Effects of High-Concentrate-Induced SARA on Antioxidant Capacity, Immune Levels and Rumen Microbiota and Function in Goats
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Treatment
2.2. Sample Collection
2.3. Measurement of Enzyme Metabolism, Antioxidant and Immune Indicators
2.4. Histological Examination
2.5. DNA Extraction and PCR Amplification
2.6. Bacterial Data Processing and Function Predication
2.7. Statistical Analysis
3. Results
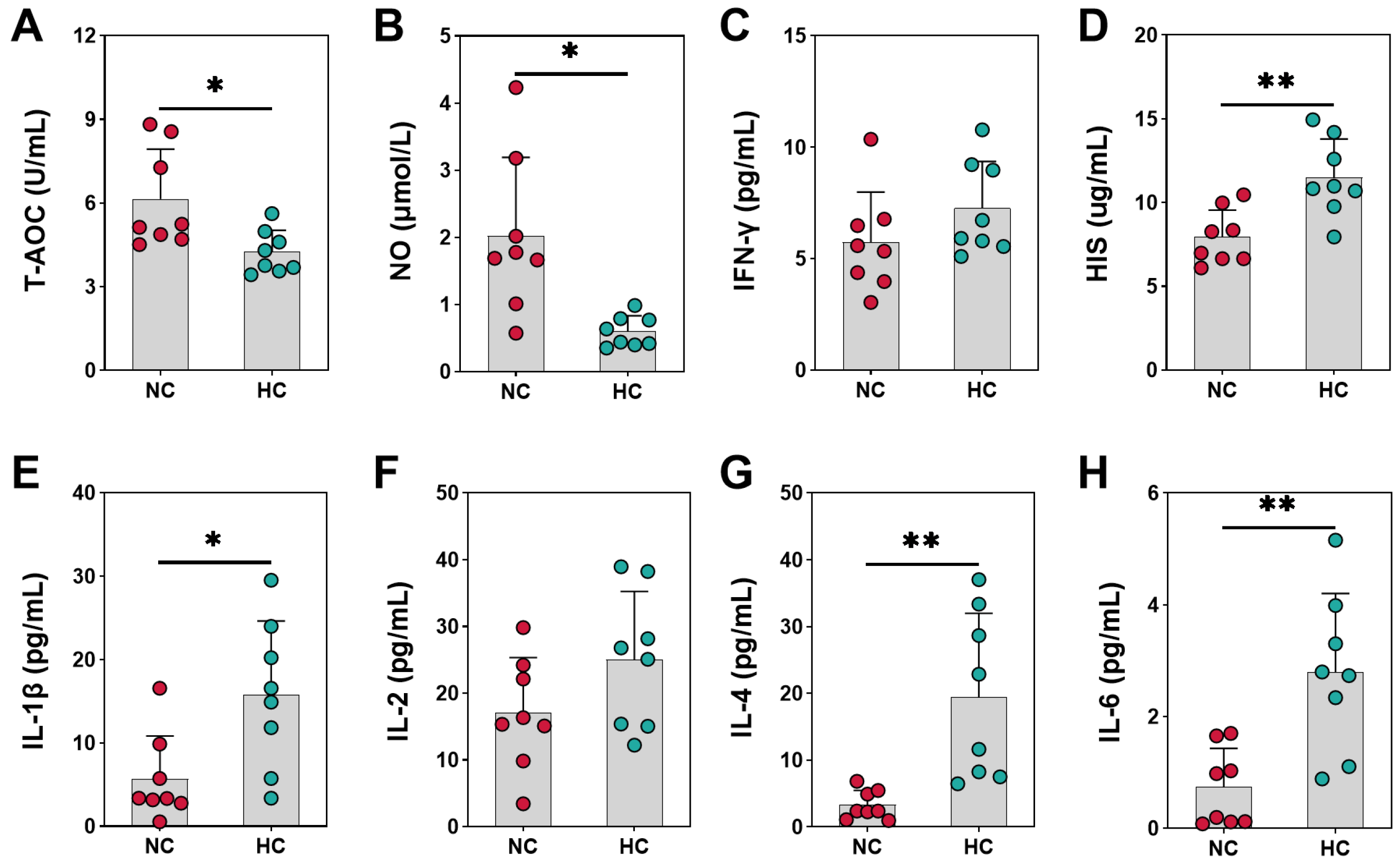
3.1. Antioxidant and Immune Indicators of Growing Goats Fed Different Diets
3.2. The Enzyme Metabolism Indicators of Growing Goats Fed Different Diets
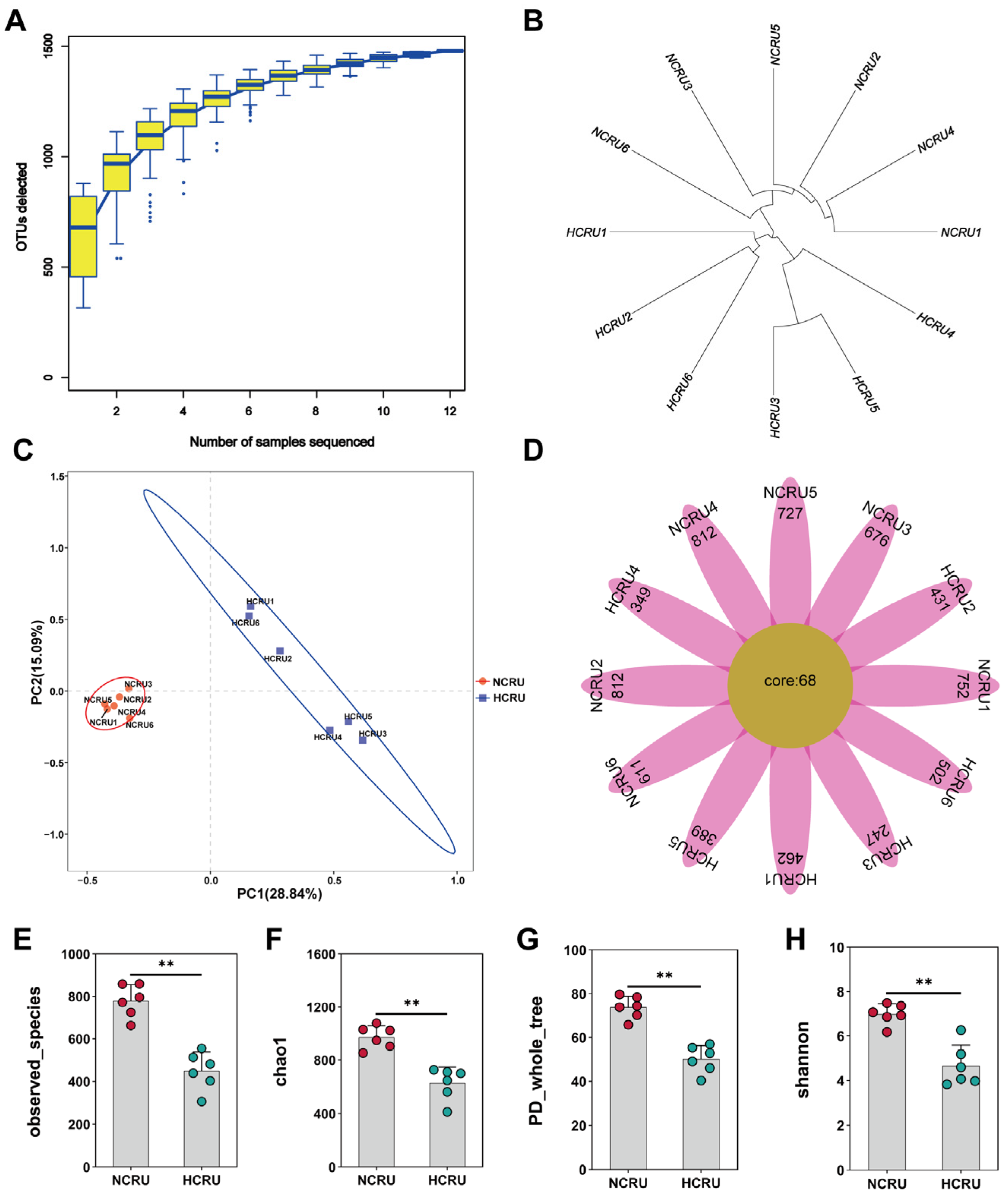
3.3. Rumen Microflora of Growing Goats Fed Different Diets
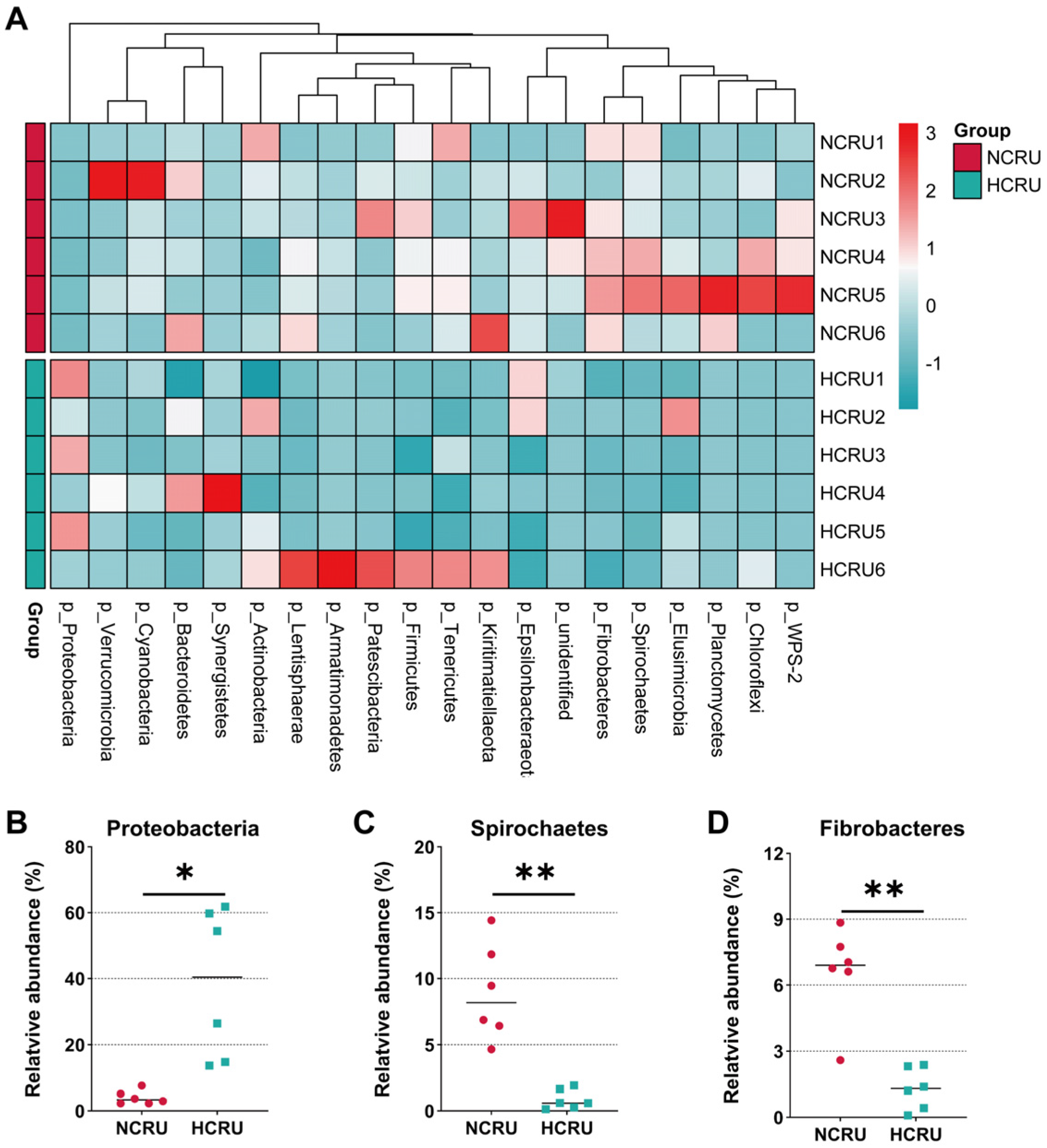
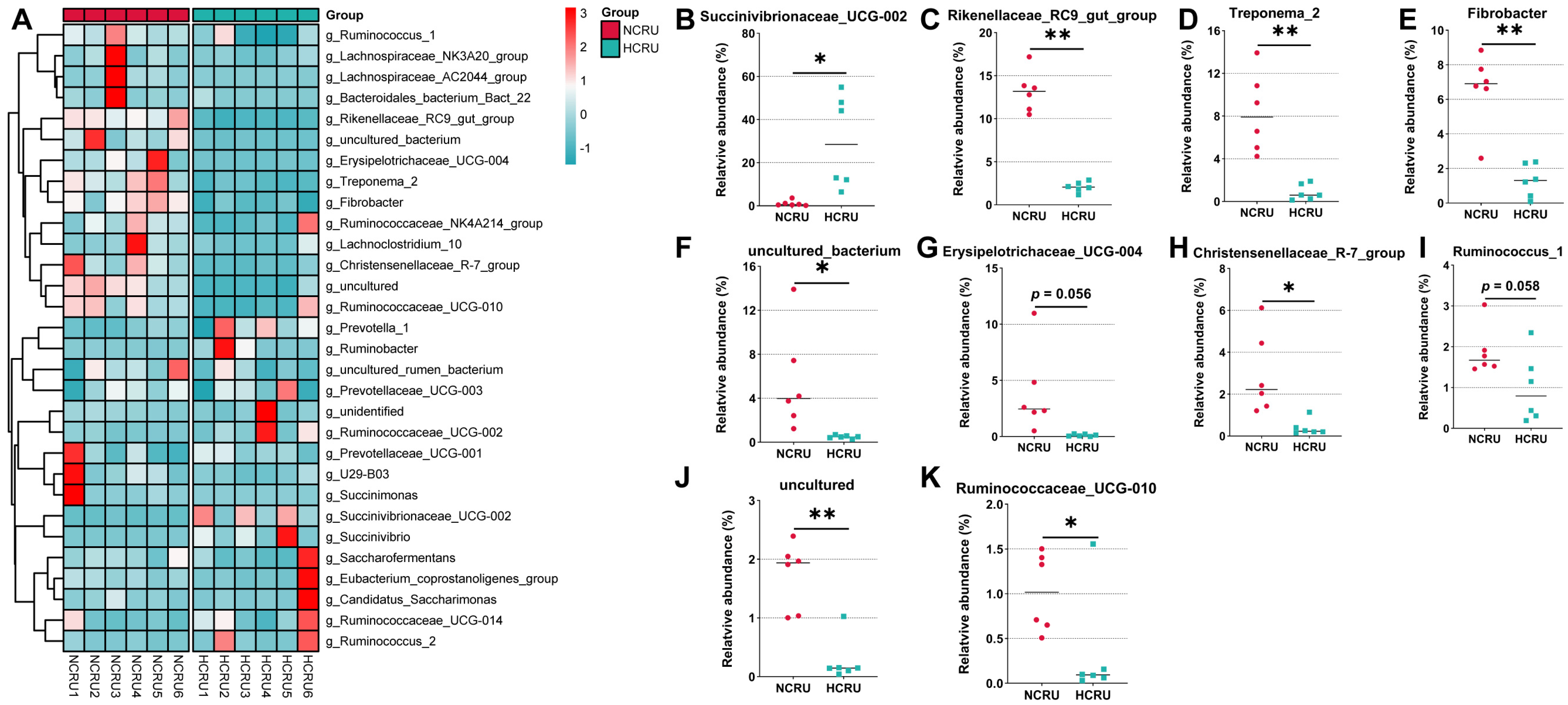
3.4. Rumen Bacterial Community Structure of Growing Goats Fed Different Diets
3.5. Function Prediction of Rumen Bacterial Community Using PICRUSt
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gozho, G.N.; Plaizier, J.C.; Krause, D.O.; Kennedy, A.D.; Wittenberg, K.M. Subacute ruminal acidosis induces ruminal lipopolysaccharide endotoxin release and triggers an inflammatory response. J. Dairy Sci. 2005, 88, 1399–1403. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Oba, M. Characteristics of dairy cows with a greater or lower risk of subacute ruminal acidosis: Volatile fatty acid absorption, rumen digestion, and expression of genes in rumen epithelial cells. J. Dairy Sci. 2016, 99, 8733–8745. [Google Scholar] [CrossRef] [PubMed]
- Abdela, N. Sub-acute Ruminal Acidosis (SARA) and its Consequence in Dairy Cattle: A Review of Past and Recent Research at Global Prospective. Achiev. Life Sci. 2016, 10, 187–196. [Google Scholar] [CrossRef]
- Kleen, J.L.; Hooijer, G.A.; Rehage, J.; Noordhuizen, J.P. Subacute ruminal acidosis (SARA): A review. J. Vet. Med. A Physiol. Pathol. Clin. Med. 2003, 50, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Bearson, S.; Bearson, B.; Foster, J.W. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 1997, 147, 173–180. [Google Scholar] [CrossRef]
- Guan, N.; Liu, L. Microbial response to acid stress: Mechanisms and applications. Appl. Microbiol. Biotechnol. 2020, 104, 51–65. [Google Scholar] [CrossRef]
- Monteiro, H.F.; Faciola, A.P. Ruminal acidosis, bacterial changes, and lipopolysaccharides. J. Anim. Sci. 2020, 98, skaa248. [Google Scholar] [CrossRef]
- Guan, N.Z.; Li, J.H.; Shin, H.D.; Du, G.C.; Chen, J.; Liu, L. Microbial response to environmental stresses: From fundamental mechanisms to practical applications. Appl. Microbiol. Biotechnol. 2017, 101, 3991–4008. [Google Scholar] [CrossRef]
- Mertens, D.R. Creating a system for meeting the fiber requirements of dairy cows. J. Dairy Sci. 1997, 80, 1463–1481. [Google Scholar] [CrossRef]
