Molecular Diversity of the Casein Gene Cluster in Bovidae: Insights from SNP Microarray Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. SNP Call Rates
3.2. SNP Polymorphism
3.3. Genetic Diversity Indices
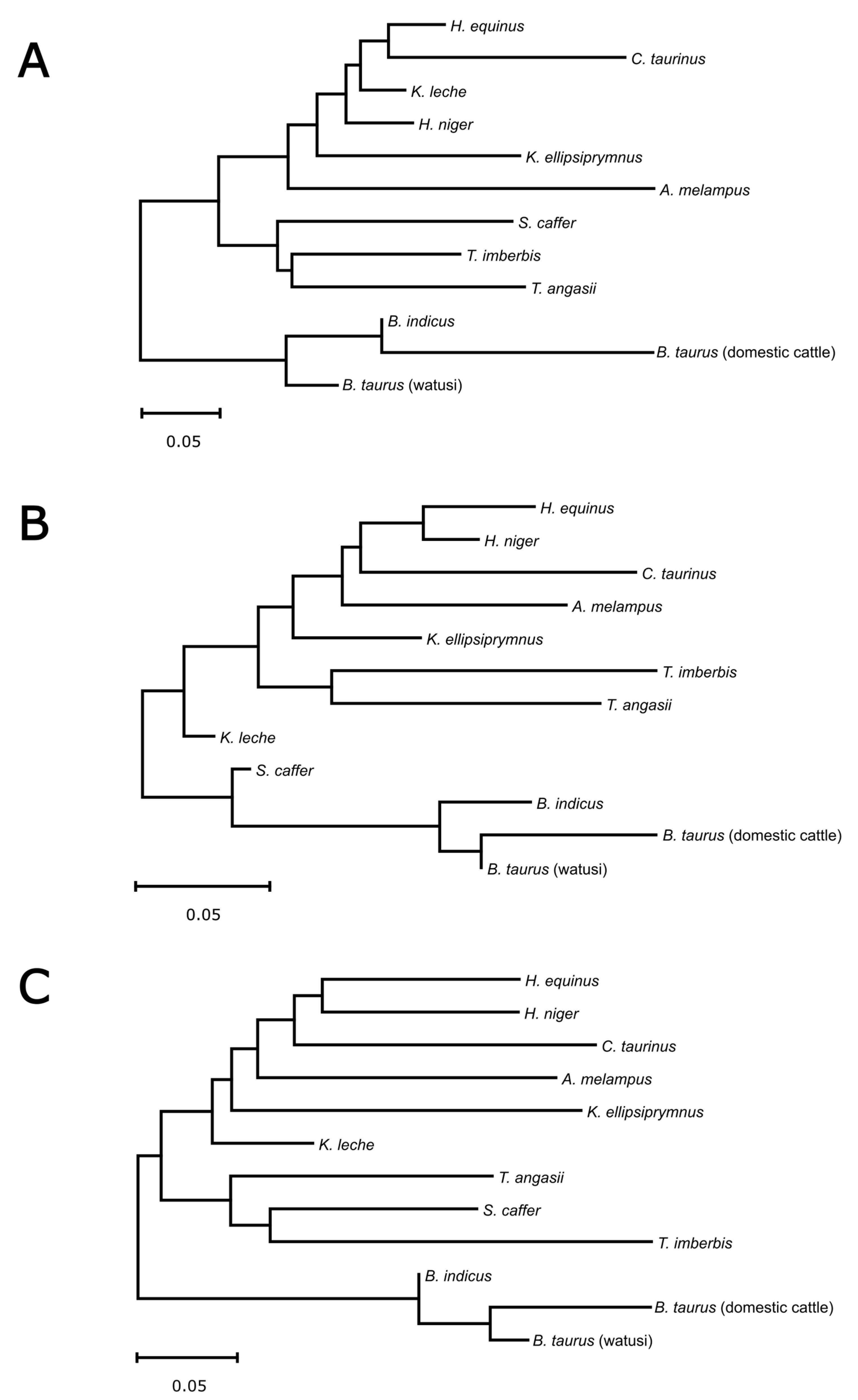
3.4. Casein Gene Cluster and Flanking Regions-Based Phylogeny
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rogiński, H. Encyclopedia of Dairy Sciences; Academic Press: London, UK, 2003. [Google Scholar]
- FAO [Food and Agriculture Organization of the United Nations]. Food Outlook—Biannual Report on Global Food Markets; FAO: Rome, Italy, 2021; pp. 52–57. [Google Scholar]
- Kawasaki, K.; Lafont, A.-G.; Sire, J.-Y. The evolution of milk casein genes from tooth genes before the origin of Mammals. Mol. Biol. Evol. 2011, 28, 2053–2061. [Google Scholar] [CrossRef] [PubMed]
- Rijnkels, M. Multispecies comparison of the casein gene loci and evolution of casein gene family. J. Mammary Gland Biol. Neoplasia 2002, 7, 327–345. [Google Scholar] [CrossRef] [PubMed]
- Kaimala, S.; Kumar, S. An evolutionarily conserved non-coding element in casein locus acts as transcriptional repressor. Gene 2015, 554, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Rijnkels, M.; Elnitski, L.; Miller, W.; Rosen, J.M. Multispecies comparative analysis of a mammalian-specific genomic domain encoding secretory proteins. Genomics 2003, 82, 417–432. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.U.; Feng, T.; Wu, S.; Luo, X.; Lei, A.; Luobu, B.; Hassan, F.U.; Liu, Q. Comparative genomics, evolutionary and gene regulatory regions analysis of casein gene family in Bubalus bubalis. Front. Genet. 2021, 12, 662609. [Google Scholar] [CrossRef]
- Cosenza, G.; Feligini, M.; Mancusi, A.; D’Avino, A.; Coletta, A.; Di Bernardino, D.; Ramunno, L. Italian Mediterranean river buffalo CSN2 gene structure and promoter analysis. Ital. J. Anim. Sci. 2009, 8 (Suppl. 2), 57–59. [Google Scholar] [CrossRef]
- Cosenza, G.; Gallo, D.; Auzino, B.; Gaspa, G.; Pauciullo, A. Complete CSN1S2 characterization, novel allele identification and association with milk fatty acid composition in river buffalo. Front. Genet. 2021, 11, 622494. [Google Scholar] [CrossRef]
- Vincent, S.T.; Momoh, O.M.; Yakubu, A. Bioinformatics analysis of beta-casein gene in some selected mammalian species. Res. Opin. Anim. Vet. Sci. 2014, 4, 564–570. [Google Scholar]
- Ward, T.J.; Honeycutt, R.L.; Derr, J.N. Nucleotide sequence evolution at the kappa-casein locus: Evidence for positive selection within the family Bovidae. Genetics 1997, 147, 1863–1872. [Google Scholar] [CrossRef]
- Caroli, A.M.; Chessa, S.; Erhardt, G.J. Invited review: Milk protein polymorphisms in cattle: Effect on animal breeding and human nutrition. J. Dairy Sci. 2009, 92, 5335–5352. [Google Scholar] [CrossRef]
- Wickramasinghe, S.; Rincon, G.; Islas-Trejo, A.; Medrano, J.F. Transcriptional profiling of bovine milk using RNA sequencing. BMC Genom. 2012, 13, 45. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Zhao, F.Q. Current major advances in the regulation of milk protein gene expression. Crit. Rev. Eukaryot. Gene Expr. 2014, 24, 357–378. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Willi, M.; Liu, C.; Hennighausen, L. Cell-specific and shared regulatory elements control a multigene locus active in mammary and salivary glands. Nat. Commun. 2023, 14, 4992. [Google Scholar] [CrossRef] [PubMed]
