Pan-Omics in Sheep: Unveiling Genetic Landscapes
Abstract
Simple Summary
Abstract
1. Introduction
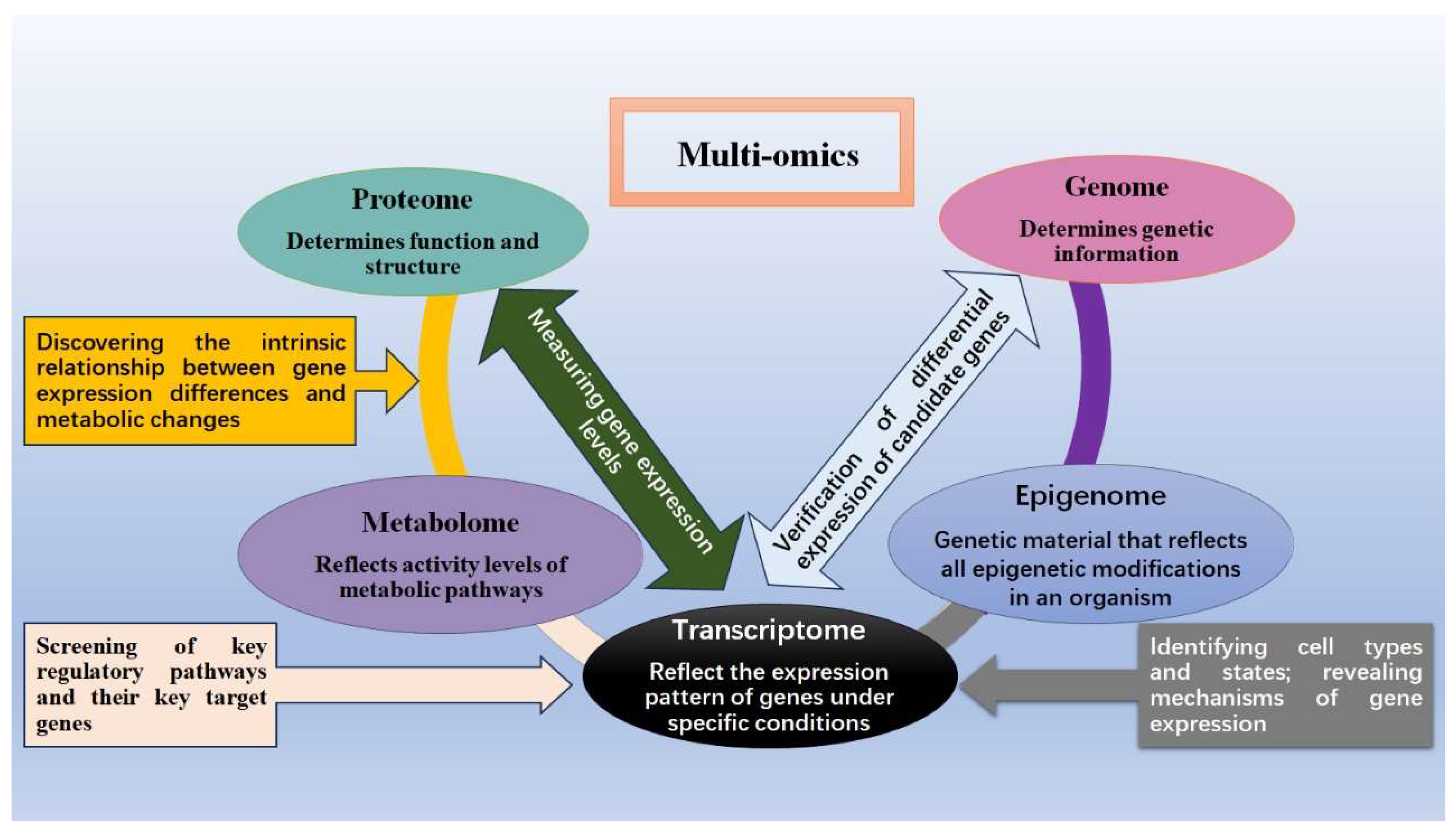
2. Main Omics Analysis Techniques
2.1. Genomics
2.2. Epigenomics
2.3. Transcriptomics
2.4. Proteomics
2.5. Metabolomics
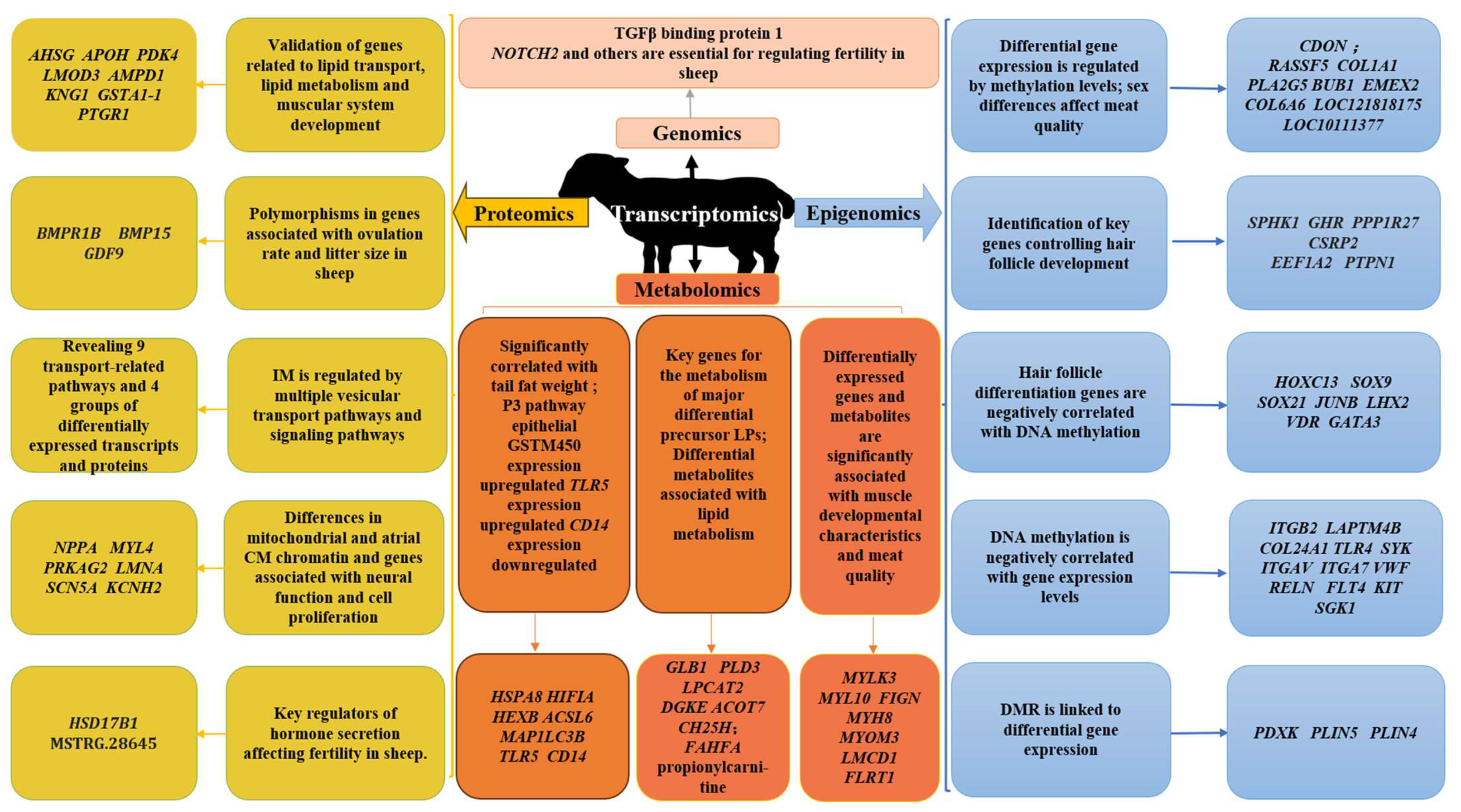
3. Research and Implementation of Multiomics Integration in Sheep Production
3.1. Meat Traits
3.2. Wool Traits
3.3. Reproductive Traits
3.4. Ovine Physiology
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kijas, J.W.; Townley, D.; Dalrymple, B.P.; Heaton, M.P.; Maddox, J.F.; McGrath, A.; Wilson, P.; Ingersoll, R.G.; McCulloch, R.; McWilliam, S.; et al. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS ONE 2009, 4, 4668. [Google Scholar] [CrossRef]
- Larkin, D.M.; Yudin, N.S. The genomes and history of domestic animals. Mol. Gen. Microbiol. Virol. 2016, 31, 197–202. [Google Scholar] [CrossRef][Green Version]
- Yang, N.; Liu, J.; Gao, Q.; Gui, S.; Chen, L.; Yang, L.; Huang, J.; Deng, T.; Luo, J.; He, L.; et al. Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nat. Genet. 2019, 51, 1052–1059. [Google Scholar] [CrossRef] [PubMed]
- Romanov, M.N.; Zinovieva, N.A.; Griffin, D.K. British Sheep Breeds as a Part of World Sheep Gene Pool Landscape: Looking into Genomic Applications. Animals 2021, 11, 994. [Google Scholar] [CrossRef] [PubMed]
- Bersanelli, M.; Mosca, E.; Remondini, D.; Giampieri, E.; Sala, C.; Castellani, G.; Milanesi, L. Methods for the integration of multi-omics data: Mathematical aspects. BMC Bioinform. 2016, 17, 15. [Google Scholar] [CrossRef]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Bansal, G.; Narang, A.; Basak, T.; Abbas, T.; Dash, D. Integrating transcriptome and proteome profiling: Strategies and applications. Proteomics 2016, 16, 2533–2544. [Google Scholar] [CrossRef]
- Kurakin, A. Scale-free flow of life: On the biology, economics, and physics of the cell. Theor. Biol. Med. Model. 2009, 6, 6. [Google Scholar] [CrossRef]
- Bensimon, A.; Heck, A.J.R.; Aebersold, R. Mass spectrometry-based proteomics and network biology. Annu. Rev. Biochem. 2012, 81, 379–405. [Google Scholar] [CrossRef]
- Gutteridge, A.; Pir, P.; Castrillo, J.I.; Charles, P.D.; Lilley, K.S.; Oliver, S.G. Nutrient control of eukaryote cell growth: A systems biology study in yeast. BMC Biol. 2010, 8, 68. [Google Scholar] [CrossRef]
- Haas, R.; Zelezniak, A.; Iacovacci, J.; Kamrad, S.; Townsend, S.; Ralser, M. Designing and interpreting ‘multi-omic’ experiments that may change our understanding of biology. Curr. Opin. Syst. Biol. 2017, 6, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Risacher, S.L.; Shen, L.; Saykin, A.J. Network approaches to systems biology analysis of complex disease: Integrative methods for multi-omics data. Brief. Bioinform. 2018, 19, 1370–1381. [Google Scholar] [CrossRef] [PubMed]
- Del Giacco, L.; Cattaneo, C. Introduction to genomics. Methods Mol. Biol. 2012, 823, 79–88. [Google Scholar] [PubMed]
- Wang, C.; Han, B. Twenty years of rice genomics research: From sequencing and functional genomics to quantitative genomics. Mol. Plant. 2022, 15, 593–619. [Google Scholar] [CrossRef]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.C.; Kuan, P.F. A Guide to Illumina BeadChip Data Analysis. Methods Mol. Biol. 2018, 1708, 303–330. [Google Scholar] [PubMed]