- Li, C.; Beauchemin, K.A.; Yang, W. Feeding diets varying in forage proportion and particle length to lactating dairy cows: I. Effects on ruminal pH and fermentation, microbial protein synthesis, digestibility, and milk production. J. Dairy Sci. 2020, 103, 4340–4354. [Google Scholar] [CrossRef]
- Yang, W.Z.; Beauchemin, K.A. Increasing physically effective fiber content of dairy cow diets through forage proportion versus forage chop length: Chewing and ruminal pH. J. Dairy Sci. 2009, 92, 1603–1615. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhu, R.; Yang, H.; Lu, Q.; Wang, W.; Song, L.; Sun, X.; Zhang, G.; Li, S.; Yang, J.; et al. Assessing the Impact of Data Preprocessing on Analyzing Next Generation Sequencing Data. Front. Bioeng. Biotechnol. 2020, 8, 817. [Google Scholar] [CrossRef]
- Wang, K.; Ma, J.; Li, Y.; Han, Q.; Yin, Z.; Zhou, M.; Luo, M.; Chen, J.; Xia, S. Effects of essential oil extracted from Artemisia argyi leaf on lipid metabolism and gut microbiota in high-fat diet-fed mice. Front. Nutr. 2022, 9, 1024722. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, K.J.; Lin, S.Z.; Zhang, Z.D.; Cheng, M.; Hu, S.S.; Hu, H.J.; Xiang, J.; Chen, F.M.; Li, G.H.; et al. Comparison of the Effects between Tannins Extracted from Different Natural Plants on Growth Performance, Antioxidant Capacity, Immunity, and Intestinal Flora of Broiler Chickens. Antioxidants 2023, 12, 441. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.M.; Wang, Y.S.; Wang, K.J.; Chen, J.Y.; Jin, K.; Peng, K.Q.; Chen, X.; Liu, Z.M.; Ouyang, J.; Wang, Y.; et al. Effects of Litsea cubeba essential oil on growth performance, blood antioxidation, immune function, apparent digestibility of nutrients, and fecal microflora of pigs. Front. Pharmacol. 2023, 14, 1166022. [Google Scholar] [CrossRef]
- Zhang, R.Y.; Ye, H.M.; Liu, J.H.; Mao, S.Y. High-grain diets altered rumen fermentation and epithelial bacterial community and resulted in rumen epithelial injuries of goats. Appl. Microbiol. Biotechnol. 2017, 101, 6981–6992. [Google Scholar] [CrossRef]
- Wang, K.; Ren, A.; Zheng, M.; Jiao, J.; Yan, Q.; Zhou, C.; Tan, Z. Diet with a High Proportion of Rice Alters Profiles and Potential Function of Digesta-Associated Microbiota in the Ileum of Goats. Animals 2020, 10, 1261. [Google Scholar] [CrossRef]
- Wang, K.J.; Yan, Q.X.; Ren, A.; Zheng, M.L.; Zhang, P.H.; Tan, Z.L.; Zhou, C.S. Novel Linkages between Bacterial Composition of Hindgut and Host Metabolic Responses to SARA Induced by High-Paddy Diet in Young Goats. Front. Vet. Sci. 2022, 8, 791482. [Google Scholar] [CrossRef]
- Wu, F.; Guo, X.; Zhang, J.; Zhang, M.; Ou, Z.; Peng, Y. Phascolarctobacterium faecium abundant colonization in human gastrointestinal tract. Exp. Ther. Med. 2017, 14, 3122–3126. [Google Scholar] [CrossRef]
