Risk of Sperm Disorders and Impaired Fertility in Frozen–Thawed Bull Semen: A Genome-Wide Association Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Sperm Evaluation
2.3. Genotyping Quality Control
2.4. Genome-Wide Association Analysis
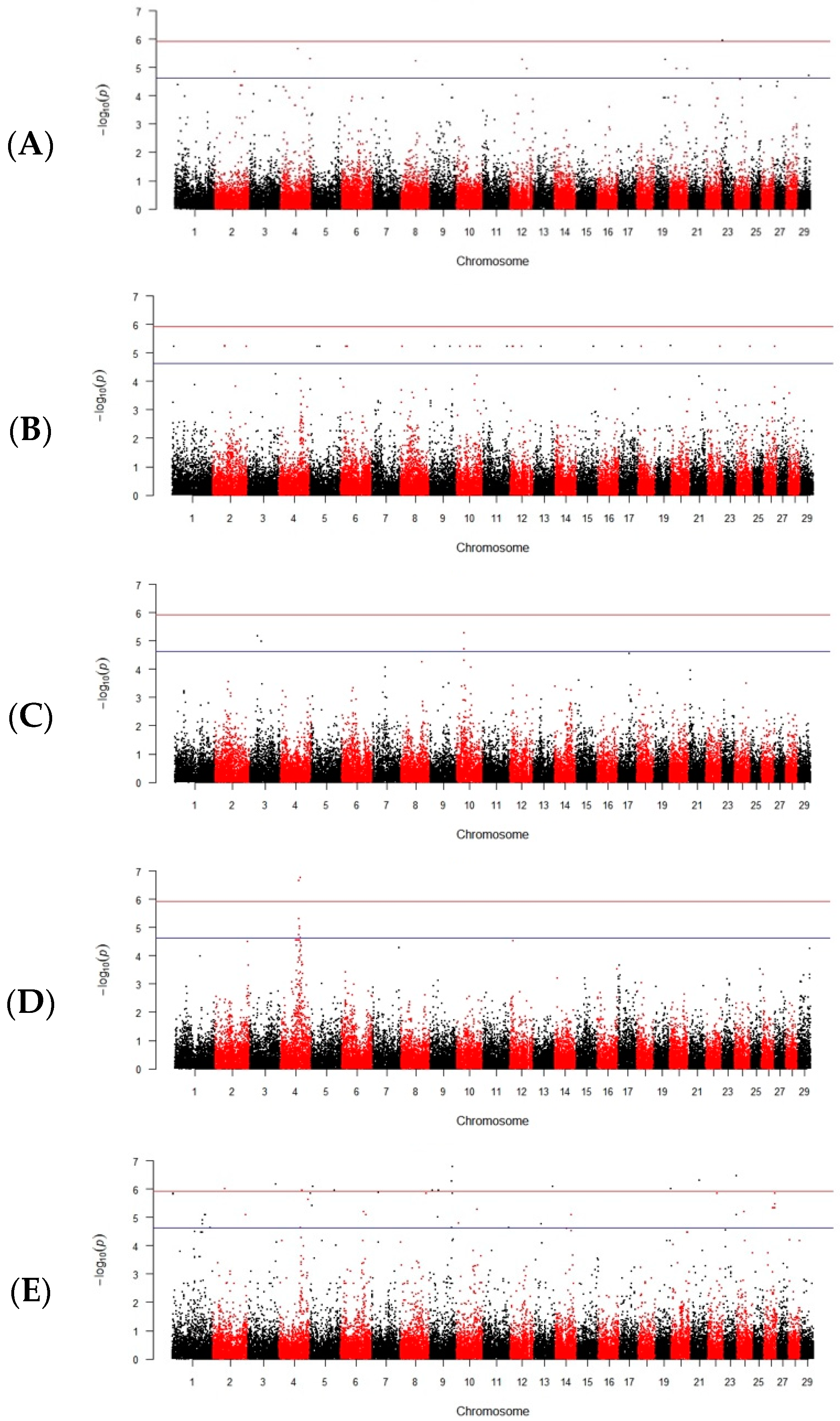
3. Results
3.1. Sperm Head Abnormalities
3.1.1. Absence of Acrosomes
3.1.2. Sperm Head Abnormalities
3.1.3. Swollen Acrosomes
3.1.4. Wrinkled Acrosomes
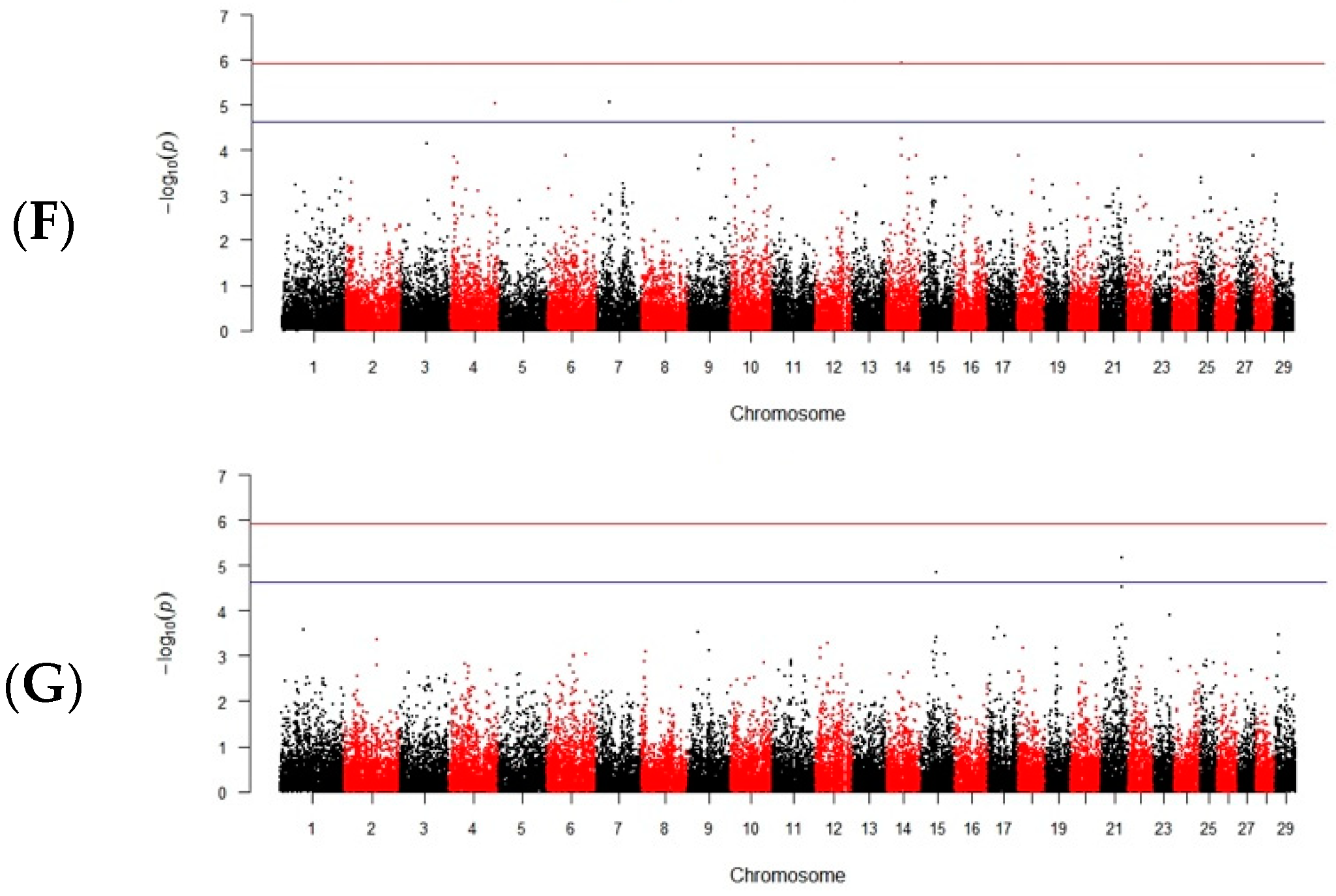
3.2. Damaged Tails and Cell Necks
3.3. Total and Progressive Motility
4. Discussion
4.1. Genomic Associations with Sperm Head Disorders after Freezing
4.1.1. Absence of Acrosomes
4.1.2. Sperm Head Abnormalities Following Cryopreservation
4.1.3. Swollen Acrosomes
4.1.4. Wrinkled Acrosomes
4.2. Damaged Tails and Cell Necks (Midpieces)
4.3. Total and Progressive Motility
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Marques, D.B.D.; Bastiaansen, J.W.M.; Broekhuijse, M.; Lopes, M.S.; Knol, E.F.; Harlizius, B.; Guimaraes, S.E.F.; Silva, F.F.; Lopes, P.S. Weighted single-step GWAS and gene network analysis reveal new candidate genes for semen traits in pigs. Genet. Sel. Evol. 2018, 50, 40. [Google Scholar] [CrossRef] [PubMed]
- Nikitkina, E.; Dementieva, N.; Shcherbakov, Y.; Musidray, A.; Krutikova, A.; Bogdanova, S.; Plemyashov, K. Search for genetic associations with semen morphology after cryopreservation in bulls. Anim. Reprod. Sci. 2022, 247, 107117. [Google Scholar] [CrossRef]
- Nikitkina, E.V.; Dementieva, N.V.; Shcherbakov, Y.S.; Atroshchenko, M.M.; Kudinov, A.A.; Samoylov, O.I.; Pozovnikova, M.V.; Dysin, A.P.; Krutikova, A.A.; Musidray, A.A.; et al. Genome-wide association study for frozen-thawed sperm motility in stallions across various horse breeds. Anim. Biosci. 2022, 35, 1827–1838. [Google Scholar] [CrossRef] [PubMed]
