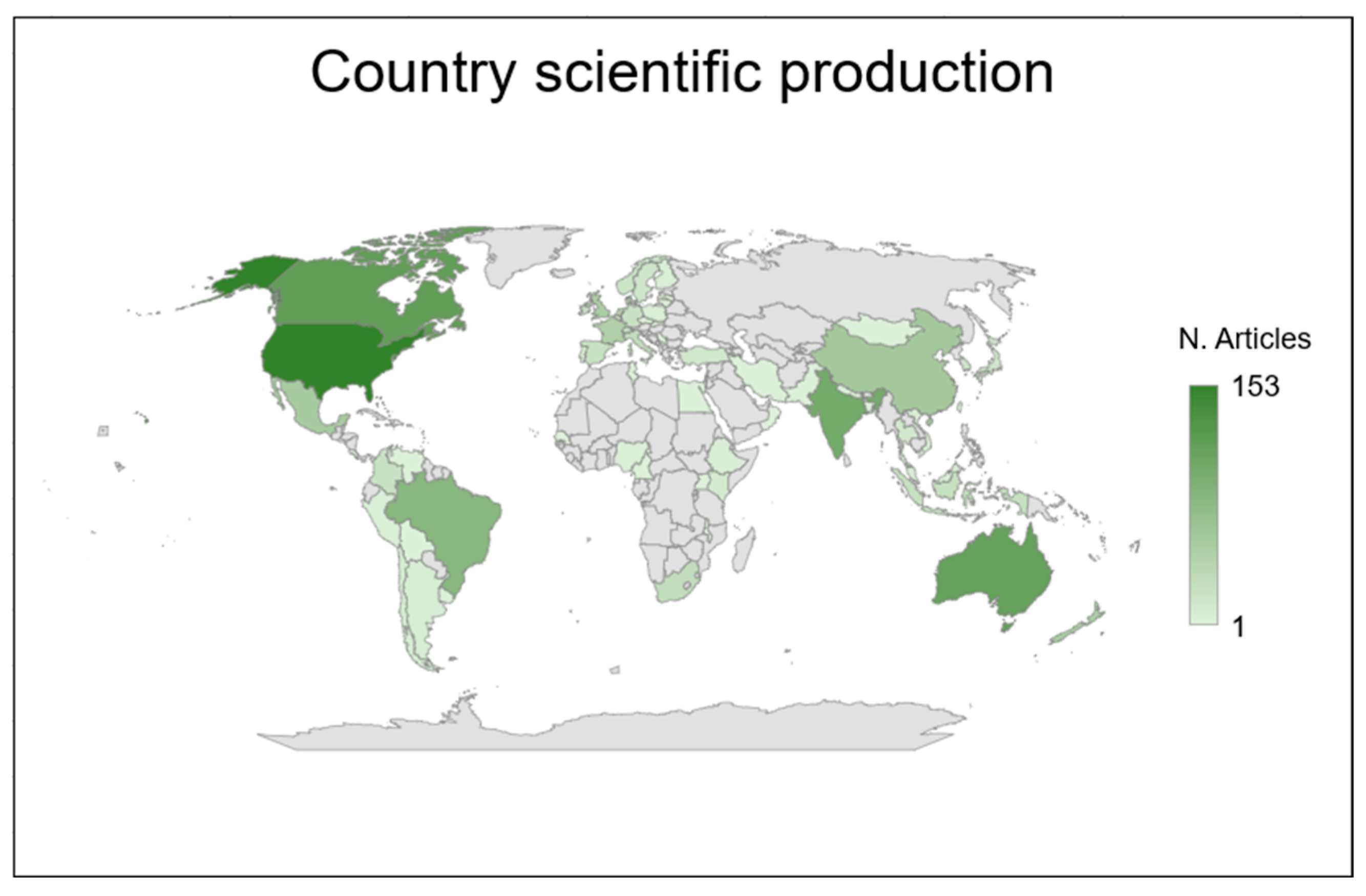
3.1. Descriptive Statistics
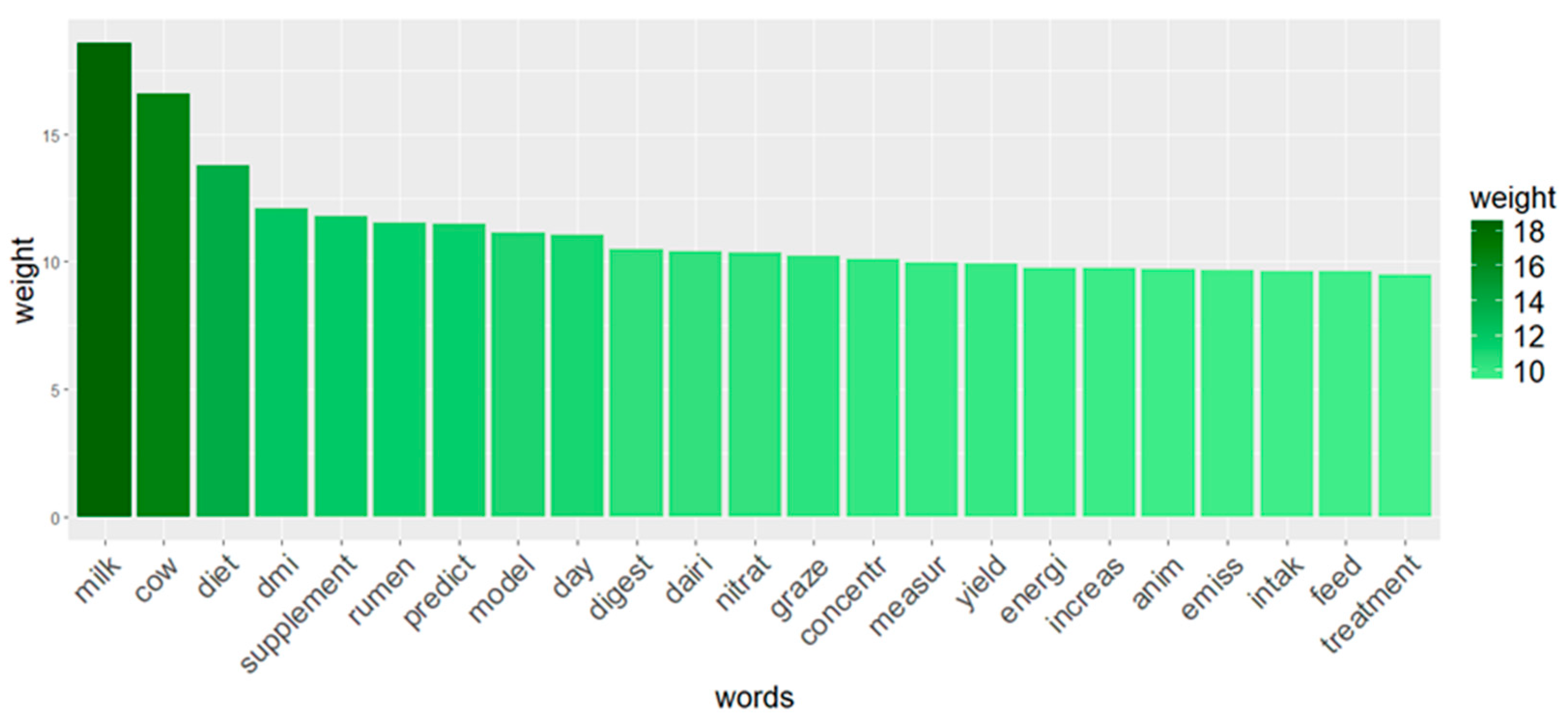
3.2. Text Mining
3.3. Topic Analysis
4. Discussion
4.1. Text Mining
4.2. Topic Analysis
5. Conclusions
Future Prospects
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zobeidi, T.; Yaghoubi, J.; Yazdanpanah, M. Farmers’ incremental adaptation to water scarcity: An application of the model of private proactive adaptation to climate change (MPPACC). Agric. Water Manag. 2022, 264, 107528. [Google Scholar] [CrossRef]
- Pörtner, H.-O.; Scholes, R.J.; Agard, J.; Archer, E.; Arneth, A.; Bai, X.; Barnes, D.; Burrows, M.; Chan, L.; Cheung, W.L.; et al. Scientific Outcome of the IPBES-IPCC Co-Sponsored Workshop on Biodiversity and Climate Change (Version 5); IPBES secretariat: Bonn, Germany, 2021. [Google Scholar] [CrossRef]
- FAO. Pathways Towards Lower Emissions—A Global Assessment of the Greenhouse Gas Emissions and Mitigation Options from Livestock Agrifood Systems. Rome. 2023. Available online: https://openknowledge.fao.org/items/b3f21d6d-bd6d-4e66-b8ca-63ce376560b5 (accessed on 20 September 2024). [CrossRef]
- Grossi, G.; Goglio, P.; Vitali, A.; Williams, A.G. Livestock and climate change: Impact of livestock on climate and mitigation strategies. Anim. Front. 2019, 9, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Gerber, P.; Hristov, A.; Henderson, B.; Makkar, H.; Oh, J.; Lee, C.; Meinen, R.; Montes, F.; Ott, T.; Firkins, J.; et al. Technical options for the mitigation of direct methane and nitrous oxide emissions from livestock: A review. Animal 2013, 7, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Balcombe, P.; Speirs, J.F.; Brandon, N.P.; Hawkes, A.D. Methane emissions: Choosing the right climate metric and time horizon. Environ. Sci. Process. Impacts 2018, 20, 1323–1339. [Google Scholar] [CrossRef] [PubMed]
- Hook, S.E.; Wright, A.-D.G.; McBride, B.W. Methanogens: Methane Producers of the Rumen and Mitigation Strategies. Archaea 2010, 2010, 945785. [Google Scholar] [CrossRef]
- McAllister, T.A.; Cheng, K.-J.; Okine, E.K.; Mathison, G.W. Dietary, environmental and microbiological aspects of methane production in ruminants. Can. J. Anim. Sci. 1996, 76, 231–243. [Google Scholar] [CrossRef]
- Appuhamy, J.A.D.R.N.; France, J.; Kebreab, E. Models for predicting enteric methane emissions from dairy cows in North America, Europe, and Australia and New Zealand. Glob. Chang. Biol. 2016, 22, 3039–3056. [Google Scholar] [CrossRef]