- Lukic, B.; Curik, I.; Drzaic, I.; Galić, V.; Shihabi, M.; Vostry, L.; Cubric-Curik, V. Genomic signatures of selection, local adaptation and production type characterisation of East Adriatic sheep breeds. J. Anim. Sci. Biotechnol. 2023, 14, 142. [Google Scholar] [CrossRef]
- Wang, H.; Zhong, L.; Dong, Y.; Meng, L.; Ji, C.; Luo, H.; Fu, M.; Qi, Z.; Mi, L. Whole-genome resequencing reveals domestication and signatures of selection in Ujimqin, Sunit, and Wu Ranke Mongolian sheep breeds. Anim. Biosci. 2022, 35, 1303–1313. [Google Scholar] [CrossRef]
- Zhao, Z.D.; Zhang, L. Applications of genome selection in sheep breeding. Yi Chuan 2019, 41, 293–303. [Google Scholar]
- Jiang, Y.; Xie, M.; Chen, W.; Talbot, R.; Maddox, J.F.; Faraut, T.; Wu, C.; Muzny, D.M.; Li, Y.; Zhang, W.; et al. The sheep genome illuminates biology of the rumen and lipid metabolism. Science 2014, 344, 1168–1173. [Google Scholar] [CrossRef]
- Su, R.; Qiao, X.; Gao, Y.; Li, X.; Jiang, W.; Chen, W.; Fan, Y.; Zheng, B.; Zhang, Y.; Liu, Z.; et al. Draft Genome of the European Mouflon (Ovis orientalis musimon). Front. Genet. 2020, 11, 533611. [Google Scholar] [CrossRef] [PubMed]
- Davenport, K.M.; Bickhart, D.M.; Worley, K.; Murali, S.C.; Salavati, M.; Clark, E.L.; Cockett, N.E.; Heaton, M.P.; Smith, T.P.L.; Murdoch, B.M.; et al. An improved ovine reference genome assembly to facilitate in-depth functional annotation of the sheep genome. Gigascience 2022, 11, 96. [Google Scholar] [CrossRef] [PubMed]
- Qiao, G.; Xu, P.; Guo, T.; Wu, Y.; Lu, X.; Zhang, Q.; He, X.; Zhu, S.; Zhao, H.; Lei, Z.; et al. Genetic Basis of Dorper Sheep (Ovis aries) Revealed by Long-Read De Novo Genome Assembly. Front. Genet. 2022, 13, 846449. [Google Scholar] [CrossRef] [PubMed]
- Norris, B.J.; Whan, V.A. A gene duplication affecting expression of the ovine ASIP gene is responsible for white and black sheep. Genome Res. 2008, 18, 1282–1293. [Google Scholar] [CrossRef]
- Wiedemar, N.; Drögemüller, C. A 1.8-kb insertion in the 3′-UTR of RXFP2 is associated with polledness in sheep. Anim. Genet. 2015, 46, 457–461. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, X.; Li, F.; Liu, T.; Hu, Z.; Gao, N.; Yuan, L.; Li, X.; Zhao, Y.; Zhao, L.; et al. Whole-genome resequencing identified candidate genes associated with the number of ribs in Hu sheep. Genomics 2021, 113, 2077–2084. [Google Scholar] [CrossRef]
- Li, R.; Gong, M.; Zhang, X.; Wang, F.; Liu, Z.; Zhang, L.; Yang, Q.; Xu, Y.; Xu, M.; Zhang, H.; et al. A sheep pangenome reveals the spectrum of structural variations and their effects on tail phenotypes. Genome Res. 2023, 33, 463–477. [Google Scholar] [CrossRef]
- Rezvannejad, E.; Asadollahpour Nanaei, H.; Esmailizadeh, A. Detection of candidate genes affecting milk production traits in sheep using whole-genome sequencing analysis. Vet. Med. Sci. 2022, 8, 1197–1204. [Google Scholar] [CrossRef]
- Yang, J.; Li, W.R.; Lv, F.H.; He, S.G.; Tian, S.L.; Peng, W.F.; Sun, Y.W.; Zhao, Y.X.; Tu, X.L.; Zhang, M.; et al. Whole-Genome Sequencing of Native Sheep Provides Insights into Rapid Adaptations to Extreme Environments. Mol. Biol. Evol. 2016, 33, 2576–2592. [Google Scholar] [CrossRef]
- Hu, X.J.; Yang, J.; Xie, X.L.; Lv, F.H.; Cao, Y.H.; Li, W.R.; Liu, M.J.; Wang, Y.T.; Li, J.Q.; Liu, Y.G.; et al. The Genome Landscape of Tibetan Sheep Reveals Adaptive Introgression from Argali and the History of Early Human Settlements on the Qinghai-Tibetan Plateau. Mol. Biol. Evol. 2019, 36, 283–303. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, Z.; Jia, J.; Du, T.; Zhang, N.; Tang, Y.; Fang, Y.; Fang, D. Overview of Histone Modification. Adv. Exp. Med. Biol. 2021, 1283, 1–16. [Google Scholar] [PubMed]