- Wang, H.; Hu, C.; Cheng, C.; Cui, J.; Ji, Y.; Hao, X.; Li, Q.; Ren, W.; Deng, B.; Yin, Y.; et al. Unraveling the association of fecal microbiota and oxidative stress with stillbirth rate of sows. Theriogenology 2019, 136, 131–137. [Google Scholar] [CrossRef]
- Yin, L.; Li, J.; Wang, H.; Yi, Z.; Wang, L.; Zhang, S.; Li, X.; Wang, Q.; Li, J.; Yang, H.; et al. Effects of vitamin B6 on the growth performance, intestinal morphology, and gene expression in weaned piglets that are fed a low-protein diet. J. Anim. Sci. 2020, 98, skaa022. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.J.; Zheng, M.L.; Ren, A.; Zhou, C.S.; Yan, Q.X.; Tan, Z.L.; Zhang, P.; Yi, K. Effects of high rice diet on growth performance, nutrients apparent digestibility, nitrogen metabolism, blood parameters and rumen fermentation in growing goats. Kafkas Univ. Vet. Fak. Derg. 2019, 25, 749–755. [Google Scholar] [CrossRef]
- Wang, K.J.; Peng, X.M.; Yang, A.Q.; Huang, Y.Q.; Tan, Y.X.; Qian, Y.J.; Lv, F.F.; Si, H.B. Effects of Diets with Different Protein Levels on Lipid Metabolism and Gut Microbes in the Host of Different Genders. Front. Nutr. 2022, 9, 940217. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.J.; Peng, X.M.; Lv, F.F.; Zheng, M.L.; Long, D.L.; Mao, H.X.; Si, H.B.; Zhang, P.H. Microbiome-Metabolites Analysis Reveals Unhealthy Alterations in the Gut Microbiota but Improved Meat Quality with a High-Rice Diet Challenge in a Small Ruminant Model. Animals 2021, 11, 2306. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.M.; He, J.Y.; Wang, X.; Lv, T.; Liu, C.J.; Liao, L.P.; Li, Z.B.; Zhou, J.; He, B.S.; Qiu, H.J.; et al. Effect of Dietary Ramie Powder at Various Levels on the Growth Performance, Meat Quality, Serum Biochemical Indices and Antioxidative Capacity of Yanling White Geese. Animals 2022, 12, 2045. [Google Scholar] [CrossRef]
- Ma, W.J.; Wei, S.S.; Peng, W.J.; Sun, T.L.; Huang, J.H.; Yu, R.; Zhang, B.K.; Li, W.Q. Antioxidant Effect of Polygonatum sibiricum Polysaccharides in D-Galactose-Induced Heart Aging Mice. Biomed. Res. Int. 2021, 2021, 6688855. [Google Scholar] [CrossRef]
- Chen, J.Y.; Chen, F.M.; Peng, S.M.; Ou, Y.J.; He, B.S.; Li, Y.H.; Lin, Q. Effects of Artemisia argyi Powder on Egg Quality, Antioxidant Capacity, and Intestinal Development of Roman Laying Hens. Front. Physiol. 2022, 13, 902568. [Google Scholar] [CrossRef]
- Zhang, H.; Xue, Y.; Xie, W.; Wang, Y.; Ma, N.; Chang, G.; Shen, X. Subacute ruminal acidosis downregulates FOXA2, changes oxidative status, and induces autophagy in the livers of dairy cows fed a high-concentrate diet. J. Dairy Sci. 2023, 106, 2007–2018. [Google Scholar] [CrossRef]
- Sato, S. Subacute ruminal acidosis (SARA) challenge, ruminal condition and cellular immunity in cattle. Jpn. J. Vet. Res. 2015, 63, S25–S36. [Google Scholar]