- Threadgill, D.W.; Womack, J.E. 1990. Genomic analysis of the major bovine milk protein genes. Nucleic Acids Res. 1990, 18, 6935–6942. [Google Scholar] [CrossRef]
- Ryskaliyeva, A.; Henry, C.; Miranda, G.; Faye, B.; Konuspayeva, G.; Martin, P. Alternative splicing events expand molecular diversity of camel CSN1S2 increasing its ability to generate potentially bioactive peptides. Sci. Rep. 2019, 9, 5243. [Google Scholar] [CrossRef]
- Pauciullo, A.; Shuiep, E.T.; Ogah, M.D.; Cosenza, G.; Di Stasio, L.; Erhardt, G. Casein gene cluster in camelids: Comparative genome analysis and new findings on haplotype variability and physical mapping. Front. Genet. 2019, 10, 748. [Google Scholar] [CrossRef]
- Keating, A.F.; Davoren, P.; Smith, T.J.; Ross, R.P.; Cairns, M.T. Bovine kappa-casein gene promoter haplotypes with potential implications for milk protein expression. J. Dairy Sci. 2007, 90, 4092–4099. [Google Scholar] [CrossRef]
- Kishore, A.; Mukesh, M.; Sobti, R.C.; Kataria, R.S.; Mishra, B.P.; Sodhi, M. Analysis of genetic variations across regulatory and coding regions of kappa-casein gene of Indian native cattle (Bos indicus) and buffalo (Bubalus bubalis). Meta Gene 2014, 2, 769–781. [Google Scholar] [CrossRef]
- Wilson, D.E.; Reeder, D.M. (Eds.) Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd ed.; Johns Hopkins University Press: Baltimore, MD, USA, 2005. [Google Scholar]
- Kuznetsov, S.B.; Solodneva, E.V.; Semina, M.T.; Beketov, S.V.; Turbina, I.S.; Stolpovsky, Y.A. New combinations of alleles in the variants of the cluster of bovine casein genes and revision of the nomenclature of these genes. Russ. J. Genet. 2022, 58, 915–926. [Google Scholar] [CrossRef]
- Ahmed, A.S.; Rahmatalla, S.; Bortfeldt, R.; Arends, D.; Reissmann, M.; Brockmann, G.A. Milk protein polymorphisms and casein haplotypes in Butana cattle. J. Appl. Genet. 2017, 58, 261–271. [Google Scholar] [CrossRef]
- Rahmatalla, S.A.; Arends, D.; Brockmann, G.A. Review: Genetic and protein variants of milk caseins in goats. Front. Genet. 2022, 13, 995349. [Google Scholar] [CrossRef] [PubMed]
- Luigi-Sierra, M.G.; Mármol-Sánchez, E.; Amills, M. Comparing the diversity of the casein genes in the Asian mouflon and domestic sheep. Anim. Genet. 2020, 51, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Osthoff, G.; Hugo, A.; De Wit, M. Milk composition of free-ranging sable antelope (Hippotragus niger). Mamm. Biol. 2007, 72, 116–122. [Google Scholar] [CrossRef]
- Osthoff, G.; Hugo, A.; De Wit, M. Comparison of the milk composition of free-ranging blesbok, black wildebeest and blue wildebeest of the subfamily Alcelaphinae (family: Bovidae). Comp. Biochem. Physiol. B 2009, 154, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Osthoff, G.; Hugo, A.; De Wit, M. Comparison of the milk composition of free-ranging eland and kudu (subfamily Bovinae, tribe Tragelaphini), and gemsbok and scimitar oryx (subfamily Hippotraginae) with observations on lechwe, okapi and Southern pudu. S. Afr. J. Wildl. Res. 2012, 42, 23–34. [Google Scholar] [CrossRef]
- Osthoff, G.; Hugo, A.; Madende, M.; Schmidt, L.; Kobeni, S.; Deacon, F. Milk composition of free-ranging impala (Aepyceros melampus) and tsessebe (Damaliscus lunatus lunatus), and comparison with other African Bovidae. Animals 2021, 11, 516. [Google Scholar] [CrossRef]
- Illumina, Inc. GenomeStudio Software, Version 2011.1; Illumina, Inc.: San Diego, CA, USA, 2011.