- Wang, K.C.; Chang, H.Y. Epigenomics: Technologies and Applications. Circ. Res. 2018, 122, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ibeagha-Awemu, E.M. Impacts of Epigenetic Processes on the Health and Productivity of Livestock. Front. Genet. 2021, 11, 613636. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Arenas, C.; Hernandez-Ferrer, C.; Vives-Usano, M.; Marí, S.; Quintela, I.; Mason, D.; Cadiou, S.; Casas, M.; Andrusaityte, S.; Gutzkow, K.B.; et al. Identification of autosomal cis expression quantitative trait methylation (cis eQTMs) in children’s blood. Elife 2022, 11, 65310. [Google Scholar] [CrossRef]
- Wang, Y.; Fischle, W.; Cheung, W.; Jacobs, S.; Khorasanizadeh, S.; Allis, C.D. Beyond the double helix: Writing and reading the histone code. Novartis Found. Symp. 2004, 259, 13–17. [Google Scholar]
- Zhu, J.; Adli, M.; Zou, J.Y.; Verstappen, G.; Coyne, M.; Zhang, X.; Durham, T.; Miri, M.; Deshpande, V.; De Jager, P.L.; et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell 2013, 152, 642–654. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, F.; Feng, X.; Yang, H.; Zhu, A.; Pang, J.; Han, L.; Zhang, T.; Yao, X.; Wang, F. Genome-wide analysis of DNA Methylation profiles on sheep ovaries associated with prolificacy using whole-genome Bisulfite sequencing. BMC Genom. 2017, 18, 759. [Google Scholar] [CrossRef]
- Fan, Y.; Liang, Y.; Deng, K.; Zhang, Z.; Zhang, G.; Zhang, Y.; Wang, F. Analysis of DNA methylation profiles during sheep skeletal muscle development using whole-genome bisulfite sequencing. BMC Genom. 2020, 21, 327. [Google Scholar] [CrossRef]
- Zhu, L.; Tillquist, N.; Scatolin, G.; Gately, R.; Kawaida, M.; Reiter, A.; Reed, S.; Zinn, S.; Govoni, K.; Jiang, Z. Maternal restricted- and over- feeding during gestation perturb offspring sperm epigenome in sheep. Reproduction 2023, 166, 311–322. [Google Scholar] [CrossRef]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, 1005457. [Google Scholar] [CrossRef]
- Harper, A.L.; He, Z.; Langer, S.; Havlickova, L.; Wang, L.; Fellgett, A.; Gupta, V.; Kumar Pradhan, A.; Bancroft, I. Validation of an Associative Transcriptomics platform in the polyploid crop species Brassica juncea by dissection of the genetic architecture of agronomic and quality traits. Plant J. 2020, 103, 1885–1893. [Google Scholar] [CrossRef]
- Sangwan, R.S.; Tripathi, S.; Singh, J.; Narnoliya, L.K.; Sangwan, N.S. De novo sequencing and assembly of Centella asiatica leaf transcriptome for mapping of structural, functional and regulatory genes with special reference to secondary metabolism. Gene 2013, 525, 58–76. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, D.J.; Winzeler, E.A. Genomics, gene expression and DNA arrays. Nature 2000, 405, 827–836. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wang, Z.; Yang, H.; Yao, X.; Yang, P.; Ren, C.; Wang, F.; Zhang, Y. Pituitary Transcriptomic Study Reveals the Differential Regulation of lncRNAs and mRNAs Related to Prolificacy in Different FecB Genotyping Sheep. Genes 2019, 10, 157. [Google Scholar] [CrossRef]