- Chen, Y.; Xiang, J.; Wang, Z.; Xiao, Y.; Zhang, D.; Chen, X.; Li, H.; Liu, M.; Zhang, Q. Associations of Bone Mineral Density with Lean Mass, Fat Mass, and Dietary Patterns in Postmenopausal Chinese Women: A 2-Year Prospective Study. PLoS ONE 2015, 10, e0137097. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yin, L.; Wang, L.; Li, J.; Huang, P.; Yang, H.; Yin, Y. Effects of vitamin B6 on growth, diarrhea rate, intestinal morphology, function, and inflammatory factors expression in a high-protein diet fed to weaned piglets. J. Anim. Sci. 2019, 97, 4865–4874. [Google Scholar] [CrossRef]
- Zhu, J.F.; Paul, W.E. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol. Rev. 2010, 238, 247–262. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Gao, R.L.; Xiong, Y.K.; Huang, Q.C.; Xu, M. Antitumor and immunomodulatory effects of a water-soluble polysaccharide from Lilii Bulbus in mice. Carbohydr. Polym. 2014, 102, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Alexander, A.F.; Kelsey, I.; Forbes, H.; Miller-Jensen, K. Single-cell secretion analysis reveals a dual role for IL-10 in restraining and resolving the TLR4-induced inflammatory response. Cell Rep. 2021, 36, 109728. [Google Scholar] [CrossRef]
- He, Z.; Tan, Z.; Sun, Z.; Beauchemin, K.A.; Tang, S.; Zhou, C.; Han, X.; Wang, M.; Wu, D. Unchanged interleukin 6 level of protein and energy restricted goats during late gestation: The role of elevated blood nitric oxide. J. Endocrinol. 2012, 213, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Trabi, E.B.; Seddik, H.E.; Xie, F.; Wang, X.F.; Liu, J.H.; Mao, S.Y. Effect of pelleted high-grain total mixed ration on rumen morphology, epithelium-associated microbiota and gene expression of proinflammatory cytokines and tight junction proteins in Hu sheep. Anim. Feed Sci. Technol. 2020, 263, 114453. [Google Scholar] [CrossRef]
- Connelly, L.; Palacios-Callender, M.; Ameixa, C.; Moncada, S.; Hobbs, A.J. Biphasic regulation of NF-kappa B activity underlies the pro- and anti-inflammatory actions of nitric oxide. J. Immunol. 2001, 166, 3873–3881. [Google Scholar] [CrossRef]
- Deng, Q.; Shao, Y.; Wang, Q.; Li, J.; Li, Y.; Ding, X.; Huang, P.; Yin, J.; Yang, H.; Yin, Y. Effects and interaction of dietary electrolyte balance and citric acid on growth performance, intestinal histomorphology, digestive enzyme activity and nutrient transporters expression of weaned piglets. J. Anim. Physiol. Anim. Nutr. 2021, 105, 272–285. [Google Scholar] [CrossRef]
- Plaizier, J.C.; Khafipour, E.; Li, S.; Gozho, G.N.; Krause, D.O. Subacute ruminal acidosis (SARA), endotoxins and health consequences. Anim. Feed Sci. Technol. 2012, 172, 9–21. [Google Scholar] [CrossRef]