- Peakall, R.; Smouse, P.E. GenALEx 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–2295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Fan, X.; Gao, S.; Fu, L.; Qiu, L.; Miao, Y. Polymorphism and molecular characteristics of the CSN1S2 gene in river and swamp buffalo. Arch. Anim. Breed. 2020, 63, 345–354. [Google Scholar] [CrossRef]
- Khan, K.; Suhail, S.M.; Khan, R.; Ahmed, I.; Khan, F.A.; Khan, M.J. Genetic polymorphism of Β-casein gene and its association with milk production and composition in Azi-Kheli buffalo. Trop. Anim. Health Prod. 2023, 55, 94. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Korkuć, P.; Arends, D.; Brockmann, G.A. DNA sequence variants and protein haplotypes of casein genes in german black pied cattle (DSN). Front. Genet. 2019, 10, 1129. [Google Scholar] [CrossRef] [PubMed]
- Pérez, M.J.; Leroux, C.; Bonastre, A.S.; Martin, P. Occurrence of a LINE sequence in the 3′ UTR of the goat alpha s1-casein E-encoding allele associated with reduced protein synthesis level. Gene 1994, 147, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Najafi, M.; Rahimi Mianji, G.; Ansari Pirsaraie, Z. Cloning and comparative analysis of gene structure in promoter site of alpha-s1 casein gene in Naeinian goat and sheep. Meta Gene 2014, 2, 854–861. [Google Scholar] [CrossRef] [PubMed]
- Cosenza, G.; Iannaccone, M.; Pico, B.A.; Ramunno, L.; Capparelli, R. The SNP g.1311T>C associated with the absence of β-casein in goat milk influences CSN2 promoter activity. Anim. Genet. 2016, 47, 615–617. [Google Scholar] [CrossRef]
- Lewerentz, F.; Vanhala, T.K.; Johansen, L.B.; Paulsson, M.; Glantz, M.; de Koning, D.J. Re-sequencing of the casein genes in Swedish Red cattle giving milk with diverse protein profiles and extreme rennet coagulation properties. JDS Commun. 2024, 5, 299–304. [Google Scholar] [CrossRef]
- Iamartino, D.; Nicolazzi, E.L.; Van Tassell, C.P.; Reecy, J.M.; Fritz-Waters, E.R.; Koltes, J.E. Design and validation of a 90K SNP genotyping assay for the water buffalo (Bubalus bubalis). PLoS ONE 2017, 12, e0185220. [Google Scholar] [CrossRef]
- Usai, M.G.; Casu, S.; Sechi, T.; Salaris, S.L.; Miari, S.; Sechi, S.; Carta, P.; Carta, A. Mapping genomic regions affecting milk traits in Sarda sheep by using the OvineSNP50 Beadchip and principal components to perform combined linkage and linkage disequilibrium analysis. Genet. Sel. Evol. 2019, 51, 65. [Google Scholar] [CrossRef]
- Michelizzi, V.N.; Wu, X.; Dodson, M.V.; Michal, J.J.; Zambrano-Varon, J.; McLean, D.J.; Jiang, Z. A global view of 54,001 single nucleotide polymorphisms (SNPs) on the Illumina Bovine SNP50 BeadChip and their transferability to water buffalo. Int. J. Biol. Sci. 2010, 7, 18–27. [Google Scholar] [CrossRef]
- Matukumalli, L.K.; Lawley, C.T.; Schnabel, R.D.; Taylor, J.F.; Allan, M.F.; Heaton, M.P.; O’Connell, J.; Moore, S.S.; Smith, T.P.; Sonstegard, T.S.; et al. Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE 2009, 4, e5350. [Google Scholar] [CrossRef]
- Pertoldi, C.; Wójcik, J.M.; Tokarska, M.; Kawałko, A.; Kristensen, T.N.; Loeschcke, V.; Gregersen, V.R.; Coltman, D.; Wilson, G.A.; Randi, E.; et al. Genome variability in European and American bison detected using BovineSNP50 BeadChip. Conserv. Genet. 2010, 11, 627–634. [Google Scholar] [CrossRef]
- Ogden, R.; Baird, J.; Senn, H.; McEwing, R. The use of cross-species genome-wide arrays to discover SNP markers for conservation genetics: A case study from Arabian and scimitar-horned oryx. Conserv. Genet. Resour. 2012, 4, 471–473. [Google Scholar] [CrossRef]
- Shafer, A.B.; Miller, J.M.; Kardos, M. Cross-species application of SNP chips is not suitable for identifying runs of homozygosity. J. Hered. 2016, 107, 193–195. [Google Scholar] [CrossRef] [PubMed]
- Giovambattista, G.; Ripoli, M.V.; Peral-Garcia, P.; Bouzat, J.L. Indigenous domestic breeds as reservoirs of genetic diversity: The Argentinean Creole cattle. Anim. Genet. 2001, 32, 240–247. [Google Scholar] [CrossRef]
- Cañas-Álvarez, J.J.; González-Rodríguez, A.; Munilla, S.; Varona, L.; Díaz, C.; Baro, J.A.; Altarriba, J.; Molina, A.; Piedrafita, J. Genetic diversity and divergence among Spanish beef cattle breeds assessed by a bovine high-density SNP chip. J. Anim. Sci. 2015, 93, 5164–5174. [Google Scholar] [CrossRef]
- Corredor, F.A.; Figueroa, D.; Estrada, R.; Salazar, W.; Quilcate, C.; Vásquez, H.V.; Gonzales, J.; Maicelo, J.L.; Medina, P.; Arbizu, C.I. Genetic diversity and population structure of a Peruvian cattle herd using SNP data. Front. Genet. 2023, 14, 1073843. [Google Scholar] [CrossRef]
- Cotterill, F.P.D. The Upemba lechwe, Kobus anselli: An antelope new to science emphasizes the conservation importance of Katange, Democratic Republic of Congo. J. Zool. 2005, 265, 113–132. [Google Scholar] [CrossRef]
- IUCN SSC Antelope Specialist Group. Kobus leche. The IUCN Red List of Threatened Species: E.T11033A50189021. 2017. Available online: https://www.iucnredlist.org/species/11033/50189021 (accessed on 3 September 2024).