- Tagirov, M.T.; Artemenko, A.B.; Tereshchenko, A.V. Preservation of the poultry gene pool by cryoconservation. Sučasne Ptahìvnictvo [Modern Poult. Farming] 2007, 1, 3–6. (In Russian) [Google Scholar]
- Tagirov, M.T.; Artemenko, A.B.; Tereshchenko, A.V.; Nalivaiko, L.I. Requirements for the preservation of the gene pool of birds through cryopreservation. Ptakhivnytstvo [Poult. Farming] 2007, 59, 145–153. (In Russian) [Google Scholar]
- Foote, R.H. The history of artificial insemination: Selected notes and notables. J. Anim. Sci. 2010, 80 (Suppl. S2), 1–10. [Google Scholar] [CrossRef]
- Ball, B.A. Oxidative stress, osmotic stress and apoptosis: Impacts on sperm function and preservation in the horse. Anim. Reprod. Sci. 2008, 107, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Linnik, T.P.; Grischenko, V.I.; Artemenko, A.B.; Tereshchenko, A.V. Effect of osmoticity of cryoprotective medium on survival of cock sperm during cryopreservation. Probl. Cryobiol. 2000, 2, 86–93. Available online: https://books.google.co.uk/books?hl=en&lr=&id=dLbWIkFjLmEC&oi=fnd&pg=PA86 (accessed on 12 December 2023).
- Longobardi, V.; Kosior, M.A.; Pagano, N.; Fatone, G.; Staropoli, A.; Vassetti, A.; Vinale, F.; Campanile, G.; Gasparrini, B. Changes in bull semen metabolome in relation to cryopreservation and fertility. Animals 2020, 10, 1065. [Google Scholar] [CrossRef]
- Pitnick, S.; Hosken, D.J.; Birkhead, T.R. 3—Sperm morphological diversity. In Sperm Biology: An Evolutionary Perspective; Birkhead, T.R., Hosken, D.J., Pitnick, S., Eds.; Academic Press: Burlington, MA, USA, 2009; pp. 69–149. [Google Scholar] [CrossRef]
- Colenbrander, B.; Feitsma, H.; Grooten, H.J. Optimizing semen production for artificial insemination in swine. J. Reprod. Fertil. Suppl. 1993, 48, 207–215. Available online: https://www.biosciproceedings.org/bp/0014/bp0014cpr14.pdf (accessed on 12 December 2023). [CrossRef]