- Gunawan, A.; Listyarini, K.; Harahap, R.S.; Jakaria Roosita, K.; Sumantri, C.; Inounu, I.; Akter, S.H.; Islam, M.A.; Uddin, M.J. Hepatic transcriptome analysis identifies genes, polymorphisms and pathways involved in the fatty acids metabolism in sheep. PLoS ONE 2021, 16, 260514. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Di, R.; Yang, Y.; Liu, Q.; Chu, M. Transcriptome Analysis of Neuroendocrine Regulation of Ovine Hypothalamus-Pituitary-Ovary Axis during Ovine Anestrus and the Breeding Season. Genes 2021, 12, 1861. [Google Scholar] [CrossRef]
- Chen, S.; Guo, X.; He, X.; Di, R.; Zhang, X.; Zhang, J.; Wang, X.; Chu, M. Transcriptome Analysis Reveals Differentially Expressed Genes and Long Non-coding RNAs Associated with Fecundity in Sheep Hypothalamus with Different FecB Genotypes. Front. Cell Dev. Biol. 2021, 9, 633747. [Google Scholar] [CrossRef]
- Wang, X.; Fang, C.; He, H.; Cao, H.; Liu, L.; Jiang, L.; Ma, Y.; Liu, W. Identification of key genes in sheep fat tail evolution Based on RNA-seq. Gene 2021, 781, 145492. [Google Scholar] [CrossRef]
- Fei, X.; Jin, M.; Wang, Y.; Li, T.; Lu, Z.; Yuan, Z.; Wang, H.; Lu, J.; Quan, K.; Di, R.; et al. Transcriptome reveals key microRNAs involved in fat deposition between different tail sheep breeds. PLoS ONE 2022, 17, 264804. [Google Scholar] [CrossRef]
- Hao, Z.; Zhou, H.; Hickford, J.G.H.; Gong, H.; Wang, J.; Hu, J.; Liu, X.; Li, S.; Zhao, M.; Luo, Y. Identification and characterization of circular RNA in lactating mammary glands from two breeds of sheep with different milk production profiles using RNA-Seq. Genomics 2020, 112, 2186–2193. [Google Scholar] [CrossRef]
- Jürgen Cox Mann, M. Is proteomics the new genomics? Cell 2007, 130, 395–398. [Google Scholar]
- Zhang, H.Y.; Lei, G.; Zhou, H.W.; He, C.; Liao, J.L.; Huang, Y.J. Quantitative iTRAQ-based proteomic analysis of rice grains to assess high night temperature stress. Proteomics 2017, 17, 1600365. [Google Scholar] [CrossRef] [PubMed]
- Tyers, M.; Mann, M. From genomics to proteomics. Nature 2003, 422, 193–197. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, S.; Stenoien, D.L.; Paša-Tolić, L. High-throughput proteomics. Annu. Rev. Anal. Chem. 2014, 7, 427–454. [Google Scholar] [CrossRef] [PubMed]
- Aslam, B.; Basit, M.; Nisar, M.A.; Khurshid, M.; Rasool, M.H. Proteomics: Technologies and Their Applications. J. Chromatogr. Sci. 2017, 55, 182–196. [Google Scholar] [CrossRef]
- Domon, B.; Aebersold, R. Mass spectrometry and protein analysis. Science 2006, 312, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Krishna, R.G.; Wold, F. Post-translational modification of proteins. Adv. Enzymol. Relat. Areas Mol. Biol. 1993, 67, 265–298. [Google Scholar]
- Wang, X.; Shi, T.; Zhao, Z.; Hou, H.; Zhang, L. Proteomic analyses of sheep (Ovis aries) embryonic skeletal muscle. Sci. Rep. 2020, 10, 1750. [Google Scholar] [CrossRef]
- Zhu, C.; Cheng, H.; Li, N.; Liu, T.; Ma, Y. Isobaric Tags for Relative and Absolute Quantification-Based Proteomics Reveals Candidate Proteins of Fat Deposition in Chinese Indigenous Sheep with Morphologically Different Tails. Front. Genet. 2021, 12, 710449. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Guo, L.; Ullah, S.; Cao, Y.; Huang, X.; Shan, H.; Jiang, J.; Wu, J.; Jiang, Y. Proteome changes of sheep rumen epithelium during postnatal development. Front. Genet. 2022, 13, 1031707. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, D.; Li, X.; Zhang, Y.; Zhao, Y.; Xu, D.; Cheng, J.; Wang, J.; Li, W.; Lin, C.; et al. Comparative proteomics reveals genetic mechanisms of body weight in Hu sheep and Dorper sheep. J. Proteom. 2022, 267, 104699. [Google Scholar] [CrossRef] [PubMed]