- Moodley, Y.; Bruford, M.W. Molecular biogeography: Towards an integrated framework for conserving pan-African biodiversity. PLoS ONE 2007, 2, e454. [Google Scholar] [CrossRef]
- Nefdt, R.J.C. Reproductive seasonality in Kafue lechwe antelope. J. Zool. 1996, 238, 155–166. [Google Scholar] [CrossRef]
- Coulon, J.B.; Dupont, D.; Pochet, S.; Pradel, P.; Duployer, H. Effect of genetic potential and level of feeding on milk protein composition. J. Dairy Res. 2001, 68, 569–577. [Google Scholar] [CrossRef]
- Li, B.; Khan, M.Z.; Khan, I.M.; Ullah, Q.; Cisang, Z.M.; Zhang, N.; Wu, D.; Huang, B.; Ma, Y.; Khan, A.; et al. Genetics, environmental stress, and amino acid supplementation affect lactational performance via mTOR signaling pathway in bovine mammary epithelial cells. Front. Genet. 2023, 14, 1195774. [Google Scholar] [CrossRef] [PubMed]
- Vesey-Fitzgerald, L.D.E.F. Lechwe pastures. Puku 1965, 3, 143–147. [Google Scholar]
- Cotterill, F.P.D. Reduncine antelope of the Zambezi basin. In Biodiversity of the Zambezi Basin Wetlands; Timberlake, J.R., Ed.; Biodiversity Foundation for Africa: Bulawayo, Zimbabwe; Zambezi Society: Harare, Zimbabwe, 2000; pp. 145–199. [Google Scholar]
- Wang, B.; Wang, Z.; Zhou, J.; Liu, W.; Lin, Z.; Zhang, C.; Liu, G.; Zhou, B.; Wan, W.; Zhao, R.; et al. The draft genome of red lechwe, Kobus leche leche. Front. Genet. 2020, 11, 582638. [Google Scholar] [CrossRef] [PubMed]
- George, L.; Alex, R.; Gowane, G.; Vohra, V.; Joshi, P.; Kumar, R.; Verma, A. Weighted single step GWAS reveals genomic regions associated with economic traits in Murrah buffaloes. Anim. Biotechnol. 2024, 35, 2319622. [Google Scholar] [CrossRef] [PubMed]
- Lázaro, S.F.; Tonhati, H.; Oliveira, H.R.; Silva, A.A.; Scalez, D.C.B.; Nascimento, A.V.; Santos, D.J.A.; Stefani, G.; Carvalho, I.S.; Sandoval, A.F.; et al. Genetic parameters and genome-wide association studies for mozzarella and milk production traits, lactation length, and lactation persistency in Murrah buffaloes. J. Dairy Sci. 2024, 107, 992–1021. [Google Scholar] [CrossRef]
- Gonçalves, M.; Siegismund, H.R.; van Vuuren, B.J.; Koepfli, K.-P.; Ferrand, N.; Godinho, R. De novo whole-genome assembly and resequencing resources for the roan (Hippotragus equinus), an iconic African antelope. G3 2021, 11, jkab002. [Google Scholar] [CrossRef]
- Koepfli, K.P.; Tamazian, G.; Wildt, D.; Dobrynin, P.; Kim, C.; Frandsen, P.B.; Godinho, R.; Yurchenko, A.A.; Komissarov, A.; Krasheninnikova, K.; et al. Whole genome sequencing and re-sequencing of the sable antelope (Hippotragus niger): A resource for monitoring diversity in ex situ and in situ populations. G3 2019, 9, 1785–1793. [Google Scholar] [CrossRef]
- Fang, Z.H.; Bovenhuis, H.; van Valenberg, H.J.F.; Martin, P.; Duchemin, S.I.; Huppertz, T.; Visker, M.H.P.W. Genome-wide association study for αS1- and αS2-casein phosphorylation in Dutch Holstein Friesian. J. Dairy Sci. 2019, 102, 1374–1385. [Google Scholar] [CrossRef]
- Mahmoudi, P.; Rostamzadeh, J.; Rashidi, A.; Zergani, E.; Razmkabir, M. A meta-analysis on association between CSN3 gene variants and milk yield and composition in cattle. Anim. Genet. 2020, 51, 369–381. [Google Scholar] [CrossRef]
- Ozdemir, M.; Motmain, Z.; Ekinci, K.; Saygılı, E. Associations between BLG, CSN3, DGAT1, GH, PIT1, and PRL gene polymorphisms and milk production traits in Holstein dairy cows: A meta-analysis. Biochem. Genet. 2024. [Google Scholar] [CrossRef]