- Kruger, T.F.; Acosta, A.A.; Simmons, K.F.; Swanson, R.J.; Matta, J.F.; Oehninger, S. Predictive value of abnormal sperm morphology in in vitro fertilization. Fertil. Steril. 1988, 49, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Walters, A.H.; Eyestone, W.E.; Saacke, R.G.; Pearson, R.E.; Gwazdauskas, F.C. Sperm morphology and preparation method affect bovine embryonic development. J. Androl. 2004, 25, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Oehninger, S.; Franken, D.R.; Ombelet, W. Sperm functional tests. Fertil. Steril. 2014, 102, 1528–1533. [Google Scholar] [CrossRef] [PubMed]
- Raetsky, A.; Moiseyeva, I. Enzyme activity in rooster sperm. Ptitsevodstvo [Poult. Farming] 1992, 5, 9–10. (In Russian) [Google Scholar]
- Bianchi, E.; Wright, G.J. Find and fuse: Unsolved mysteries in sperm–egg recognition. PLoS Biol. 2020, 18, e3000953. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, V.R.; Ramesh, V.; Dewry, R.K.; Kumar, G.; Raval, K.; Patoliya, P. Implications of cryopreservation on structural and functional attributes of bovine spermatozoa: An overview. Andrologia 2021, 53, e14154. [Google Scholar] [CrossRef] [PubMed]
- Gravance, C.G.; Vishwnath, R.; Pitt, C.; Garner, L.; Casey, J. Effects of cryopreservation on bull sperm head morphometry. J. Androl. 1998, 19, 704–709. [Google Scholar] [CrossRef]
- Davies, R.; Jayasena, C.N.; Minhas, S. Chapter 24—Sperm quality evaluation and cryopreservation. In Management of Infertility; Academic Press; Elsevier: Cambridge, MA, USA, 2023; pp. 241–249. [Google Scholar] [CrossRef]
- Zhu, W.J.; Liu, X.G. Cryodamage to plasma membrane integrity in head and tail regions of human sperm. Asian J. Androl. 2000, 2, 135–138. Available online: http://www.corc.ac.cn/handle/1471x/3329037?mode=full (accessed on 12 December 2023).