- Palangi, V.; Lackner, M. Management of Enteric Methane Emissions in Ruminants Using Feed Additives: A Review. Animals 2022, 12, 3452. [Google Scholar] [CrossRef]
- VijayGaikwad, S.; Chaugule, A.; Patil, P. Text Mining Methods and Techniques. Int. J. Comput. Appl. 2014, 85, 42–45. [Google Scholar] [CrossRef]
- Wang, S.-H.; Ding, Y.; Zhao, W.; Huang, Y.-H.; Perkins, R.; Zou, W.; Chen, J.J. Text mining for identifying topics in the literatures about adolescent substance use and depression. BMC Public Health 2016, 16, 279. [Google Scholar] [CrossRef]
- Marino, R.; Petrera, F.; Abeni, F. Scientific Productions on Precision Livestock Farming: An Overview of the Evolution and Current State of Research Based on a Bibliometric Analysis. Animals 2023, 13, 2280. [Google Scholar] [CrossRef] [PubMed]
- Gislon, G.; Bava, L.; Zucali, M.; Tamburini, A.; Sandrucci, A. Unlocking insights: Text mining analysis on the health, welfare, and behavior of cows in automated milking systems. J. Anim. Sci. 2024, 102, skae159. [Google Scholar] [CrossRef]
- Nalon, E.; Contiero, B.; Gottardo, F.; Cozzi, G. The Welfare of Beef Cattle in the Scientific Literature From 1990 to 2019: A Text Mining Approach. Front. Vet. Sci. 2021, 7, 588749. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, B.; Felici, M.; Costa, L.N.; Padalino, B. A review of horse welfare literature from 1980 to 2023 with a text mining and topic analysis approach. Ital. J. Anim. Sci. 2023, 22, 1095–1109. [Google Scholar] [CrossRef]
- Trapanese, L.; Jasinski, F.P.; Bifulco, G.; Pasquino, N.; Bernabucci, U.; Salzano, A. Buffalo welfare: A literature review from 1992 to 2023 with a text mining and topic analysis approach. Ital. J. Anim. Sci. 2024, 23, 570–584. [Google Scholar] [CrossRef]
- R Core Team. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. Available online: https://www.r-project.org (accessed on 15 June 2024).
- Feinerer, I.; Hornik, K.; Meyer, D. Text Mining Infrastructure in R. J. Stat. Softw. 2008, 25, 1–54. [Google Scholar] [CrossRef]
- Bouchet-Valat, M. SnowballC: Snowball Stemmers Based on the C Libstemmer UTF-8 Library. R Package Version 0.6.0. Available online: https://cran.r-project.org/web/packages/SnowballC/SnowballC.pdf (accessed on 15 June 2024).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 15 June 2024).