- Wang, X.B.; Ogawa, T.; Suda, S.; Taniguchi, K.; Uike, H.; Kumagai, H.; Mitani, K. Effects of nutritional level on digestive enzyme activities in the pancreas and small intestine of calves slaughtered at same body weight. Asian-Australas J. Anim. Sci. 1998, 11, 375–380. [Google Scholar] [CrossRef]
- Owsley, W.F.; Orr, D.E.; Tribble, L.F. Effects of age and diet on the development of the pancreas and the synthesis and secretion of pancreatic enzymes in the young pig. J. Anim. Sci. 1986, 63, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.Z.; Ke, Y.T.; Shang, Y.Z.; Wang, J.H.; Wen, Y.Q.; Wei, J.T. Effects of different crude protein levels of formulated pellet diets on growth performance and blood biochemical parameters of Matou goats. Chin. J. Anim. Sci. 2019, 55, 108–110, 125. [Google Scholar]
- Wang, Z.W.; Mou, X.L.; Yang, G.W.; Li, J.K.; Liu, D.S. Effects of dietary nutritional levels on calcium, phosphorus concentrations and ALK activity in serum and calcium, phosphorus contents in tibia for geese. China Poult. 2009, 31, 16–20. [Google Scholar]
- Zhang, C.L.; Meng, H.J.; Zhu, S.X.; Wu, L.; Liu, Z. Effect of alkaline phosphatase on intestinal health of weaning piglets. Swine Prod. 2017, 6, 13–16. [Google Scholar]
- DiLorenzo, N.; Smith, D.R.; Quinn, M.J.; May, M.L.; Ponce, C.H.; Steinberg, W.; Engstrom, M.A.; Galyean, M.L. Effects of grain processing and supplementation with exogenous amylase on nutrient digestibility in feedlot diets. Livest. Sci. 2011, 137, 178–184. [Google Scholar] [CrossRef]
- McCarthy, M.M.; Engstrom, M.A.; Azem, E.; Gressley, T.F. The effect of an exogenous amylase on performance and total-tract digestibility in lactating dairy cows fed a high-byproduct diet. J. Dairy Sci. 2013, 96, 3075–3084. [Google Scholar] [CrossRef]
- Chen, C.; Wang, Z.; Li, J.; Li, Y.; Huang, P.; Ding, X.; Yin, J.; He, S.; Yang, H.; Yin, Y. Dietary vitamin E affects small intestinal histomorphology, digestive enzyme activity, and the expression of nutrient transporters by inhibiting proliferation of intestinal epithelial cells within jejunum in weaned piglets. J. Anim. Sci. 2019, 97, 1212–1221. [Google Scholar] [CrossRef]
- Luo, W.W.; Tian, L.; Tan, B.; Shen, Z.H.; Xiao, M.W.; Wu, S.; Meng, X.R.; Wu, X.; Wang, X.Y. Update: Innate Lymphoid Cells in Inflammatory Bowel Disease. Dig. Dis. Sci. 2022, 67, 56–66. [Google Scholar] [CrossRef]
- Wu, J.; He, C.; Bu, J.; Luo, Y.; Yang, S.; Ye, C.; Yu, S.; He, B.; Yin, Y.; Yang, X. Betaine attenuates LPS-induced downregulation of Occludin and Claudin-1 and restores intestinal barrier function. BMC Vet. Res. 2020, 16, 75. [Google Scholar] [CrossRef]
- Rashid, S.; Irshadullah, M. Evaluation of antioxidant and oxidant status of goats (Capra aegagrus hircus) naturally infected with Haemonchus contortus. J. Helminthol. 2020, 94, e36. [Google Scholar] [CrossRef] [PubMed]