- Tereshchenko, A.V. Damage to Rooster Sperm During Freezing. In Contribution of Young Scientists of Ukraine to the Intensification of Agricultural Production: Abstracts of the 2nd Republican Scientific and Industrial Conference of Young Scientists and Specialists, Kharkov, Ukraine, 24–26 September 1986; USSR: Kharkov, Ukraine, 1986; p. 160. (In Russian) [Google Scholar]
- Bychko, S.V.; Dunayeva, O.V.; Artemenko, O.B.; Tereschenko, O.V. Revealing of damages in poultry spermatozoa during low-temperature preservation. Probl. Cryobiol. 2005, 15, 272–275. Available online: http://cryo.org.ua/journal/index.php/probl-cryobiol-cryomed/article/view/1101 (accessed on 12 December 2023). (In Ukrainian).
- Gallo, A.; Esposito, M.C.; Tosti, E.; Boni, R. Sperm motility, oxidative status, and mitochondrial activity: Exploring correlation in different species. Antioxidants 2021, 10, 1131. [Google Scholar] [CrossRef]
- Torres, M.A.; Pedrosa, A.C.; Novais, F.J.; Alkmin, D.V.; Cooper, B.R.; Yasui, G.S.; Fukumasu, H.; Machaty, Z.; de Andrade, A.F.C. Metabolomic signature of spermatozoa established during holding time is responsible for differences in boar sperm freezability. Biol. Reprod. 2021, 106, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Gürler, H.; Malama, E.; Heppelmann, M.; Calisici, O.; Leiding, C.; Kastelic, J.P.; Bollwein, H. Effects of cryopreservation on sperm viability, synthesis of reactive oxygen species, and DNA damage of bovine sperm. Theriogenology 2016, 86, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Attia, S.; Katila, T.; Andersson, M. The effect of sperm morphology and sire fertility on calving rate of Finnish Ayrshire AI bulls. Reprod. Dom. Anim. 2016, 51, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Crisóstomo, L.; Videira, R.A.; Jarak, I.; Starčević, K.; Mašek, T.; Rato, L.; Raposo, J.F.; Batterham, R.L.; Oliveira, P.F.; Alves, M.G. Inherited metabolic memory of high-fat diet impairs testicular fatty acid content and sperm parameters. Mol. Nutr. Food Res. 2022, 66, 2100680. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.; Coutton, C.; Loeuillet, C.; Cazin, C.; Muroňová, J.; Boguenet, M.; Lambert, E.; Dhellemmes, M.; Chevalier, G.; Hograindleur, J.-P.; et al. Oligogenic heterozygous inheritance of sperm abnormalities in mouse. eLife 2022, 11, e75373. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef]
- Fowler, K.E.; Pong-Wong, R.; Bauer, J.; Clemente, E.J.; Reitter, C.P.; Affara, N.A.; Waite, S.; Walling, G.A.; Griffin, D.K. Genome wide analysis reveals single nucleotide polymorphisms associated with fatness and putative novel copy number variants in three pig breeds. BMC Genom. 2013, 14, 784. [Google Scholar] [CrossRef]
- Abril-Parreño, L.; Carthy, T.R.; Keogh, K.; Štiavnická, M.; O’Meara, C.; Lonergan, P.; Kenny, D.A.; Fair, S. Genome-wide association study reveals candidate markers related to field fertility and semen quality traits in Holstein-Friesian bulls. Animal 2023, 17, 100841. [Google Scholar] [CrossRef]
- Kamiński, S.; Hering, D.M.; Oleński, K.; Lecewicz, M.; Kordan, W. Genome-wide association study for sperm membrane integrity in frozen-thawed semen of Holstein-Friesian bulls. Anim. Reprod. Sci. 2016, 170, 135–140. [Google Scholar] [CrossRef]
- Ramirez-Diaz, J.; Cenadelli, S.; Bornaghi, V.; Bongioni, G.; Montedoro, S.M.; Achilli, A.; Capelli, C.; Rincon, J.C.; Milanesi, M.; Passamonti, M.M.; et al. Identification of genomic regions associated with total and progressive sperm motility in Italian Holstein bulls. J. Dairy Sci. 2023, 106, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, D.; Mortimer, S.T. Computer-Aided Sperm Analysis (CASA) of sperm motility and hyperactivation. In Spermatogenesis. Methods in Molecular Biology; Carrell, D., Aston, K., Eds.; Humana Press: Totowa, NJ, USA, 2013; Volume 927, pp. 77–87. [Google Scholar] [CrossRef]
- Matukumalli, L.K.; Lawley, C.T.; Schnabel, R.D.; Taylor, J.F.; Allan, M.F.; Heaton, M.P.; O’Connell, J.; Moore, S.S.; Smith, T.P.; Sonstegard, T.S.; et al. Development and characterization of a high density SNP genotyping assay for cattle. PLoS ONE 2009, 4, e5350. [Google Scholar] [CrossRef] [PubMed]
- Nikitkina, E.; Krutikova, A.; Musidray, A.; Plemyashov, K. Search for associations of FSHR, INHA, INHAB, PRL, TNP2 and SPEF2 genes polymorphisms with semen quality in Russian Holstein bulls (pilot study). Animals 2021, 11, 2882. [Google Scholar] [CrossRef]
- Illumina. BovineSNP50 v3 BeadChip. Ref. 370-2007-029-B. 2020, pp. 1–3. Available online: https://www.illumina.com/content/dam/illumina-marketing/documents/products/datasheets/datasheet_bovine_snp5O.pdf (accessed on 12 December 2023).