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. Dplyr: A Grammar of Data Manipulation. R Package Version 1.1.4. Available online: https://dplyr.tidyverse.org/reference/dplyr-package.html (accessed on 15 June 2024).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.A.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Sebastiani, F. Machine learning in automated text categorization. ACM Comput. Surv. 2002, 34, 1–47. [Google Scholar] [CrossRef]
- Adamaκopoulou, C.; Benedetti, B.; Zappaterra, M.; Felici, M.; Masebo, N.T.; Previti, A.; Passantino, A.; Padalino, B. Cats’ and dogs’ welfare: Text mining and topics modeling analysis of the scientific literature. Front. Vet. Sci. 2023, 10, 1268821. [Google Scholar] [CrossRef]
- Salton, G.; Buckley, C. Term-weighting approaches in automatic text retrieval. Inf. Process. Manag. 1988, 24, 513–523. [Google Scholar] [CrossRef]
- Blei, D.M.; Ng, A.Y.; Jordan, M.I. Latent Dirichlet Allocation. J. Mach. Learn. Res. 2003, 3, 993–1022. [Google Scholar]
- Grün, B.; Hornik, K. Topicmodels: An R Package for Fitting Topic Models. J. Stat. Softw. 2011, 40, 1–30. [Google Scholar] [CrossRef]
- Moss, A.R.; Jouany, J.-P.; Newbold, J. Methane production by ruminants: Its contribution to global warming. Ann. Zootech. 2000, 49, 231–253. [Google Scholar] [CrossRef]
- Beauchemin, K.A.; Kreuzer, M.; O’Mara, F.; McAllister, T.A. Nutritional management for enteric methane abatement: A review. Aust. J. Exp. Agric. 2008, 48, 21–27. [Google Scholar] [CrossRef]
- Crutzen, P.J.; Aselmann, I.; Seiler, W. Methane production by domestic animals, wild ruminants, other herbivorous fauna, and humans. Tellus B Chem. Phys. Meteorol. 1986, 38, 271. [Google Scholar] [CrossRef]
- Della Rosa, M.M.; Waghorn, G.C.; Vibart, R.E.; Jonker, A. An assessment of global ruminant methane-emission measurements shows bias relative to contributions of farmed species, populations and among continents. Anim. Prod. Sci. 2022, 63, 201–212. [Google Scholar] [CrossRef]
- Negussie, E.; de Haas, Y.; Dehareng, F.; Dewhurst, R.; Dijkstra, J.; Gengler, N.; Morgavi, D.; Soyeurt, H.; van Gastelen, S.; Yan, T.; et al. Invited review: Large-scale indirect measurements for enteric methane emissions in dairy cattle: A review of proxies and their potential for use in management and breeding decisions. J. Dairy Sci. 2017, 100, 2433–2453. [Google Scholar] [CrossRef]
- Haque, M.N. Dietary Manipulation: A Sustainable Way to Mitigate Methane Emissions from Ruminants. J. Anim. Sci. Technol. 2018, 60, 15. [Google Scholar] [CrossRef]
- van Gastelen, S.; Bannink, A.; Dijkstra, J. Effect of silage characteristics on enteric methane emission from ruminants. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2019, 1–9. [Google Scholar] [CrossRef]
- Beauchemin, K.A.; Ungerfeld, E.M.; Abdalla, A.L.; Alvarez, C.; Arndt, C.; Becquet, P.; Benchaar, C.; Berndt, A.; Mauricio, R.M.; McAllister, T.A.; et al. Invited review: Current enteric methane mitigation options. J. Dairy Sci. 2022, 105, 9297–9326. [Google Scholar] [CrossRef]
- Benchaar, C.; Pomar, C.; Chiquette, J. Evaluation of dietary strategies to reduce methane production in ruminants: A modelling approach. Can. J. Anim. Sci. 2001, 81, 563–574. [Google Scholar] [CrossRef]
- Bielak, A.; Derno, M.; Tuchscherer, A.; Hammon, H.M.; Susenbeth, A.; Kuhla, B. Body fat mobilization in early lactation influences methane production of dairy cows. Sci. Rep. 2016, 6, 28135. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Kebreab, E.; Hristov, A.N.; Oh, J.; Arndt, C.; Bannink, A.; Bayat, A.R.; Brito, A.F.; Boland, T.; Casper, D.; et al. Prediction of enteric methane production, yield, and intensity in dairy cattle using an intercontinental database. Glob. Chang. Biol. 2018, 24, 3368–3389. [Google Scholar] [CrossRef] [PubMed]
- Benaouda, M.; Martin, C.; Li, X.; Kebreab, E.; Hristov, A.N.; Yu, Z.; Yáñez-Ruiz, D.R.; Reynolds, C.K.; Crompton, L.A.; Dijkstra, J.; et al. Evaluation of the performance of existing mathematical models predicting enteric methane emissions from ruminants: Animal categories and dietary mitigation strategies. Anim. Feed. Sci. Technol. 2019, 255, 114207. [Google Scholar] [CrossRef]
- Mills, J.A.N.; Kebreab, E.; Yates, C.M.; Crompton, L.A.; Cammell, S.B.; Dhanoa, M.S.; Agnew, R.E.; France, J. Alternative approaches to predicting methane emissions from dairy cows1. J. Anim. Sci. 2003, 81, 3141–3150. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.; Kebreab, E.; Odongo, N.; McBride, B.; Okine, E.; France, J. Prediction of Methane Production from Dairy and Beef Cattle. J. Dairy Sci. 2007, 90, 3456–3466. [Google Scholar] [CrossRef]
- Thoma, G.; Popp, J.; Nutter, D.; Shonnard, D.; Ulrich, R.; Matlock, M.; Kim, D.S.; Neiderman, Z.; Kemper, N.; East, C.; et al. Greenhouse gas emissions from milk production and consumption in the United States: A cradle-to-grave life cycle assessment circa 2008. Int. Dairy J. 2013, 31, S3–S14. [Google Scholar] [CrossRef]
- Vitali, A.; Grossi, G.; Martino, G.; Bernabucci, U.; Nardone, A.; Lacetera, N. Carbon footprint of organic beef meat from farm to fork: A case study of short supply chain. J. Sci. Food Agric. 2018, 98, 5518–5524. [Google Scholar] [CrossRef]
- Rossi, C.; Grossi, G.; Lacetera, N.; Vitali, A. Carbon Footprint and Carbon Sink of a Local Italian Dairy Supply Chain. Dairy 2024, 5, 201–216. [Google Scholar] [CrossRef]
- Soder, K.J.; Brito, A.F. Enteric methane emissions in grazing dairy systems. JDS Commun. 2023, 4, 324–328. [Google Scholar] [CrossRef]
- Hristov, A.N.; Oh, J.; Firkins, J.L.; Dijkstra, J.; Kebreab, E.; Waghorn, G.; Makkar, H.P.S.; Adesogan, A.T.; Yang, W.; Lee, C.; et al. SPECIAL TOPICS-Mitigation of Methane and Nitrous Oxide Emissions from Animal Operations: I. A Review of Enteric Methane Mitigation Options. J. Anim. Sci. 2013, 91, 5045–5069. [Google Scholar] [CrossRef] [PubMed]
- Mapfumo, L.; Grobler, S.M.; Mupangwa, J.F.; Scholtz, M.M.; Muchenje, V. Enteric methane output from selected herds of beef cattle raised under extensive arid rangelands. Pastoralism 2018, 8, 15. [Google Scholar] [CrossRef]
- Avetisyan, M.; Golub, A.; Hertel, T.; Rose, S.; Henderson, B. Why a Global Carbon Policy Could Have a Dramatic Impact on the Pattern of the Worldwide Livestock Production. Appl. Econ. Perspect. Policy 2011, 33, 584–605. [Google Scholar] [CrossRef]
- Schuman, G.; Janzen, H.; Herrick, J. Soil carbon dynamics and potential carbon sequestration by rangelands. Environ. Pollut. 2002, 116, 391–396. [Google Scholar] [CrossRef]
- Agethen, K.; Mauricio, R.M.; Deblitz, C. Economics of Greenhouse Gas Mitigation Strategies in a North-Eastern Brazilian Beef Production System. Available online: https://uknowledge.uky.edu/igc/24/3-2/3 (accessed on 21 July 2024).