- Newgard, C.B. Metabolomics and Metabolic Diseases: Where Do We Stand? Cell Metab. 2017, 25, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Lonati, C.; Berezhnoy, G.; Lawler, N.; Masuda, R.; Kulkarni, A.; Sala, S.; Nitschke, P.; Zizmare, L.; Bucci, D.; Cannet, C.; et al. Urinary phenotyping of SARS-CoV-2 infection connects clinical diagnostics with metabolomics and uncovers impaired NAD+ pathway and SIRT1 activation. Clin. Chem. Lab. Med. 2023. [Google Scholar] [CrossRef]
- González-Gomariz, J.; Guruceaga, E.; López-Sánchez, M.; Segura, V. Proteogenomics in the context of the Human Proteome Project (HPP). Expert Rev. Proteom. 2019, 16, 267–275. [Google Scholar] [CrossRef]
- Goodacre, R.; Vaidyanathan, S.; Dunn, W.B.; Harrigan, G.G.; Kell, D.B. Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 2004, 22, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J. Am. Soc. Mass. Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef]
- Fraga-Corral, M.; Carpena, M.; Garcia-Oliveira, P.; Pereira, A.G.; Prieto, M.A.; Simal-Gandara, J. Analytical Metabolomics and Applications in Health, Environmental and Food Science. Crit. Rev. Anal. Chem. 2022, 52, 712–734. [Google Scholar] [CrossRef]
- Chacko, S.; Haseeb, Y.B.; Haseeb, S. Metabolomics Work Flow and Analytics in Systems Biology. Curr. Mol. Med. 2022, 22, 870–881. [Google Scholar] [CrossRef]
- Baharum, S.N.; Azizan, K.A. Metabolomics in Systems Biology. Adv. Exp. Med. Biol. 2018, 1102, 51–68. [Google Scholar]
- Palma, M.; Scanlon, T.; Kilminster, T.; Milton, J.; Oldham, C.; Greeff, J.; Matzapetakis, M.; Almeida, A.M. The hepatic and skeletal muscle ovine metabolomes as affected by weight loss: A study in three sheep breeds using NMR-metabolomics. Sci. Rep. 2016, 6, 39120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Han, L.; Hou, S.; Raza, S.H.A.; Wang, Z.; Yang, B.; Sun, S.; Ding, B.; Gui, L.; Simal-Gandara, J.; et al. Effects of different feeding regimes on muscle metabolism and its association with meat quality of Tibetan sheep. Food Chem. 2022, 374, 131611. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Sun, L.; Su, R.; Corazzin, M.; Yang, Z.; Dou, L.; Hu, G.; Zhang, Y.; Liu, T.; Guo, Y.; et al. Widely targeted metabolomic analysis reveals the dynamic changes of metabolites during postmortem chilled aging in Mongolian sheep. Food Chem. 2023, 431, 137035. [Google Scholar] [CrossRef] [PubMed]
- Caboni, P.; Murgia, A.; Porcu, A.; Manis, C.; Ibba, I.; Contu, M.; Scano, P. A metabolomics comparison between sheep’s and goat’s milk. Food Res. Int. 2019, 119, 869–875. [Google Scholar] [CrossRef]
- Zhang, R.; Pavan, E.; Ross, A.B.; Deb-Choudhury, S.; Dixit, Y.; Mungure, T.E.; Realini, C.E.; Cao, M.; Farouk, M.M. Molecular insights into quality and authentication of sheep meat from proteomics and metabolomics. J. Proteom. 2023, 276, 104836. [Google Scholar] [CrossRef]
- Hegarty, R.S.; Warner, R.D.; Pethick, D.W. Genetic and nutritional regulation of lamb growth and muscle characteristics. Aust. J. Agric. Res. 2006, 57, 721–730. [Google Scholar] [CrossRef]