- Asim, M.; Saif-ur Rehman, M.; Hassan, F.-u.; Awan, F.S. Genetic variants of CSN1S1, CSN2, CSN3, and BLG genes and their association with dairy production traits in Sahiwal cattle and Nili-Ravi buffaloes. Anim. Biotechnol. 2023, 34, 2951–2962. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, N.B.C.; Guerreiro, S.L.M.; Pereira, R.S.R.; Sá, A.L.; Rodrigues, M.; Mezzomo, R.; Maciel, R.P.; Hamoy, I.G.; Rodrigues, M.D.N. CSN1S1 and CSN3 gene variants in female Murrah buffaloes in the Brazilian Amazon. An. Acad. Bras. Cienc. 2022, 94 (Suppl. 3), e20211394. [Google Scholar] [CrossRef] [PubMed]
- Pauciullo, A.; Gaspa, G.; Zhang, Y.; Liu, Q.; Cosenza, G. CSN1S1, CSN3 and LPL: Three validated gene polymorphisms useful for more sustainable dairy production in the Mediterranean river buffalo. Animals 2024, 14, 1414. [Google Scholar] [CrossRef] [PubMed]
- Venturini, G.C.; Cardoso, D.F.; Baldi, F.; Freitas, A.C.; Aspilcueta-Borquis, R.R.; Santos, D.J.; Camargo, G.M.; Stafuzza, N.B.; Albuquerque, L.G.; Tonhati, H. Association between single-nucleotide polymorphisms and milk production traits in buffalo. Genet. Mol. Res. 2014, 13, 10256–10268. [Google Scholar] [CrossRef] [PubMed]
- Malewski, T.; Zwierzchowski, L. Computer-aided analysis of potential transcription–factor binding sites in the rabbit β–casein gene promoter. BioSystems 1995, 36, 109–119. [Google Scholar] [CrossRef]
- Malewski, T. Computer analysis of distribution of putative cis- and trans-regulatory elements in milk protein gene promoters. BioSystems 1998, 45, 29–44. [Google Scholar] [CrossRef]
- Parveen, S.; Zhu, P.; Shafique, L.; Lan, H.; Xu, D.; Ashraf, S.; Sherazi, M.; Liu, Q. Molecular characterization and phylogenetic analysis of casein gene family in Camelus ferus. Genes 2023, 14, 256. [Google Scholar] [CrossRef]
- Kappeler, S.R.; Farah, Z.; Puhan, Z. 5′-flanking regions of camel milk genes are highly similar to homologue regions of other species and can be divided into two distinct groups. J. Dairy Sci. 2003, 86, 498–508. [Google Scholar] [CrossRef]
- Kang, Y.K.; Lee, C.S.; Chung, A.S.; Lee, K.K. Prolactin-inducible enhancer activity of the first intron of the bovine beta-casein gene. Mol. Cells. 1998, 8, 259–265. [Google Scholar] [CrossRef]
- Kolb, A. The first intron of the murine beta-casein gene contains a functional promoter. Biochem. Biophys. Res. Commun. 2003, 306, 1099–1105. [Google Scholar] [CrossRef]
- Song, N.; Luo, J.; Huang, L.; Tian, H.; Chen, Y.; He, Q. miR-204-5p and miR-211 synergistically downregulate the αS1-casein content and contribute to the lower allergy of goat milk. J. Agric. Food Chem. 2021, 69, 5353–5362. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Haneda, S.; Imakawa, K.; Sakai, S.; Nagaoka, K. A microRNA, miR-101a, controls mammary gland development by regulating cyclooxygenase-2 expression. Differentiation 2009, 77, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Li, C.; Li, J.; Song, J.; Zhang, S. Integrated Small RNA Sequencing, Transcriptome and GWAS Data reveal microRNA regulation in response to milk protein traits in Chinese Holstein cattle. Front. Genet. 2021, 12, 726706. [Google Scholar] [CrossRef] [PubMed]
- Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes. Off. J. Eur. Union 2010, L 267, 33–79.


| Probe Name | Sequence |
|---|---|
| EuroG10K_Hapmap 60224 | CCAACCAGAGGTCCACCAGAGTCACCCTAGAGAGAAAAGGATATATATAA |
| Hapmap 59186 | CCATGTGAGTGTCAATGTTGGCTTGAACAGTGTTGTTTTCCTGGCAACCA |
| Hapmap 25708-BTC-043671 | CTAGAAGTCCTCCTGAGAAGACATAAGAGAAAAGCCAAAGACACTGTCAT |
| Hapmap 24184-BTC-070077 | GAGCACAACAAATTATAACTGCAAAGCATCAAAAAGACCACATCAAACAC |
| BTA-111108-no-rs | TTAATAAACAGTAAAGGTCAGGCAGATGGTCCCTCTCCCTCACATATTCA |
| Hapmap 52348 | TACTTTCCACCTTATTTATTACCCAGAGCCACACAGTTAAGCAAGCGATT |
| BTA-77173-no-rs | GCATACAGGATAAATTATAAATGTGTGCCCATTATAATAAGGCCTGCAAA |
| ARS-BFGL-NGS-2418 | GTTTTTGTTTTTTATTTCACTTTCAGGAAGGCCCACTACAATGGCAGAGT |