- Conant, R.T.; Cerri, C.E.P.; Osborne, B.B.; Paustian, K. Grassland management impacts on soil carbon stocks: A new synthesis. Ecol. Appl. 2017, 27, 662–668. [Google Scholar] [CrossRef]
- Godde, C.M.; Boone, R.; Ash, A.J.; Waha, K.; Sloat, L.; Thornton, P.K.; Herrero, M. Global rangeland production systems and livelihoods at threat under climate change and variability. Environ. Res. Lett. 2020, 15, 044021. [Google Scholar] [CrossRef]
- Hammond, K.J.; Crompton, L.A.; Bannink, A.; Dijkstra, J.; Yáñez-Ruiz, D.R.; O’Kiely, P.; Kebreab, E.; Eugène, M.; Yu, Z.; Shingfield, K.J.; et al. Review of current in vivo measurement techniques for quantifying enteric methane emission from ruminants. Anim. Feed. Sci. Technol. 2016, 219, 13–30. [Google Scholar] [CrossRef]
- Della Rosa, M.; Jonker, A.; Waghorn, G. A review of technical variations and protocols used to measure methane emissions from ruminants using respiration chambers, SF6 tracer technique and GreenFeed, to facilitate global integration of published data. Anim. Feed. Sci. Technol. 2021, 279, 115018. [Google Scholar] [CrossRef]
- Storm, I.M.L.D.; Hellwing, A.L.F.; Nielsen, N.I.; Madsen, J. Methods for Measuring and Estimating Methane Emission from Ruminants. Animals 2012, 2, 160–183. [Google Scholar] [CrossRef]
- Bekele, W.; Guinguina, A.; Zegeye, A.; Simachew, A.; Ramin, M. Contemporary Methods of Measuring and Estimating Methane Emission from Ruminants. Methane 2022, 1, 82–95. [Google Scholar] [CrossRef]
- Nejad, J.G.; Ju, M.-S.; Jo, J.-H.; Oh, K.-H.; Lee, Y.-S.; Lee, S.-D.; Kim, E.-J.; Roh, S.; Lee, H.-G. Advances in Methane Emission Estimation in Livestock: A Review of Data Collection Methods, Model Development and the Role of AI Technologies. Animals 2024, 14, 435. [Google Scholar] [CrossRef] [PubMed]
- Pickering, N.K.; Oddy, V.H.; Basarab, J.; Cammack, K.; Hayes, B.; Hegarty, R.S.; Lassen, J.; McEwan, J.C.; Miller, S.; Pinares-Patiño, C.S.; et al. Animal board invited review: Genetic possibilities to reduce enteric methane emissions from ruminants. Animal 2015, 9, 1431–1440. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.L.; Bannink, A.; France, J.; Kebreab, E.; Dijkstra, J. Evaluation of enteric methane prediction equations for dairy cows used in whole farm models. Glob. Chang. Biol. 2010, 16, 3246–3256. [Google Scholar] [CrossRef]
- Vlaeminck, B.; Fievez, V.; Tamminga, S.; Dewhurst, R.; van Vuuren, A.; De Brabander, D.; Demeyer, D. Milk Odd- and Branched-Chain Fatty Acids in Relation to the Rumen Fermentation Pattern. J. Dairy Sci. 2006, 89, 3954–3964. [Google Scholar] [CrossRef]
- Dijkstra, J.; van Zijderveld, S.; Apajalahti, J.; Bannink, A.; Gerrits, W.; Newbold, J.; Perdok, H.; Berends, H. Relationships between methane production and milk fatty acid profiles in dairy cattle. Anim. Feed. Sci. Technol. 2011, 166–167, 590–595. [Google Scholar] [CrossRef]
- van Gastelen, S.; Dijkstra, J. Prediction of Methane Emission from Lactating Dairy Cows Using Milk Fatty Acids and Mid-Infrared Spectroscopy. J. Sci. Food Agric. 2016, 96, 3963–3968. [Google Scholar] [CrossRef]
- Bougouin, A.; Appuhamy, J.A.D.R.N.; Ferlay, A.; Kebreab, E.; Martin, C.; Moate, P.; Benchaar, C.; Lund, P.; Eugène, M. Individual milk fatty acids are potential predictors of enteric methane emissions from dairy cows fed a wide range of diets: Approach by meta-analysis. J. Dairy Sci. 2019, 102, 10616–10631. [Google Scholar] [CrossRef]
- Jonker, A.; Molano, G.; Koolaard, J.; Muetzel, S. Methane emissions from lactating and non-lactating dairy cows and growing cattle fed fresh pasture. Anim. Prod. Sci. 2017, 57, 643. [Google Scholar] [CrossRef]