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Gao, X. Multiple testing corrections for imputed SNPs. Genet. Epidemiol. 2011, 35, 154–158. [Google Scholar] [CrossRef]
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- Li, Y.-F.; He, W.; Jha, K.N.; Klotz, K.; Kim, Y.-H.; Mandal, A.; Pulido, S.; Digilio, L.; Flickinger, C.J.; Herr, J.C. FSCB, a novel protein kinase A-phosphorylated calcium-binding protein, is a CABYR-binding partner involved in late steps of fibrous sheath biogenesis. J. Biol. Chem. 2007, 282, 34104–34119. [Google Scholar] [CrossRef]
- Perotti, D.; Doneda, L.; Radice, P. POU6F2 (POU domain, class 6, transcription factor). Atlas Genet. Cytogenet. Oncol. Haematol. 2005, 9, 544–549. [Google Scholar] [CrossRef]
- Cho, H.J.; Gurbuz, F.; Stamou, M.; Kotan, L.D.; Farmer, S.M.; Can, S.; Tompkins, M.F.; Mammadova, J.; Altincik, S.A.; Gokce, C.; et al. POU6F2 mutation identified in humans with pubertal failure shifts isoform formation and alters GnRH transcript expression. bioRxiv 2022, 2022, 10.12.511883. [Google Scholar] [CrossRef]
- Cho, H.J.; Gurbuz, F.; Stamou, M.; Kotan, L.D.; Farmer, S.M.; Can, S.; Tompkins, M.F.; Mammadova, J.; Altincik, S.A.; Gokce, C.; et al. POU6F2 mutation in humans with pubertal failure alters GnRH transcript expression. Front. Endocrinol. 2023, 14, 1203542. [Google Scholar] [CrossRef]
- Miao, Y.; Li, C.; Guo, J.; Wang, H.; Gong, L.; Xie, W.; Zhang, Y. Identification of a novel somatic mutation of POU6F2 by whole-genome sequencing in prolactinoma. Mol. Genet. Genomic Med. 2019, 7, e1022. [Google Scholar] [CrossRef] [PubMed]
- Stovall, D.W.; Shabanowitz, R.B. The effects of prolactin on human sperm capacitation and acrosome reaction. Fertil. Steril. 1991, 56, 960–966. [Google Scholar] [CrossRef] [PubMed]
- Chauvigné, F.; Ducat, C.; Ferré, A.; Hansen, T.; Carrascal, M.; Abián, J.; Finn, R.N.; Cerdà, J. A multiplier peroxiporin signal transduction pathway powers piscine spermatozoa. Proc. Natl. Acad. Sci. USA 2021, 118, e2019346118. [Google Scholar] [CrossRef] [PubMed]
- Edgar, A.J. The gene structure and expression of human ABHD1: Overlapping polyadenylation signal sequence with Sec12. BMC Genom. 2003, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Rolland, A.D.; Lareyre, J.; Goupil, A.; Montfort, J.; Ricordel, M.; Esquerre, D.; Hugot, K.; Houlgatte, R.; Chalmel, F.; Gac, F.L. Expression profiling of rainbow trout testis development identifies evolutionary conserved genes involved in spermatogenesis. BMC Genom. 2009, 10, 546. [Google Scholar] [CrossRef]
- Tu, Z.; Bayazit, M.B.; Liu, H.; Zhang, J.; Busayavalasa, K.; Risal, S.; Shao, J.; Satyanarayana, A.; Coppola, V.; Tessarollo, L.; et al. Speedy A–Cdk2 binding mediates initial telomere–nuclear envelope attachment during meiotic prophase I independent of Cdk2 activation. Proc. Natl. Acad. Sci. USA 2017, 114, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wang, H.; Hu, Z.; Sahlu, B.W.; Heng, N.; Gong, J.; Wang, H.; Zhu, H. Identification of spermatogenesis-related lncRNA in Holstein bull testis after sexual maturity based on transcriptome analysis. Anim. Reprod. Sci. 2022, 247, 107146. [Google Scholar] [CrossRef]
- Beato, M.; Klug, J. Steroid hormone receptors: An update. Hum. Reprod. Update 2000, 6, 225–236. [Google Scholar] [CrossRef]
- Seite, P.; Huebner, K.; Rousseau-Merck, M.F.; Berger, R.; Thiesen, H.J. Two human genes encoding zinc finger proteins, ZNF12 (KOX 3) and ZNF 26 (KOX 20), map to chromosomes 7p22-p21 and 12q24.33, respectively. Hum. Genet. 1991, 86, 585–590. [Google Scholar] [CrossRef]