- Maughan, H.; Wang, P.W.; Diaz Caballero, J.; Fung, P.; Gong, Y.; Donaldson, S.L.; Yuan, L.; Keshavjee, S.; Zhang, Y.; Yau, Y.C.; et al. Analysis of the cystic fibrosis lung microbiota via serial Illumina sequencing of bacterial 16S rRNA hypervariable regions. PLoS ONE 2012, 7, e45791. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.T.; Peng, X.; Deng, G.H.; Sheng, H.F.; Wang, Y.; Zhou, H.W.; Tam, N.F. Illumina sequencing of 16S rRNA tag revealed spatial variations of bacterial communities in a mangrove wetland. Microb. Ecol. 2013, 66, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Nagata, R.; Ohkubo, A.; Ohtani, N.; Kushibiki, S.; Ichijo, T.; Sato, S. Changes in ruminal and reticular pH and bacterial communities in Holstein cattle fed a high-grain diet. BMC Vet. Res. 2018, 14, 310. [Google Scholar] [CrossRef] [PubMed]
- Ji, F.J.; Wang, L.X.; Yang, H.S.; Hu, A.; Yin, Y.L. Review: The roles and functions of glutamine on intestinal health and performance of weaning pigs. Animal 2019, 13, 2727–2735. [Google Scholar] [CrossRef]
- Wang, K.; Zhou, M.; Gong, X.; Zhou, Y.; Chen, J.; Ma, J.; Zhang, P. Starch–protein interaction effects on lipid metabolism and gut microbes in host. Front. Nutr. 2022, 9, 1018026. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, S.; Zhao, H.; Li, L.; Li, Y.; Liu, M.; Jiang, L. Integrated multi-omics analysis reveals the positive leverage of citrus flavonoids on hindgut microbiota and host homeostasis by modulating sphingolipid metabolism in mid-lactation dairy cows consuming a high-starch diet. Microbiome 2023, 11, 236. [Google Scholar] [CrossRef]
- Akkermans, S.; Logist, F.; Van Impe, J.F. An interaction model for the combined effect of temperature, pH and water activity on the growth rate of E. coli K12. Food Res. Int. 2018, 106, 1123–1131. [Google Scholar] [CrossRef]
- Ren, X.D.; Chen, X.S.; Tang, L.; Zeng, X.; Wang, L.; Mao, Z.G. Physiological mechanism of the overproduction of epsilon-poly-L-lysine by acidic pH shock in fed-batch fermentation. Bioprocess Biosyst. Eng. 2015, 38, 2085–2094. [Google Scholar] [CrossRef]
- Li, Y.Q.; Xi, Y.M.; Zeng, H.F.; Zhang, L.; Chen, M.J.; Han, Z.Y. Diversity Changes of Bacterial Community in Rumen of Dairy Cows in Vitro Fermentation Analyzed by 16S rRNA Sequencing Technology. Chin. J. Anim. Nutr. 2018, 30, 4059–4070. [Google Scholar] [CrossRef]
- Nagaraja, T.G.; Titgemeyer, E.C. Ruminal acidosis in beef cattle: The current microbiological and nutritional outlook. J. Dairy Sci. 2007, 90 (Suppl. 1), E17–E38. [Google Scholar] [CrossRef]
- Chiquette, J.; Allison, M.J.; Rasmussen, M.A. Prevotella bryantii 25A used as a probiotic in early-lactation dairy cows: Effect on ruminal fermentation characteristics, milk production, and milk composition. J. Dairy Sci. 2008, 91, 3536–3543. [Google Scholar] [CrossRef] [PubMed]