| BTA-118117 | AAATAAGATGTGATGTTTAAGAGACACTGATGGAACCTGGTGCTTATTTA |
| BTB-00268331 | CGCCTGGCAAGAACTGCCATCCTAAAACATACCCAAAGTTGCCTTGGATT |
| Species | GenBank Accession No | EuroG10KHapmap 60224 | Hapmap 59186 | Hapmap 25708-BTC-043671 | Hapmap 24184-BTC-070077 | Hapmap 25779-BTC-072896 | Hapmap 52348 | BTA-77173-no-rs | ARS-BFGL-NGS-2418 | BTA-118117 | BTB-00268331 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| B. indicus | GCF_000247795.1 | 100 | 100 | 100 | 96 | 98 | 100 | 100 | 100 | 100 | 100 |
| A. melampus | GCA_006408695.1 | 100 | 96 | 96 | 94 | ns | 92 | 96 ** | 90 | 86 ** | 92 |
| C. taurinus | GCA_006408615.1 | 100 | 96 | 90 ** | 94 ** | 92 | 93 * (80) | 96 | 92 * (60) | 90 ** | 92 |
| S. caffer | GCA_006408785.2 | 98 | 96 | 96 | 100 | 98 | 98 | 100 | 98 | 100 | 88 |
| T. imberbis | GCA_006410775.1 | 98 | 88 | 94 | 94 | 92 | 94 | 98 | 92 | 94 | 92 * (80) |
| H. niger | GCA_006942125.1 | 100 | 98 | 92 | 92 | 90 | 98 | 92 | 92 | 94 * (60) | 92 |
| K. leche | GCA_014926565.1 | 100 | 96 | 94 | 92 | 94 * (80) | 98 | 98 | 92 * (80) | 94 * (40) | 90 |
| K. ellipsiprymnus | GCA_006410655.1 | 100 | 96 | 94 | 92 * (80) | 94 | 98 | 100 | 92 | 94 * (20) | 88 |
| Species | Probe Sequence |
|---|---|
| B. taurus | GAGCACAACAAATTATAACTGCAAAGCATCAAAAAGACCACATCAAACAC |
| B. indicus | ........................................Y.W....... |
| A. melampus | ...G............G.........................C....... |
| C. taurinus | ...G............G................................T |
| S. caffer | .................................................. |
| T. imberbis | ...............CG..A.............................. |
| H. niger | ...G....A.......G...T............................. |
| K. leche | ................G..........T..........TG.......... |
| K. ellipsiprymnus | ................G..........T..........TG.......... |
| Species | n | Call Rate | |
|---|---|---|---|
| All SNPs | Casein Gene Cluster | ||
| A. melampus | 5 | 83.0 | 82.9 |
| C. taurinus | 5 | 85.1 | 84.2 |
| Domestic cattle | 12 | 100.0 | 100.0 |
| Watusi | 5 | 98.3 | 98.0 |
| B. indicus (zebu) | 5 | 99.5 | 99.5 |
| S. caffer | 9 | 95.0 | 94.5 |
| T. angasii | 5 | 88.7 | 87.5 |
| T. imberbis | 6 | 89.6 | 87.6 |
| H. equinus | 5 | 87.1 | 87.1 |
| H. niger | 5 | 87.9 | 87.6 |
| K. leche | 5 | 83.1 | 84.0 |
| K. ellipsiprymnus | 5 | 83.3 | 83.6 |
| Taxon | 5′-Upstream Total | Casein Gene Cluster | 3′-Downstream Total | |||
|---|---|---|---|---|---|---|
| Total | Exons | Introns | Intergenic | |||
| Species | ||||||
| A. melampus | 13.64 | 8.89 | 8.57 | 0.00 | 28.57 | 8.51 |
| C. taurinus | 27.27 | 17.78 | 20.00 | 16.67 | 14.29 | 6.38 |
| Domestic cattle | 68.18 | 37.78 | 34.29 | 50.00 | 71.43 | 89.36 |
| Watusi | 68.18 | 40.00 | 37.14 | 33.33 | 57.14 | 53.19 |
| B. indicus (zebu) | 63.64 | 35.56 | 28.57 | 50.00 | 42.86 | 40.43 |
| S. caffer | 18.18 | 15.56 | 14.29 | 16.67 | 28.57 | 8.51 |
| T. imberbis | 9.09 | 11.11 | 14.29 | 0.00 | 14.29 | 8.51 |
| T. angasii | 18.18 | 20.00 | 22.86 | 16.67 | 0.00 | 10.64 |
| H. equinus | 22.73 | 15.56 | 17.14 | 0.00 | 28.57 | 6.38 |
| H. niger | 18.18 | 15.56 | 17.14 | 0.00 | 28.57 | 6.38 |
| K. leche | 54.55 | 55.56 | 57.14 | 33.33 | 71.43 | 36.17 |
| K. ellipsiprymnus | 9.09 | 13.33 | 14.29 | 0.00 | 28.57 | 4.26 |
| Genus | ||||||
| Bos | 90.91 | 55.56 | 51.43 | 50.00 | 85.71 | 91.49 |
| Tragelaphus | 18.18 | 26.67 | 28.57 | 16.67 | 28.57 | 17.02 |
| Hippotragus | 22.72 | 20.00 | 22.86 | 0.00 | 28.57 | 8.51 |
| Kobus | 54.54 | 60.00 | 62.86 | 33.33 | 71.43 | 38.30 |
| Mean | 30.42 | 22.56 | 22.64 | 16.67 | 31.87 | 21.27 |
| SE | 6.62 | 4.07 | 3.78 | 5.34 | 6.30 | 7.20 |
| Taxon | Ho | He | uHe | Fst |