- Goopy, J.P.; Donaldson, A.; Hegarty, R.; Vercoe, P.E.; Haynes, F.; Barnett, M.; Oddy, V.H. Low-methane yield sheep have smaller rumens and shorter rumen retention time. Br. J. Nutr. 2013, 111, 578–585. [Google Scholar] [CrossRef]
- Nkrumah, J.D.; Okine, E.K.; Mathison, G.W.; Schmid, K.; Li, C.; Basarab, J.A.; Price, M.A.; Wang, Z.; Moore, S.S. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle1. J. Anim. Sci. 2006, 84, 145–153. [Google Scholar] [CrossRef]
- Jonker, A.; Molano, G.; Antwi, C.; Waghorn, G. Feeding lucerne silage to beef cattle at three allowances and four feeding frequencies affects circadian patterns of methane emissions, but not emissions per unit of intake. Anim. Prod. Sci. 2014, 54, 1350–1353. [Google Scholar] [CrossRef]
- Watt, L.; Clark, C.; Krebs, G.; Petzel, C.; Nielsen, S.; Utsumi, S. Differential rumination, intake, and enteric methane production of dairy cows in a pasture-based automatic milking system. J. Dairy Sci. 2015, 98, 7248–7263. [Google Scholar] [CrossRef]
- Lovarelli, D.; Bacenetti, J.; Guarino, M. A review on dairy cattle farming: Is precision livestock farming the compromise for an environmental, economic and social sustainable production? J. Clean. Prod. 2020, 262, 121409. [Google Scholar] [CrossRef]
- Murray, R.M.; Bryant, A.M.; Leng, R.A. Rates of production of methane in the rumen and large intestine of sheep. Br. J. Nutr. 1976, 36, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Key, N.; Tallard, G. Mitigating methane emissions from livestock: A global analysis of sectoral policies. Clim. Chang. 2011, 112, 387–414. [Google Scholar] [CrossRef]
- Pereira, A.M.; de Lurdes Nunes Enes Dapkevicius, M.; Borba, A.E.S. Alternative Pathways for Hydrogen Sink Originated from the Ruminal Fermentation of Carbohydrates: Which Microorganisms Are Involved in Lowering Methane Emission? Anim. Microbiome 2022, 4, 5. [Google Scholar] [CrossRef]
- Wallace, R.J.; Rooke, J.A.; McKain, N.; Duthie, C.-A.; Hyslop, J.J.; Ross, D.W.; Waterhouse, A.; Watson, M.; Roehe, R. The rumen microbial metagenome associated with high methane production in cattle. BMC Genom. 2015, 16, 1–14. [Google Scholar] [CrossRef]
- Bouchard, K.; Wittenberg, K.M.; Legesse, G.; Krause, D.O.; Khafipour, E.; Buckley, K.E.; Ominski, K.H. Comparison of feed intake, body weight gain, enteric methane emission and relative abundance of rumen microbes in steers fed sainfoin and lucerne silages under western Canadian conditions. Grass Forage Sci. 2013, 70, 116–129. [Google Scholar] [CrossRef]
- Tseten, T.; Sanjorjo, R.A.; Kwon, M.; Kim, S.-W. Strategies to Mitigate Enteric Methane Emissions from Ruminant Animals. J. Microbiol. Biotechnol. 2022, 32, 269–277. [Google Scholar] [CrossRef]
- van Wyngaard, J.; Meeske, R.; Erasmus, L. Effect of concentrate feeding level on methane emissions, production performance and rumen fermentation of Jersey cows grazing ryegrass pasture during spring. Anim. Feed. Sci. Technol. 2018, 241, 121–132. [Google Scholar] [CrossRef]
- Van Kessel, J.A.S.; Russell, J.B. The effect of pH on ruminal methanogenesis. FEMS Microbiol. Ecol. 1996, 20, 205–210. [Google Scholar] [CrossRef]
- Hünerberg, M.; McGinn, S.M.; Beauchemin, K.A.; Entz, T.; Okine, E.K.; Harstad, O.M.; McAllister, T.A. Impact of ruminal pH on enteric methane emissions1. J. Anim. Sci. 2015, 93, 1760–1766. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.A.; Johnson’, D.E. Methane Emissions from Cattle. J. Anim. Sci. 1995, 73, 2483–2493. [Google Scholar] [CrossRef] [PubMed]
- Grainger, C.; Beauchemin, K. Can enteric methane emissions from ruminants be lowered without lowering their production? Anim. Feed. Sci. Technol. 2011, 166–167, 308–320. [Google Scholar] [CrossRef]