- Zhao, L.; Li, F.; Zhang, X.; Zhang, D.; Li, X.; Zhang, Y.; Zhao, Y.; Song, Q.; Huang, K.; Xu, D.; et al. Integrative analysis of transcriptomics and proteomics of longissimus thoracis of the Hu sheep compared with the Dorper sheep. Meat Sci. 2022, 193, 108930. [Google Scholar] [CrossRef]
- Fonseca, P.A.S.; Alonso-García, M.; Pelayo, R.; Marina, H.; Esteban-Blanco, C.; Mateo, J.; Gutiérrez-Gil, B.; Arranz, J.J.; Suárez-Vega, A. Integrated analyses of the methylome and transcriptome to unravel sex differences in the perirenal fat from suckling lambs. Front. Genet. 2022, 13, 1035063. [Google Scholar] [CrossRef]
- Zhang, B.; Sun, Z.; Yu, Z.; Li, H.; Luo, H.; Wang, B. Transcriptome and targeted metabolome analysis provide insights into bile acids’ new roles and mechanisms on fat deposition and meat quality in lamb. Food Res. Int. 2022, 162, 111941. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Yuan, Z.H.; Li, F.D.; Yue, X.P. Integrating transcriptome and metabolome to identify key genes regulating important muscular flavour precursors in sheep. Animals 2022, 16, 100679. [Google Scholar] [CrossRef]
- Chen, B.; Yue, Y.; Li, J.; Liu, J.; Yuan, C.; Guo, T.; Zhang, D.; Yang, B.; Lu, Z. Transcriptome-metabolome analysis reveals how sires affect meat quality in hybrid sheep populations. Front. Nutr. 2022, 9, 967985. [Google Scholar] [CrossRef] [PubMed]
- Paus, R.; Cotsarelis, G. The biology of hair follicles. N. Engl. J. Med. 1999, 341, 491–497. [Google Scholar] [CrossRef]
- Schneider, M.R.; Schmidt-Ullrich, R.; Paus, R. The hair follicle as a dynamic miniorgan. Curr. Biol. 2009, 19, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Wang, S.H.; Sun, B.; Zhang, Y.L.; Shen, W.; Khatib, H.; Wang, X. Melatonin promotes Cashmere goat (Capra hircus) secondary hair follicle growth: A view from integrated analysis of long non-coding and coding RNAs. Cell Cycle 2018, 17, 1255–1267. [Google Scholar] [CrossRef]
- Wang, S.; Ge, W.; Luo, Z.; Guo, Y.; Jiao, B.; Qu, L.; Zhang, Z.; Wang, X. Integrated analysis of coding genes and non-coding RNAs during hair follicle cycle of cashmere goat (Capra hircus). BMC Genom. 2017, 18, 767. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, E. Scratching the surface of skin development. Nature 2007, 445, 834–842. [Google Scholar] [CrossRef] [PubMed]
- Driskell, R.R.; Clavel, C.; Rendl, M.; Watt, F.M. Hair follicle dermal papilla cells at a glance. J. Cell Sci. 2011, 124, 1179–1182. [Google Scholar] [CrossRef]
- Enshell-Seijffers, D.; Lindon, C.; Kashiwagi, M.; Morgan, B.A. Beta-catenin activity in the dermal papilla regulates morphogenesis and regeneration of hair. Dev. Cell. 2010, 18, 633–642. [Google Scholar] [CrossRef]
- Wang, S.; Li, F.; Liu, J.; Zhang, Y.; Zheng, Y.; Ge, W.; Qu, L.; Wang, X. Integrative Analysis of Methylome and Transcriptome Reveals the Regulatory Mechanisms of Hair Follicle Morphogenesis in Cashmere Goat. Cells 2020, 9, 969. [Google Scholar] [CrossRef]
- Zhao, B.; Luo, H.; He, J.; Huang, X.; Chen, S.; Fu, X.; Zeng, W.; Tian, Y.; Liu, S.; Li, C.J.; et al. Comprehensive transcriptome and methylome analysis delineates the biological basis of hair follicle development and wool-related traits in Merino sheep. BMC Biol. 2021, 19, 197. [Google Scholar] [CrossRef]
- Abebe, A.; Berhane, G.; Getachew, T.; Gizaw, S.; Haile, A. Reproductive performance and productivity of local and Dorper x local crossbred ewes under community-based management system, Ethiopia. Heliyon 2023, 9, 19906. [Google Scholar] [CrossRef] [PubMed]