- Bekele, A.Z.; Koike, S.; Kobayashi, Y. Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rRNA gene-based analysis. FEMS Microbiol. Lett. 2010, 305, 49–57. [Google Scholar] [CrossRef]
- Bekele, A.Z.; Koike, S.; Kobayashi, Y. Phylogenetic diversity and dietary association of rumen Treponema revealed using group-specific 16S rRNA gene-based analysis. FEMS Microbiol. Lett. 2011, 316, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Ransom-Jones, E.; Jones, D.L.; McCarthy, A.J.; McDonald, J.E. The Fibrobacteres: An Important Phylum of Cellulose-Degrading Bacteria. Microb. Ecol. 2012, 63, 267–281. [Google Scholar] [CrossRef] [PubMed]
- Plaizier, J.C.; Li, S.C.; Danscher, A.M.; Derakshani, H.; Andersen, P.H.; Khafipour, E. Changes in Microbiota in Rumen Digesta and Feces Due to a Grain-Based Subacute Ruminal Acidosis (SARA) Challenge. Microb. Ecol. 2017, 74, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Mickdam, E.; Khiaosa-ard, R.; Metzler-Zebeli, B.U.; Klevenhusen, F.; Chizzola, R.; Zebeli, Q. Rumen microbial abundance and fermentation profile during severe subacute ruminal acidosis and its modulation by plant derived alkaloids in vitro. Anaerobe 2016, 39, 4–13. [Google Scholar] [CrossRef]
- Wang, X.; Martin, G.B.; Wen, Q.; Liu, S.; Li, Y.; Shi, B.; Guo, X.; Zhao, Y.; Guo, Y.; Yan, S. Palm oil protects alpha-linolenic acid from rumen biohydrogenation and muscle oxidation in cashmere goat kids. J. Anim. Sci. Biotechnol. 2020, 11, 100. [Google Scholar] [CrossRef]
- Su, X.L.; Tian, Q.; Zhang, J.; Yuan, X.Z.; Shi, X.S.; Guo, R.B.; Qiu, Y.L. Acetobacteroides hydrogenigenes gen. nov., sp. nov., an anaerobic hydrogen-producing bacterium in the family Rikenellaceae isolated from a reed swamp. Int. J. Syst. Evol. Microbiol. 2014, 64, 2986–2991. [Google Scholar] [CrossRef]
- Zhao, Y.; Xie, B.; Gao, J.; Zhao, G. Dietary supplementation with sodium sulfate improves rumen fermentation, fiber digestibility, and the plasma metabolome through modulation of rumen bacterial communities in steers. Appl. Environ. Microbiol. 2020, 86, e01412-20. [Google Scholar] [CrossRef]
- Seshadri, R.; Leahy, S.C.; Attwood, G.T.; Teh, K.H.; Lambie, S.C.; Cookson, A.L.; Eloe-Fadrosh, E.A.; Pavlopoulos, G.A.; Hadjithomas, M.; Varghese, N.J.; et al. Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection. Nat. Biotechnol. 2018, 36, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Holman, D.B.; Gzyl, K.E. A meta-analysis of the bovine gastrointestinal tract microbiota. FEMS Microbiol. Ecol. 2019, 95, fiz072. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Su, X.; Li, J.; Yang, Y.; Wang, P.; Yan, F.; Yao, J.; Wu, S. Real-time monitoring of ruminal microbiota reveals their roles in dairy goats during subacute ruminal acidosis. NPJ Biofilms Microbiomes 2021, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; He, Y.; Xiang, K.; Zhao, C.; He, Z.; Qiu, M.; Hu, X.; Zhang, N. The role of rumen microbiota and its metabolites in subacute ruminal acidosis (SARA)-induced inflammatory diseases of ruminants. Microorganisms 2022, 10, 1495. [Google Scholar] [CrossRef]