- van Gastelen, S.; Dijkstra, J.; Heck, J.M.; Kindermann, M.; Klop, A.; de Mol, R.; Rijnders, D.; Walker, N.; Bannink, A. Methane mitigation potential of 3-nitrooxypropanol in lactating cows is influenced by basal diet composition. J. Dairy Sci. 2022, 105, 4064–4082. [Google Scholar] [CrossRef]
- Bodas, R.; Prieto, N.; García-González, R.; Andrés, S.; Giráldez, F.J.; López, S. Manipulation of rumen fermentation and methane production with plant secondary metabolites. Anim. Feed Sci. Technol. 2012, 176, 78–93. [Google Scholar] [CrossRef]
- Kebreab, E.; Bannink, A.; Pressman, E.M.; Walker, N.; Karagiannis, A.; van Gastelen, S.; Dijkstra, J. A meta-analysis of effects of 3-nitrooxypropanol on methane production, yield, and intensity in dairy cattle. J. Dairy Sci. 2023, 106, 927–936. [Google Scholar] [CrossRef]
- Beauchemin, K.A.; McGinn, S.M. Methane emissions from feedlot cattle fed barley or corn diets1. J. Anim. Sci. 2005, 83, 653–661. [Google Scholar] [CrossRef]
- Hales, K.E.; Cole, N.A.; MacDonald, J.C. Effects of corn processing method and dietary inclusion of wet distillers grains with solubles on energy metabolism, carbon−nitrogen balance, and methane emissions of cattle1,2. J. Anim. Sci. 2012, 90, 3174–3185. [Google Scholar] [CrossRef]
- Herrera-Saldana, R.; Huber, J.; Poore, M. Dry Matter, Crude Protein, and Starch Degradability of Five Cereal Grains. J. Dairy Sci. 1990, 73, 2386–2393. [Google Scholar] [CrossRef]
- Sun, F.; Hu, W.; Cao, J.; Wang, X.; Zhang, Z.; Ramezani, J.; Shen, S. Sustained and intensified lacustrine methane cycling during Early Permian climate warming. Nat. Commun. 2022, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Milich, L. The role of methane in global warming: Where might mitigation strategies be focused? Glob. Environ. Chang. 1999, 9, 179–201. [Google Scholar] [CrossRef]
- Evangelista, C.; Basiricò, L.; Bernabucci, U. An Overview on the Use of Near Infrared Spectroscopy (NIRS) on Farms for the Management of Dairy Cows. Agriculture 2021, 11, 296. [Google Scholar] [CrossRef]
- Boadi, D.; Benchaar, C.; Chiquette, J.; Massé, D. Mitigation strategies to reduce enteric methane emissions from dairy cows: Update review. Can. J. Anim. Sci. 2004, 84, 319–335. [Google Scholar] [CrossRef]
- Arndt, C.; Hristov, A.N.; Price, W.J.; McClelland, S.C.; Pelaez, A.M.; Cueva, S.F.; Oh, J.; Dijkstra, J.; Bannink, A.; Bayat, A.R.; et al. Full adoption of the most effective strategies to mitigate methane emissions by ruminants can help meet the 1.5 °C target by 2030 but not 2050. Proc. Natl. Acad. Sci. USA 2022, 119, e2111294119. [Google Scholar] [CrossRef]
- Eugène, M.; Massé, D.; Chiquette, J.; Benchaar, C. Meta-analysis on the effects of lipid supplementation on methane production in lactating dairy cows. Can. J. Anim. Sci. 2008, 88, 331–337. [Google Scholar] [CrossRef]
- Knapp, J.R.; Laur, G.L.; Vadas, P.A.; Weiss, W.P.; Tricarico, J.M. Invited review: Enteric methane in dairy cattle production: Quantifying the opportunities and impact of reducing emissions. J. Dairy Sci. 2014, 97, 3231–3261. [Google Scholar] [CrossRef]
- Almeida, A.K.; Hegarty, R.S.; Cowie, A. Meta-analysis quantifying the potential of dietary additives and rumen modifiers for methane mitigation in ruminant production systems. Anim. Nutr. 2021, 7, 1219–1230. [Google Scholar] [CrossRef]
- Beauchemin, K.A.; Ungerfeld, E.M.; Eckard, R.J.; Wang, M. Review: Fifty years of research on rumen methanogenesis: Lessons learned and future challenges for mitigation. Animal 2020, 14, s2–s16. [Google Scholar] [CrossRef]