- Walkom, S.F.; Brien, F.D.; Hebart, M.L.; Fogarty, N.M.; Hatcher, S.; Pitchford, W.S. Season and reproductive status rather than genetics factors influence change in ewe weight and fat over time. 4. Genetic relationships of ewe weight and fat with fleece, reproduction and milk traits. Anim. Prod. Sci. 2015, 56, 708–715. [Google Scholar] [CrossRef]
- Fogarty, N.M.; Snowder, G.D. Composite trait selection to improve reproduction and ewe productivity: A review. Anim. Prod. Sci. 2008, 49, 9–16. [Google Scholar]
- Miao, X.; Luo, Q. Genome-wide transcriptome analysis between small-tail Han sheep and the Surabaya fur sheep using high-throughput RNA sequencing. Reproduction 2013, 145, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhao, Y.; Yuan, Z.; Wu, Y.; Zhao, Z.; Wu, C.; Hou, J.; Zhang, M. Genome-Wide Identification of mRNAs, lncRNAs, and Proteins, and Their Relationship with Sheep Fecundity. Front. Genet. 2022, 12, 750947. [Google Scholar] [CrossRef]
- Romero, J.J.; Liebig, B.E.; Broeckling, C.D.; Prenni, J.E.; Hansen, T.R. Pregnancy-induced changes in metabolome and proteome in ovine uterine flushings. Biol. Reprod. 2017, 97, 273–287. [Google Scholar] [CrossRef]
- Namous, H.; Peñagaricano, F.; Del Corvo, M.; Capra, E.; Thomas, D.L.; Stella, A.; Williams, J.L.; Marsan, P.A.; Khatib, H. Integrative analysis of methylomic and transcriptomic data in fetal sheep muscle tissues in response to maternal diet during pregnancy. BMC Genom. 2018, 19, 123. [Google Scholar] [CrossRef] [PubMed]
- Cheung, C.Y.; Anderson, D.F.; Brace, R.A. Multiomics analyses of vesicular transport pathway-specific transcripts and proteins in ovine amnion: Responses to altered intramembranous transport. Physiol. Genom. 2019, 51, 267–278. [Google Scholar] [CrossRef]
- Wang, X.; Guo, X.; He, X.; Di, R.; Zhang, X.; Zhang, J.; Chu, M. Integrated Proteotranscriptomics of the Hypothalamus Reveals Altered Regulation Associated with the FecB Mutation in the BMPR1B Gene That Affects Prolificacy in Small Tail Han Sheep. Biology 2022, 12, 72. [Google Scholar] [CrossRef]
- Yao, X.; Li, F.; Wei, Z.; Ei-Samahy, M.A.; Feng, X.; Yang, F.; Wang, F. Integrative Genome-Wide DNA Methylome and Transcriptome Analysis of Ovaries from Hu Sheep with High and Low Prolific. Front. Cell Dev. Biol. 2022, 10, 820558. [Google Scholar] [CrossRef]
- Goering, A.W.; McClure, R.A.; Doroghazi, J.R.; Albright, J.C.; Haverland, N.A.; Zhang, Y.; Ju, K.S.; Thomson, R.J.; Metcalf, W.W.; Kelleher, N.L. Metabologenomics: Correlation of Microbial Gene Clusters with Metabolites Drives Discovery of a Nonribosomal Peptide with an Unusual Amino Acid Monomer. ACS Cent. Sci. 2016, 2, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Paulus, C.; Rebets, Y.; Tokovenko, B.; Nadmid, S.; Terekhova, L.P.; Myronovskyi, M.; Zotchev, S.B.; Rückert, C.; Braig, S.; Zahler, S.; et al. New natural products identified by combined genomics-metabolomics profiling of marine Streptomyces sp. MP131-18. Sci. Rep. 2017, 7, 42382. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Franco, A.; Rouco, R.; Ramirez, R.J.; Guerrero-Serna, G.; Tiana, M.; Cogliati, S.; Kaur, K.; Saeed, M.; Magni, R.; Enriquez, J.A.; et al. Transcriptome and proteome mapping in the sheep atria reveal molecular featurets of atrial fibrillation progression. Cardiovasc. Res. 2021, 117, 1760–1775. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Sha, Y.; Lv, W.; Cao, G.; Guo, X.; Pu, X.; Wang, J.; Li, S.; Hu, J.; Luo, Y. Multi-Omics Reveals That the Rumen Transcriptome, Microbiome, and Its Metabolome Co-regulate Cold Season Adaptability of Tibetan Sheep. Front. Microbiol. 2022, 13, 859601. [Google Scholar] [CrossRef]