- Rosati, M.; Rocchi, M.; Storlazzi, C.T.; Grimaldi, G. Assignment to chromosome 12q24.33, gene organization and splicing of the human KRAB/FPB containing zinc finger gene ZNF84. Cytogenet. Cell Genet. 2001, 94, 127–130. [Google Scholar] [CrossRef]
- Van Mil, S.W.C.; Klomp, L.W.J.; Bull, L.N.; Houwen, R.H.J. FIC1 disease: A spectrum of intrahepatic cholestatic disorders. Semin. Liver Dis. 2001, 21, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-R.; Liu, Y.-X. Regulation of spermatogonial stem cell self-renewal and spermatocyte meiosis by Sertoli cell signaling. Reproduction 2015, 149, R159–R167. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhuang, J.; Zhao, D.; Zhang, F.; Ma, J.; Xu, C. Cyclic stretch-induced the cytoskeleton rearrangement and gene expression of cytoskeletal regulators in human periodontal ligament cells. Acta Odontol. Scand. 2017, 75, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, M.; Hawse, J.R.; Rajamannan, N.M.; Ingle, J.N.; Spelsberg, T.C. Functional role of KLF10 in multiple disease processes. Bioactors 2010, 36, 8–18. [Google Scholar] [CrossRef]
- Galaviz-Hernandez, C.; Stagg, C.; de Ridder, G.; Tanaka, T.S.; Ko, M.S.; Schlessinger, D.; Nagaraja, R. Plac8 and Plac9, novel placental-enriched genes identified through microarray analysis. Gene 2003, 309, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Zalkin, H.; Dixon, J.E. De novo purine nucleotide biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 1992, 42, 259–287. [Google Scholar] [CrossRef]
- Zhang, L.-L.; Wang, J.-J.; Liu, Y.; Lu, X.-B.; Kuang, Y.; Wan, Y.-H.; Chen, Y.; Yan, H.-M.; Fei, J.; Wang, Z.-G. GPR26-deficient mice display increased anxiety- and depression-like behaviors accompanied by reduced phosphorylated cyclic AMP responsive element-binding protein level in central amygdala. Neuroscience 2011, 196, 203–214. [Google Scholar] [CrossRef]
- Tang, Y.; Zhou, J.; Hooi, S.C.; Jiang, Y.M.; Lu, G.D. Fatty acid activation in carcinogenesis and cancer development: Essential roles of long-chain acyl-CoA synthetases. Oncol. Lett. 2018, 16, 1390–1396. [Google Scholar] [CrossRef]
- Ducharme, N.A.; Ham, A.J.L.; Lapierre, L.A.; Goldenring, J.R. Rab11-FIP2 influences multiple components of the endosomal system in polarized MDCK cells. Cell Logist. 2011, 1, 57–68. [Google Scholar] [CrossRef]
- Muraleedharan, A.; Vanderperre, B. The endo-lysosomal system in Parkinson’s disease: Expanding the horizon. J. Mol. Biol. 2023, 435, 168140. [Google Scholar] [CrossRef]
- Qifeng, L. Proteomics Identification of Intracellular and Secreted Proteins Involved in Metastasis from a Pair of Isogenic Colorectal Cancer Cell Lines. Ph.D. Thesis, National University of Singapore, Singapore, 2014. Available online: https://scholarbank.nus.edu.sg/handle/10635/117494 (accessed on 12 December 2023).
- Heikkilä, M. Development of the Adreno-genital System: Female Sex Determination, Ovarian and Adrenal Gland Ontogeny Regulated by Wnt-4 in Mice. Academic Dissertation, University of Oulu, Oulu, Finland, 2002. Available online: https://urn.fi/URN:ISBN:951426844X (accessed on 12 December 2023).
- Kikuchi, A.; Yamamoto, H.; Sato, A.; Matsumoto, S. Wnt5a: Its signalling, functions and implication in diseases. Acta Physiol. 2012, 204, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Dawson, P.A. Sulfate in fetal development. Semin. Cell Dev. Biol. 2011, 22, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Doolittle, M.L. Dissecting the Genetic Influences on Osteoporosis Reveals Zbtb40 as a Novel Regulator of Osteoblast Function. Ph.D. Thesis, University of Rochester, Rochester, NY, USA, 2020. Available online: https://urresearch.rochester.edu/institutionalPublicationPublicView.action?institutionalItemVersionId=35298 (accessed on 12 December 2023).