- Plaizier, J.C.; Mulligan, F.J.; Neville, E.W.; Guan, L.L.; Steele, M.A.; Penner, G.B. Invited review: Effect of subacute ruminal acidosis on gut health of dairy cows. J. Dairy Sci. 2022, 105, 7141–7160. [Google Scholar] [CrossRef]


| Item | NC 1 | HC 2 |
|---|---|---|
| Ingredients composition (%) | ||
| Forage | ||
| Rice straw | 45.0 | 10.0 |
| Concentrate | ||
| Rice with shell | 33.2 | 54.3 |
| Soybean meal | 9.60 | 15.7 |
| Wheat bran | 6.00 | 9.80 |
| Fat powder | 3.20 | 5.20 |
| Calcium carbonate | 0.50 | 0.80 |
| Calcium bicarbonate | 1.10 | 1.80 |
| Sodium chloride | 0.60 | 1.00 |
| Premix 3 | 1.00 | 1.40 |
| Nutrient levels 4, % of DM | ||
| Crude protein | 13.5 | 17.6 |
| Crude ash | 9.34 | 9.12 |
| Crude fat | 4.18 | 6.01 |
| Neutral detergent fiber | 49.8 | 38.4 |
| Acid detergent fiber | 36.5 | 9.51 |
| Starch | 26.57 | 38.57 |
| Level 2 | Level 3 | Pathway ID | NC 1 | HC 2 | p-Value |
|---|---|---|---|---|---|
| Amino acid metabolism | Histidine metabolism | ko00340 | 0.69 ± 0.03 | 0.65 ± 0.05 | 0.081 |
| Lysine degradation | ko00310 | 0.16 ± 0.02 | 0.13 ± 0.02 | 0.030 | |
| Tryptophan metabolism | ko00380 | 0.19 ± 0.01 | 0.16 ± 0.02 | 0.003 | |
| Valine, leucine and isoleucine degradation | ko00280 | 0.29 ± 0.02 | 0.25 ± 0.02 | 0.006 | |
| Carbohydrate metabolism | Citrate cycle (TCA cycle) | ko00020 | 0.73 ± 0.05 | 0.80 ± 0.04 | 0.028 |
| Metabolism of other amino acids | Glutathione metabolism | ko00480 | 0.17 ± 0.004 | 0.24 ± 0.05 | 0.008 |
| Selenocompound metabolism | ko00450 | 0.38 ± 0.01 | 0.34 ± 0.02 | 0.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, S.; Zheng, M.; Ren, A.; Mao, H.; Long, D.; Yang, L. Effects of High-Concentrate-Induced SARA on Antioxidant Capacity, Immune Levels and Rumen Microbiota and Function in Goats. Animals 2024, 14, 263. https://doi.org/10.3390/ani14020263
Fan S, Zheng M, Ren A, Mao H, Long D, Yang L. Effects of High-Concentrate-Induced SARA on Antioxidant Capacity, Immune Levels and Rumen Microbiota and Function in Goats. Animals. 2024; 14(2):263. https://doi.org/10.3390/ani14020263
Chicago/Turabian StyleFan, Siqin, Mengli Zheng, Ao Ren, Hongxiang Mao, Donglei Long, and Lingyuan Yang. 2024. "Effects of High-Concentrate-Induced SARA on Antioxidant Capacity, Immune Levels and Rumen Microbiota and Function in Goats" Animals 14, no. 2: 263. https://doi.org/10.3390/ani14020263
APA StyleFan, S., Zheng, M., Ren, A., Mao, H., Long, D., & Yang, L. (2024). Effects of High-Concentrate-Induced SARA on Antioxidant Capacity, Immune Levels and Rumen Microbiota and Function in Goats. Animals, 14(2), 263. https://doi.org/10.3390/ani14020263