|---|---|---|---|---|
| A. 5′-upstream region | ||||
| A. melampus | 0.121 ± 0.068 | 0.066 ± 0.036 | 0.077 ± 0.042 | −0.833 ± 0.062 |
| C. taurinus | 0.205 ± 0.085 | 0.123 ± 0.044 | 0.186 ± 0.072 | −0.556 ± 0.172 |
| Domestic cattle | 0.223 ± 0.051 | 0.230 ± 0.045 | 0.240 ± 0.047 | 0.028 ± 0.078 |
| Watusi | 0.373 ± 0.075 | 0.275 ± 0.045 | 0.309 ± 0.051 | −0.322 ± 0.104 |
| B. indicus (zebu) | 0.232 ± 0.050 | 0.197 ± 0.039 | 0.220 ± 0.043 | −0.164 ± 0.058 |
| S. caffer | 0.091 ± 0.063 | 0.083 ± 0.039 | 0.109 ± 0.055 | 0.000 ± 0.246 |
| T. angasii | 0.066 ± 0.047 | 0.055 ± 0.028 | 0.062 ± 0.031 | −0.063 ± 0.175 |
| T. imberbis | 0.045 ± 0.045 | 0.037 ± 0.026 | 0.041 ± 0.029 | 0.000 ± 0.302 |
| H. equinus | 0.152 ± 0.075 | 0.105 ± 0.043 | 0.144 ± 0.062 | −0.333 ± 0.201 |
| H. niger | 0.125 ± 0.065 | 0.072 ± 0.035 | 0.106 ± 0.055 | −0.619 ± 0.095 |
| K. leche | 0.232 ± 0.077 | 0.196 ± 0.044 | 0.249 ± 0.061 | −0.069 ± 0.163 |
| K. ellipsiprymnus | 0.091 ± 0.063 | 0.045 ± 0.031 | 0.052 ± 0.036 | −1.000 ± 0.000 |
| Total | 0.163 ± 0.019 | 0.124 ± 0.012 | 0.150 ± 0.015 | −0.230 ± 0.039 |
| B. Total casein gene cluster | ||||
| A. melampus | 0.089 ± 0.043 | 0.044 ± 0.021 | 0.074 ± 0.037 | −1.000 ± 0.000 |
| C. taurinus | 0.172 ± 0.056 | 0.088 ± 0.029 | 0.141 ± 0.048 | −0.950 ± 0.021 |
| Domestic cattle | 0.122 ± 0.029 | 0.112 ± 0.026 | 0.117 ± 0.027 | −0.082 ± 0.023 |
| Watusi | 0.188 ± 0.042 | 0.151 ± 0.030 | 0.169 ± 0.034 | −0.227 ± 0.065 |
| B. indicus (zebu) | 0.151 ± 0.036 | 0.124 ± 0.027 | 0.138 ± 0.030 | −0.200 ± 0.050 |
| S. caffer | 0.077 ± 0.035 | 0.047 ± 0.019 | 0.057 ± 0.024 | −0.429 ± 0.065 |
| T. angasii | 0.093 ± 0.038 | 0.061 ± 0.021 | 0.079 ± 0.030 | −0.349 ± 0.077 |
| T. imberbis | 0.042 ± 0.024 | 0.033 ± 0.016 | 0.040 ± 0.019 | −0.196 ± 0.075 |
| H. equinus | 0.104 ± 0.044 | 0.064 ± 0.023 | 0.094 ± 0.037 | −0.492 ± 0.113 |
| H. niger | 0.144 ± 0.052 | 0.075 ± 0.026 | 0.122 ± 0.044 | −0.905 ± 0.038 |
| K. leche | 0.150 ± 0.040 | 0.175 ± 0.026 | 0.207 ± 0.032 | 0.196 ± 0.106 |
| K. ellipsiprymnus | 0.122 ± 0.048 | 0.064 ± 0.025 | 0.104 ± 0.042 | −0.889 ± 0.041 |
| Total | 0.118 ± 0.011 | 0.083 ± 0.007 | 0.110 ± 0.010 | −0.307 ± 0.024 |
| C. Exon casein gene cluster | ||||
| A. melampus | 0.086 ± 0.048 | 0.043 ± 0.024 | 0.067 ± 0.038 | −1.000 ± 0.000 |
| C. taurinus | 0.193 ± 0.066 | 0.099 ± 0.034 | 0.153 ± 0.055 | −0.943 ± 0.026 |
| Domestic cattle | 0.088 ± 0.026 | 0.089 ± 0.027 | 0.093 ± 0.028 | −0.013 ± 0.015 |
| Watusi | 0.171 ± 0.047 | 0.138 ± 0.033 | 0.154 ± 0.037 | −0.225 ± 0.076 |
| B. indicus (zebu) | 0.109 ± 0.033 | 0.093 ± 0.027 | 0.104 ± 0.030 | −0.160 ± 0.045 |
| S. caffer | 0.085 ± 0.044 | 0.048 ± 0.024 | 0.059 ± 0.030 | −0.545 ± 0.079 |
| T. angasii | 0.114 ± 0.048 | 0.073 ± 0.026 | 0.095 ± 0.037 | −0.379 ± 0.092 |
| T. imberbis | 0.054 ± 0.030 | 0.042 ± 0.020 | 0.051 ± 0.024 | −0.196 ± 0.085 |
| H. equinus | 0.106 ± 0.049 | 0.068 ± 0.027 | 0.092 ± 0.039 | −0.407 ± 0.134 |
| H. niger | 0.157 ± 0.061 | 0.082 ± 0.031 | 0.138 ± 0.054 | −0.889 ± 0.046 |
| K. leche | 0.155 ± 0.048 | 0.187 ± 0.030 | 0.222 ± 0.037 | 0.239 ± 0.126 |
| K. ellipsiprymnus | 0.129 ± 0.056 | 0.068 ± 0.029 | 0.106 ± 0.047 | −0.867 ± 0.050 |
| Total | 0.121 ± 0.014 | 0.086 ± 0.008 | 0.111 ± 0.011 | −0.284 ± 0.030 |
| D. Intron casein gene cluster | ||||
| A. melampus | 0.000 | 0.000 | 0.000 | |
| C. taurinus | 0.167 ± 0.167 | 0.083 ± 0.083 | 0.167 ± 0.167 | −1.000 ± 0.167 |
| Domestic cattle | 0.250 ± 0.120 | 0.190 ± 0.089 | 0.198 ± 0.093 | −0.302 ± 0.037 |
| Watusi | 0.200 ± 0.126 | 0.140 ± 0.089 | 0.156 ± 0.098 | −0.429 ± 0.000 |
| B. indicus (zebu) | 0.333 ± 0.161 | 0.213 ± 0.098 | 0.237 ± 0.109 | −0.528 ± 0.098 |