- Li, R.; Wu, F.; Ruonala, R.; Sapkota, D.; Hu, Z.; Mu, X. Isl1 and Pou4f2 form a complex to regulate target genes in developing retinal ganglion cells. PLoS ONE 2014, 9, e92105. [Google Scholar] [CrossRef] [PubMed]
- Uronen, R.-L.E.; Huttunen, H.J. Genetic risk factors of Alzheimer’s disease and cell-to-cell transmission of Tau. J. Neurol. Neuromed. 2016, 1, 17–22. [Google Scholar] [CrossRef][Green Version]
- Malhotra, V. Genetic basis of sperm morphologic defects: Head defects and body and tail defects. In Genetics of Male Infertility: A Case-Based Guide for Clinicians; Arafa, M., Elbardisi, H., Majzoub, A., Agarwal, A., Eds.; Springer: Cham, Switzerland, 2020; pp. 121–136. [Google Scholar] [CrossRef]
- Yatsenko, A.N.; O’Neil, D.S.; Roy, A.; Arias-Mendoza, P.A.; Chen, R.; Murthy, L.J.; Lamb, D.J.; Matzuk, M.M. Association of mutations in the zona pellucida binding protein 1 (ZPBP1) gene with abnormal sperm head morphology in infertile men. Mol. Hum. Reprod. 2012, 18, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Duncker, B.P.; Chesnokov, I.N.; McConkey, B.J. The origin recognition complex protein family. Genome Biol. 2009, 10, 214. [Google Scholar] [CrossRef] [PubMed]
- Phillips, D.M.; Yanagimachi, R. Difference in the manner of association of acrosome-intact and acrosome-reacted hamster spermatozoa with egg microvilli as revealed by scanning electron microscopy: (hamster/sperm-egg association/scanning electron microscopy). Dev. Growth Differ. 1982, 24, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Asimakopoulos, B.; Tiptiri-Kourpeti, A.; Metallinou, C. IGF-I and NGFβ enhance in vitro progressive motility and vitality of human spermatozoa. Reprod. Med. Biol. 2021, 20, 361–367. [Google Scholar] [CrossRef]
- Selvaraju, S.; Reddy, I.J.; Nandi, S.; Rao, S.B.N.; Ravindra, J.P. Influence of IGF-I on buffalo (Bubalus bubalis) spermatozoa motility, membrane integrity, lipid peroxidation and fructose uptake in vitro. Anim. Reprod. Sci. 2009, 113, 60–70. [Google Scholar] [CrossRef]
- Cheng, Q.; Wang, W.; Liu, J.; Lv, Z.; Ji, W.; Yu, J.; Zhang, W.; Yang, Y. Elevated MPP6 expression correlates with an unfavorable prognosis, angiogenesis and immune evasion in hepatocellular carcinoma. Front. Immunol. 2023, 14, 1173848. [Google Scholar] [CrossRef]
- Terada, N.; Saitoh, Y.; Ohno, N.; Komada, M.; Yamauchi, J.; Ohno, S. Involvement of Src in the membrane skeletal complex, MPP6–4.1G, in Schmidt–Lanterman incisures of mouse myelinated nerve fibers in PNS. Histochem. Cell Biol. 2013, 140, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Gewaify, M.S.; Mansour, A.A.; Fayed, M.H.; Wakayama, T.; lseki, S. Immunohistochemical expression of membrane protein palmitoylated 6 (MPP6) in mice testis. Kafrelsheikh Vet. Med. J. 2014, 12, 179–194. Available online: https://web.p.ebscohost.com/abstract?direct=true&profile=ehost&scope=site&authtype=crawler&jrnl=16871456&AN=121993303&h=fYZWYLADOa3kzxtcwg2tGt5ivvEbKTLN3hwOVlTi6%2f3WOPKDuN7oUEbmPl7mQHqoZpEUePhSJ0Yzup11s7Jzow%3d%3d&crl=c (accessed on 12 December 2023).