- Chen, W.; Lv, X.; Cao, X.; Yuan, Z.; Wang, S.; Getachew, T.; Mwacharo, J.M.; Haile, A.; Quan, K.; Li, Y.; et al. Integration of the Microbiome, Metabolome and Transcriptome Reveals Escherichia coli F17 Susceptibility of Sheep. Animals 2023, 13, 1050. [Google Scholar] [CrossRef]
| Breed | Genome Versions | Sequence Size/Gb | Contig N50 | Scaffolds N50/Mb | Sequencing Technology | Time | Reference |
|---|---|---|---|---|---|---|---|
| Texel | Oar_v3.1 | 2.61 | 40 KB | 100 | Illumina | 2014 | [20] |
| Texel | Oar_v4.0 | 2.6 | 150.5 KB | 100 | Illumina; 454 PacBio | 2015 | GCA_000298735.2 |
| Rambouillet | Oar_rambouillet_v1.0 | 2.9 | 2.6 Mb | 107.7 | HiSeq X× Ten PacBio | 2017 | GCA_002742125.1 |
| Mouflon | Platanus | 2.69 | 110.1 KB | 10.4 | Illumina | 2020 | [21] |
| Tibetan | CAU_O.aries_1.0 | 2.7 | 74.6 Mb | 105.2 | PacBio | 2021 | GCA_017524585.1 |
| Rambouillet | ARS-UI_Ramb_v2.0 | 2.63 | 43.2 Mb | 101.3 | Illumina | 2022 | [22] |
| Dorper | Oar_v4.0; ARS-UI_Ramb_v2.0 | 2.64 | 73.33 Mb | -- | Illumina PacBio | 2022 | [23] |
| Kazakh | ASM2243284v1 | 2.9 | 73.4 Mb | 96.2 | PromethION | 2022 | GCA_022432845.1 |
| Dorset | ASM2241691v1 | 2.9 | 92.4 Mb | 96.5 | Ilumina | 2022 | GCA_022416915.1 |
| Romanov x Dorper | Oar_ARS-UKY_WhiteDorper_v1.0 | 2.6 | 61.8 Mb | 95.6 | PacBio | 2022 | GCA_022244695.1 |
| Genes/Metabolites | Omics Type | Mutant Site/Metabolic Pathway | Traits | Reference |
|---|---|---|---|---|
| ASIP | Genomics | g.100-105delAGGAA g.10-19delAGCCGCCTC g.5172T > A | Hair color | [24] |
| RXFP2 | Genomics | 3′UTR | Hornless | [25] |
| CPOX KCNH1 CPQ | Genomics | g.178,730,623 T > G g.75,716,237 C > G g.88,323,841 A > G | Ribcage | [26] |
| HOXB13 | Genomics | 5′UTR | Tailed | [27] |
| MAPT DLK1 DIAPH1 NR4A1 | Epigenomics | promoter region | Muscle growth metabolism | [38] |
| HOXC9 | Transcriptomics | 3′UTR | Caudal fat deposition | [49] |
| APOA2 GALK1 ADIPOQ NDUFS4 | Proteomics | --- | Caudal fat deposition | [59] |
| Amino acids, MMA Methylmalonic acid | Metabolomics | biosynthesis of amino acids; biosynthesis of unsaturated fatty acids | meat quality | [72] |
| Amino acids fatty acyl groups glycerophospholipids | Metabolomics | protein digestion and absorption; aminoacyl-trNA biosynthesis; carbon metabolism | Succulent | [73] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Lu, Y.; Gao, Z.; Yue, D.; Hong, J.; Wu, J.; Xi, D.; Deng, W.; Chong, Y. Pan-Omics in Sheep: Unveiling Genetic Landscapes. Animals 2024, 14, 273. https://doi.org/10.3390/ani14020273
Li M, Lu Y, Gao Z, Yue D, Hong J, Wu J, Xi D, Deng W, Chong Y. Pan-Omics in Sheep: Unveiling Genetic Landscapes. Animals. 2024; 14(2):273. https://doi.org/10.3390/ani14020273
Chicago/Turabian StyleLi, Mengfei, Ying Lu, Zhendong Gao, Dan Yue, Jieyun Hong, Jiao Wu, Dongmei Xi, Weidong Deng, and Yuqing Chong. 2024. "Pan-Omics in Sheep: Unveiling Genetic Landscapes" Animals 14, no. 2: 273. https://doi.org/10.3390/ani14020273
APA StyleLi, M., Lu, Y., Gao, Z., Yue, D., Hong, J., Wu, J., Xi, D., Deng, W., & Chong, Y. (2024). Pan-Omics in Sheep: Unveiling Genetic Landscapes. Animals, 14(2), 273. https://doi.org/10.3390/ani14020273