- van Gastelen, S.; Burgers, E.E.; Dijkstra, J.; de Mol, R.; Muizelaar, W.; Walker, N.; Bannink, A. Long-term effects of 3-nitrooxypropanol on methane emission and milk production characteristics in Holstein-Friesian dairy cows. J. Dairy Sci. 2024, 107, 5556–5573. [Google Scholar] [CrossRef]
- Duin, E.C.; Wagner, T.; Shima, S.; Prakash, D.; Cronin, B.; Yáñez-Ruiz, D.R.; Duval, S.; Rümbeli, R.; Stemmler, R.T.; Thauer, R.K.; et al. Mode of action uncovered for the specific reduction of methane emissions from ruminants by the small molecule 3-nitrooxypropanol. Proc. Natl. Acad. Sci. USA 2016, 113, 6172–6177. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, J.; Bannink, A.; France, J.; Kebreab, E.; van Gastelen, S. Short communication: Antimethanogenic effects of 3-nitrooxypropanol depend on supplementation dose, dietary fiber content, and cattle type. J. Dairy Sci. 2018, 101, 9041–9047. [Google Scholar] [CrossRef]
- Melgar, A.; Welter, K.; Nedelkov, K.; Martins, C.; Harper, M.; Oh, J.; Räisänen, S.; Chen, X.; Cueva, S.; Duval, S.; et al. Dose-response effect of 3-nitrooxypropanol on enteric methane emissions in dairy cows. J. Dairy Sci. 2020, 103, 6145–6156. [Google Scholar] [CrossRef] [PubMed]
- Jayanegara, A.; Sarwono, K.A.; Kondo, M.; Matsui, H.; Ridla, M.; Laconi, E.B. Nahrowi Use of 3-nitrooxypropanol as feed additive for mitigating enteric methane emissions from ruminants: A meta-analysis. Ital. J. Anim. Sci. 2017, 17, 650–656. [Google Scholar] [CrossRef]
- DSM Receives Landmark EU Market Approval for Its Methane-Reducing Feed Additive Bovaer®. Available online: https://our-company.dsm-firmenich.com/en/our-company/news/press-releases/legacy-archive/2022/dsm-receives-eu-approval-Bovaer.html (accessed on 25 May 2024).
- Jentsch, W.; Schweigel, M.; Weissbach, F.; Scholze, H.; Pitroff, W.; Derno, M. Methane production in cattle calculated by the nutrient composition of the diet. Arch. Anim. Nutr. 2007, 61, 10–19. [Google Scholar] [CrossRef]
- Holter, J.; Young, A. Methane Prediction in Dry and Lactating Holstein Cows. J. Dairy Sci. 1992, 75, 2165–2175. [Google Scholar] [CrossRef]
- Ricci, P.; Rooke, J.A.; Nevison, I.; Waterhouse, A. Methane emissions from beef and dairy cattle: Quantifying the effect of physiological stage and diet characteristics1. J. Anim. Sci. 2013, 91, 5379–5389. [Google Scholar] [CrossRef]
- Moraes, L.E.; Strathe, A.B.; Fadel, J.G.; Casper, D.P.; Kebreab, E. Prediction of enteric methane emissions from cattle. Glob. Chang. Biol. 2014, 20, 2140–2148. [Google Scholar] [CrossRef]
- Lyons, T.; Bielak, A.; Doyle, E.; Kuhla, B. Variations in methane yield and microbial community profiles in the rumen of dairy cows as they pass through stages of first lactation. J. Dairy Sci. 2018, 101, 5102–5114. [Google Scholar] [CrossRef]
- Garnsworthy, P.; Craigon, J.; Hernandez-Medrano, J.; Saunders, N. On-farm methane measurements during milking correlate with total methane production by individual dairy cows. J. Dairy Sci. 2012, 95, 3166–3180. [Google Scholar] [CrossRef]
- Oddy, V.H.; Donaldson, A.J.; Cameron, M.; Bond, J.; Dominik, S.; Robinson, D.L. Variation in methane production over time and physiological state in sheep. Anim. Prod. Sci. 2019, 59, 441–448. [Google Scholar] [CrossRef]
- Dong, L.; Li, B.; Diao, Q. Effects of Dietary Forage Proportion on Feed Intake, Growth Performance, Nutrient Digestibility, and Enteric Methane Emissions of Holstein Heifers at Various Growth Stages. Animals 2019, 9, 725. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Restrepo, C.A.; Clark, H.; Muetzel, S. Methane emissions from young and mature dairy cattle. Anim. Prod. Sci. 2016, 56, 1897–1905. [Google Scholar] [CrossRef]