| S. caffer | 0.056 ± 0.056 | 0.046 ± 0.046 | 0.056 ± 0.056 | −0.200 ± 0.056 |
| T. angasii | 0.033 ± 0.033 | 0.030 ± 0.030 | 0.033 ± 0.033 | −0.111 ± 0.033 |
| T. imberbis | 0.000 | 0.000 | 0.000 | |
| H. equinus | 0.000 | 0.000 | 0.000 | |
| H. niger | 0.000 | 0.000 | 0.000 | |
| K. leche | 0.042 ± 0.042 | 0.090 ± 0.058 | 0.101 ± 0.065 | 0.429 ± 0.330 |
| K. ellipsiprymnus | 0.000 | 0.000 | 0.000 | |
| Total | 0.090 ± 0.027 | 0.066 ± 0.018 | 0.079 ± 0.022 | −0.292 ± 0.055 |
| E. Intergenic casein gene cluster | ||||
| A. melampus | 0.286 ± 0.184 | 0.143 ± 0.092 | 0.238 ± 0.158 | −1.000 ± 0.000 |
| C. taurinus | 0.143 ± 0.143 | 0.071 ± 0.071 | 0.143 ± 0.143 | −1.000 ± 0.143 |
| Domestic cattle | 0.226 ± 0.077 | 0.219 ± 0.074 | 0.228 ± 0.077 | −0.056 ± 0.080 |
| Watusi | 0.207 ± 0.088 | 0.198 ± 0.078 | 0.223 ± 0.088 | −0.081 ± 0.146 |
| B. indicus (zebu) | 0.086 ± 0.040 | 0.111 ± 0.061 | 0.124 ± 0.067 | 0.101 ± 0.139 |
| S. caffer | 0.286 ± 0.184 | 0.143 ± 0.092 | 0.190 ± 0.123 | −1.000 ± 0.000 |
| T. angasii | 0.000 | 0.000 | 0.000 | |
| T. imberbis | 0.036 ± 0.036 | 0.031 ± 0.031 | 0.036 ± 0.036 | −0.143 ± 0.036 |
| H. equinus | 0.286 ± 0.184 | 0.143 ± 0.092 | 0.229 ± 0.154 | −1.000 ± 0.000 |
| H. niger | 0.286 ± 0.184 | 0.143 ± 0.092 | 0.190 ± 0.123 | −1.000 ± 0.000 |
| K. leche | 0.195 ± 0.106 | 0.234 ± 0.068 | 0.278 ± 0.083 | 0.211 ± 0.277 |
| K. ellipsiprymnus | 0.214 ± 0.149 | 0.125 ± 0.082 | 0.214 ± 0.149 | −0.667 ± 0.178 |
| Total | 0.167 ± 0.034 | 0.121 ± 0.020 | 0.162 ± 0.030 | −0.271 ± 0.063 |
| F. 3′-downstream region | ||||
| A. melampus | 0.080 ± 0.039 | 0.040 ± 0.019 | 0.072 ± 0.036 | −1.000 ± 0.000 |
| C. taurinus | 0.056 ± 0.032 | 0.030 ± 0.017 | 0.051 ± 0.030 | −0.889 ± 0.027 |
| Domestic cattle | 0.350 ± 0.031 | 0.313 ± 0.024 | 0.326 ± 0.025 | −0.101 ± 0.033 |
| Watusi | 0.235 ± 0.038 | 0.205 ± 0.029 | 0.230 ± 0.032 | −0.160 ± 0.061 |
| B. indicus (zebu) | 0.143 ± 0.032 | 0.149 ± 0.027 | 0.166 ± 0.030 | 0.052 ± 0.068 |
| S. caffer | 0.067 ± 0.035 | 0.037 ± 0.018 | 0.050 ± 0.027 | −0.723 ± 0.057 |
| T. angasii | 0.084 ± 0.039 | 0.044 ± 0.020 | 0.067 ± 0.032 | −0.822 ± 0.056 |
| T. imberbis | 0.080 ± 0.039 | 0.040 ± 0.019 | 0.071 ± 0.035 | −1.000 ± 0.000 |
| H. equinus | 0.041 ± 0.025 | 0.025 ± 0.015 | 0.038 ± 0.023 | −0.583 ± 0.054 |
| H. niger | 0.060 ± 0.034 | 0.030 ± 0.017 | 0.052 ± 0.030 | −1.000 ± 0.000 |
| K. leche | 0.074 ± 0.030 | 0.103 ± 0.022 | 0.132 ± 0.033 | 0.314 ± 0.102 |
| K. ellipsiprymnus | 0.040 ± 0.028 | 0.020 ± 0.014 | 0.032 ± 0.023 | −1.000 ± 0.000 |
| Total | 0.110 ± 0.011 | 0.086 ± 0.007 | 0.108 ± 0.010 | −0.202 ± 0.023 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malewski, T.; Kamiński, S.; Śmiełowski, J.; Oleński, K.; Bogdanowicz, W. Molecular Diversity of the Casein Gene Cluster in Bovidae: Insights from SNP Microarray Analysis. Animals 2024, 14, 3034. https://doi.org/10.3390/ani14203034
Malewski T, Kamiński S, Śmiełowski J, Oleński K, Bogdanowicz W. Molecular Diversity of the Casein Gene Cluster in Bovidae: Insights from SNP Microarray Analysis. Animals. 2024; 14(20):3034. https://doi.org/10.3390/ani14203034
Chicago/Turabian StyleMalewski, Tadeusz, Stanisław Kamiński, Jan Śmiełowski, Kamil Oleński, and Wiesław Bogdanowicz. 2024. "Molecular Diversity of the Casein Gene Cluster in Bovidae: Insights from SNP Microarray Analysis" Animals 14, no. 20: 3034. https://doi.org/10.3390/ani14203034
APA StyleMalewski, T., Kamiński, S., Śmiełowski, J., Oleński, K., & Bogdanowicz, W. (2024). Molecular Diversity of the Casein Gene Cluster in Bovidae: Insights from SNP Microarray Analysis. Animals, 14(20), 3034. https://doi.org/10.3390/ani14203034