- Salas-Riega, C.Y.; Osorio, S.; Gamarra, J.d.P.; Alvarado-Bolovich, V.; Osorio, C.M.; Gomez, C.A. Enteric methane emissions by lactating and dry cows in the high Andes of Peru. Trop. Anim. Heal. Prod. 2022, 54, 1–11. [Google Scholar] [CrossRef]
- Demment, M.W.; Van Soest, P.J. A nutritional explanation for body-size patterns of ruminant and nonruminant herbivores. Am. Nat. 1985, 125, 641–672. [Google Scholar] [CrossRef]

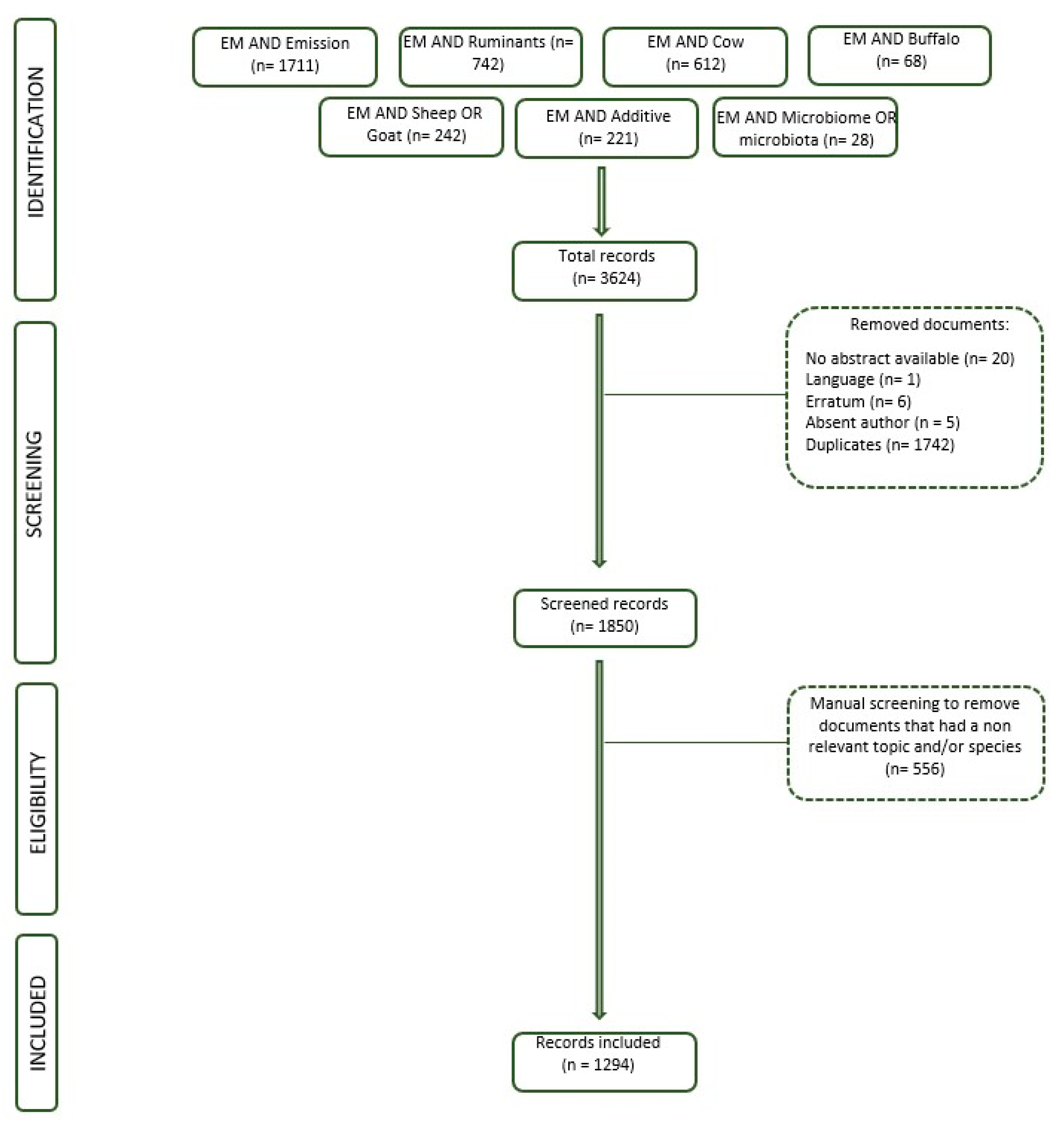



| Search Words | Original No. of Records |
|---|---|
| Enteric Methane AND Emission | 1711 |
| Enteric Methane AND Ruminants | 742 |
| Enteric Methane AND Cow | 612 |
| Enteric Methane AND Sheep OR Goat | 242 |
| Enteric Methane AND Additive | 221 |
| Enteric Methane AND Buffalo | 68 |
| Enteric Methane AND Microbiome OR Microbiota | 28 |
| Total | 3624 |
| Words (TF-IDF ≥ 9.5) | Associated Words (Grade of Correlation ≥ 0.4) |
|---|---|
| Cow | Lactat (0.47) |
| Model | Error (0.46) |
| Digest | Nutrient (0.42) |
| Energi | Gross (0.46); Metaboliz (0.41) |
| Graze | Pastur (0.43) |
| Increas | Linear (0.45) |
| Measur | Chamber (0.41) |
| Predict | Error (0.50); Equat (0.47); Extant (0.42) |
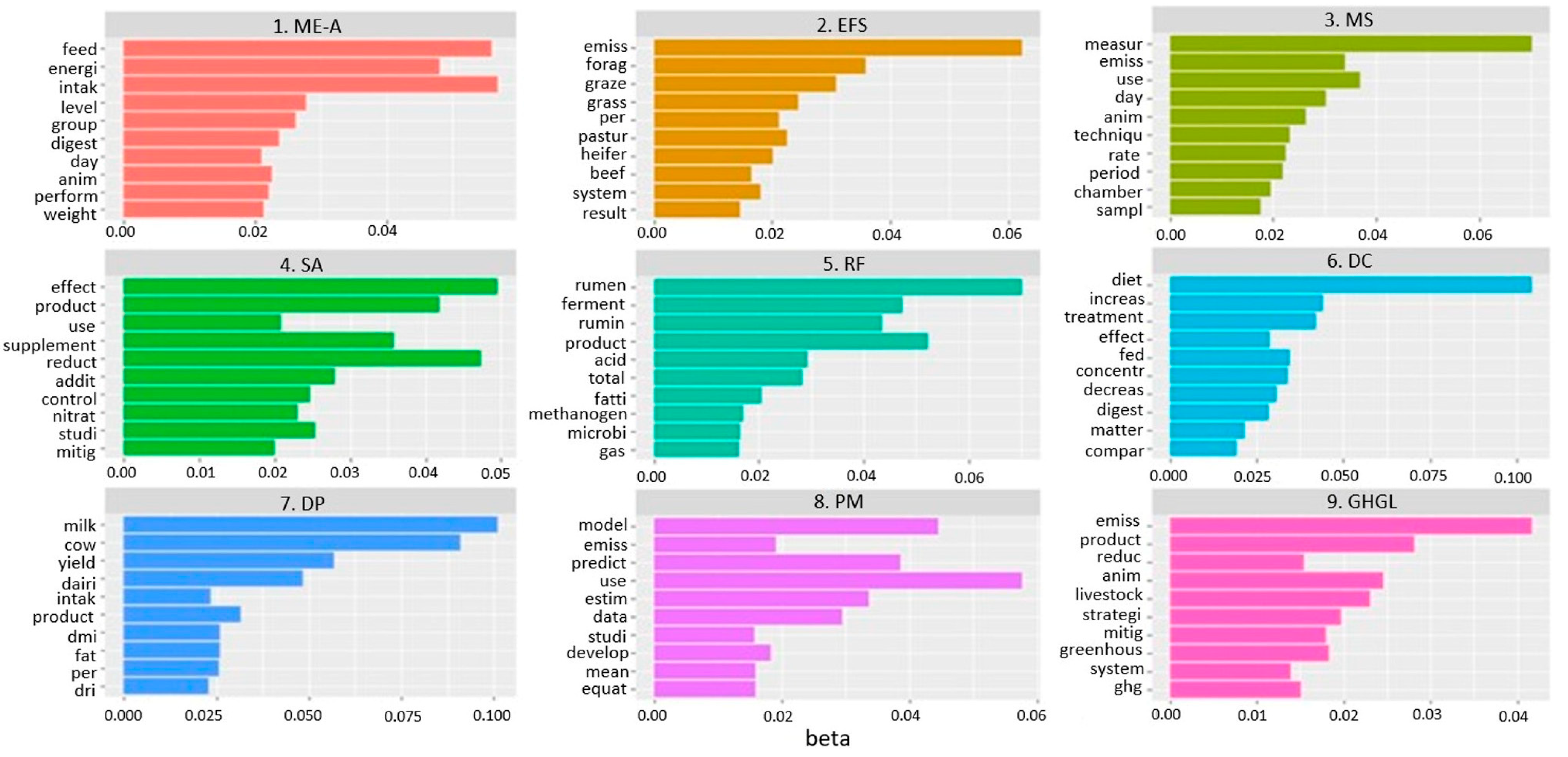
| Topic | Label of Topic | Acronyms | No. of Records per Topic (%) | Year of First Publication |
|---|---|---|---|---|
| 1 | Methane emission-animal | ME-A | 100 (7.73%) | 1998 |
| 2 | Extensive farming system | EFS | 120 (9.27%) | 2005 |
| 3 | In vivo measurement system | MS | 127 (9.81%) | 2001 |
| 4 | Supplement and additive | SA | 142 (10.97%) | 2006 |
| 5 | Ruminal fermentation | RF | 132 (10.20%) | 2006 |
| 6 | Diet composition | DC | 174 (13.45%) | 2005 |
| 7 | Dairy production | DP | 119 (9.20%) | 2005 |
| 8 | Prediction model | PM | 146 (11.28%) | 2005 |
| 9 | Greenhouse gas emission from livestock | GHGL | 234 (18.08%) | 1986 |
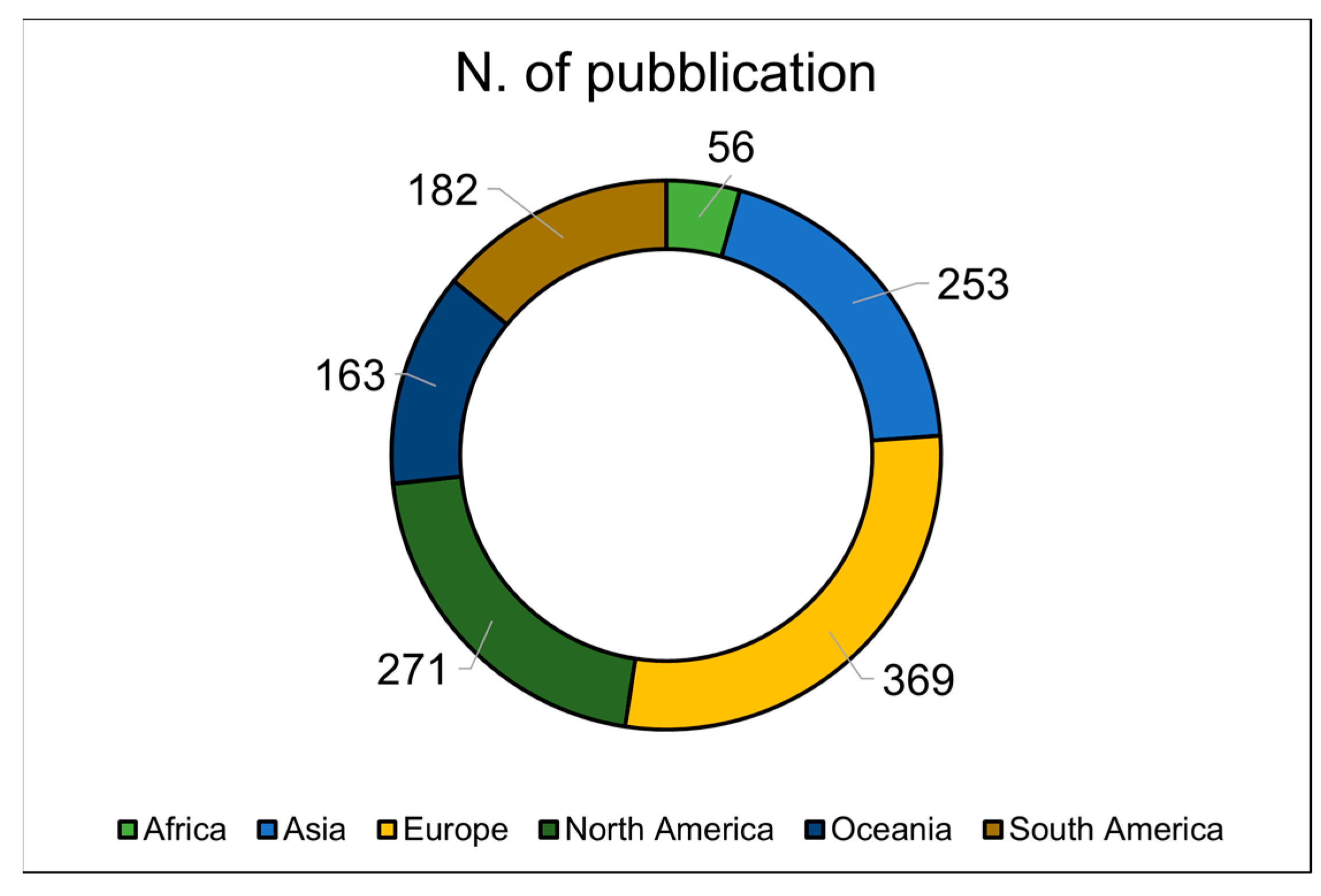
| Continent | Studies (%) | Species 1 | EME Technique 2 |
|---|---|---|---|
| Europe | 32 | Cattle, goat, sheep, and other | RC, SF6, GF, and other |
| Oceania | 23 | Cattle, sheep, and other | RC, SF6, GF, and other |
| North America | 20 | Cattle, goat, sheep, and other | RC, SF6, and GF |
| Asia | 15 | Buffalo, cattle, goat, sheep, and other | RC, SF6, GF, and other |
| South America | 9 | Cattle, goat, and sheep | RC, SF6, and other |
| Africa | 1 | Cattle and goat | SF6 and other |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Evangelista, C.; Milanesi, M.; Pietrucci, D.; Chillemi, G.; Bernabucci, U. Enteric Methane Emission in Livestock Sector: Bibliometric Research from 1986 to 2024 with Text Mining and Topic Analysis Approach by Machine Learning Algorithms. Animals 2024, 14, 3158. https://doi.org/10.3390/ani14213158
Evangelista C, Milanesi M, Pietrucci D, Chillemi G, Bernabucci U. Enteric Methane Emission in Livestock Sector: Bibliometric Research from 1986 to 2024 with Text Mining and Topic Analysis Approach by Machine Learning Algorithms. Animals. 2024; 14(21):3158. https://doi.org/10.3390/ani14213158
Chicago/Turabian StyleEvangelista, Chiara, Marco Milanesi, Daniele Pietrucci, Giovanni Chillemi, and Umberto Bernabucci. 2024. "Enteric Methane Emission in Livestock Sector: Bibliometric Research from 1986 to 2024 with Text Mining and Topic Analysis Approach by Machine Learning Algorithms" Animals 14, no. 21: 3158. https://doi.org/10.3390/ani14213158
APA StyleEvangelista, C., Milanesi, M., Pietrucci, D., Chillemi, G., & Bernabucci, U. (2024). Enteric Methane Emission in Livestock Sector: Bibliometric Research from 1986 to 2024 with Text Mining and Topic Analysis Approach by Machine Learning Algorithms. Animals, 14(21), 3158. https://doi.org/10.3390/ani14213158