- Schilders, G.; Raijmakers, R.; Raats, J.M.; Pruijn, G.J. MPP6 is an exosome-associated RNA-binding protein involved in 5.8S rRNA maturation. Nucleic Acids Res. 2005, 33, 6795–6804. [Google Scholar] [CrossRef] [PubMed]
- Nee Priyadarshini, P.S.; Lal, B. Seasonal variations in cellular expression of neuropeptide Y (NPY) in testis of the catfish, Clarias batrachus and its potential role in regulation of steroidogenesis. Peptides 2018, 103, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, D.; Boh, B.K.; Zhou, Y.; Chen, L.; Cornvik, T.C.; Hong, W.; Lu, L. Mammalian Mon2/Ysl2 regulates endosome-to-Golgi trafficking but possesses no guanine nucleotide exchange activity toward Arl1 GTPase. Sci. Rep. 2013, 3, 3362. [Google Scholar] [CrossRef] [PubMed]
- Baccetti, B. The role of the Golgi complex during spermiogenesis. Curr. Top. Dev. Biol. 1975, 10, 103–122. [Google Scholar] [CrossRef]
- Ferreira, J.D.B. Isolation and Purification of STEAP1 Protein Fragment in Escherichia coli Cells: A Potential Target for Prostate Cancer. Master’s Thesis, University of Beira Interior, Covilhã, Portugal, 2016. Available online: http://hdl.handle.net/10400.6/5817 (accessed on 12 December 2023).
- Sironen, A.; Uimari, P.; Nagy, S.; Paku, S.; Andersson, M.; Vilkki, J. Knobbed acrosome defect is associated with a region containing the genes STK17b and HECW2 on porcine chromosome 15. BMC Genom. 2010, 11, 699. [Google Scholar] [CrossRef]
- Yanagimachi, R. Mammalian sperm acrosome reaction: Where does it begin before fertilization? Biol. Reprod. 2011, 85, 4–5. [Google Scholar] [CrossRef]
- Wong, J.; Gasperoni, J.; Fuller, J.; Grommen, S.V.H.; De Groef, B.; Hogarth, C.; Dworkin, S. Crucial convolution: Genetic and molecular mechanisms of coiling during epididymis formation and development in embryogenesis. J. Dev. Biol. 2022, 10, 25. [Google Scholar] [CrossRef]
- Fraser, L.; Brym, P.; Pareek, C.S.; Mogielnicka-Brzozowska, M.; Paukszto, Ł.; Jastrzębski, J.P.; Wasilewska-Sakowska, K.; Mańkowska, A.; Sobiech, P.; Żukowski, K. Transcriptome analysis of boar spermatozoa with different freezability using RNA-Seq. Theriogenology 2020, 142, 400–413. [Google Scholar] [CrossRef]
- Gao, H.H.; Wen, H.; Cao, C.C.; Dong, D.Q.; Yang, C.H.; Xie, S.S.; Zhang, J.; Huang, X.B.; Huang, X.X.; Yuan, S.Q.; et al. Overexpression of microRNA-10a in germ cells causes male infertility by targeting Rad51 in mouse and human. Front. Physiol. 2019, 10, 765. [Google Scholar] [CrossRef] [PubMed]
- Hornecker, J.L.; Samollow, P.B.; Robinson, E.S.; Vandeberg, J.L.; McCarrey, J.R. Meiotic sex chromosome inactivation in the marsupial Monodelphis domestica. Genesis 2007, 45, 696–708. [Google Scholar] [CrossRef] [PubMed]
- Nunn, C.; Mao, H.; Chidiac, P.; Albert, P.R. RGS17/RGSZ2 and the RZ/A family of regulators of G-protein signaling. Semin. Cell Dev. Biol. 2006, 17, 390–399. [Google Scholar] [CrossRef] [PubMed]
- He, Y.D.; Wohlford, E.M.; Uhle, F.; Buturovic, L.; Liesenfeld, O.; Sweeney, T.E. The optimization and biological significance of a 29-host-immune-mRNA panel for the diagnosis of acute infections and sepsis. J. Pers. Med. 2021, 11, 735. [Google Scholar] [CrossRef] [PubMed]
- Shults, C.L.; Dingwall, C.B.; Kim, C.K.; Pinceti, E.; Rao, Y.S.; Pak, T.R. 17β-estradiol regulates the RNA-binding protein Nova1, which then regulates the alternative splicing of estrogen receptor β in the aging female rat brain. Neurobiol. Aging 2018, 61, 13–22. [Google Scholar] [CrossRef]
- Clements, C.M.; Henen, M.A.; Vögeli, B.; Shellman, Y.G. The structural dynamics, complexity of interactions, and functions in cancer of Multi-SAM containing proteins. Cancers 2023, 15, 3019. [Google Scholar] [CrossRef]
- Jaufmann, J.; Franke, F.C.; Sperlich, A.; Blumendeller, C.; Kloos, I.; Schneider, B.; Sasaki, D.; Janssen, K.P.; Beer-Hammer, S. The emerging and diverse roles of the SLy/SASH1-protein family in health and disease—Overview of three multifunctional proteins. FASEB J. 2021, 35, e21470. [Google Scholar] [CrossRef]
- Zhang, X.; Walczak, C.E. Chromosome segregation: Correcting improperly attached chromosomes. Curr. Biol. 2006, 16, R677–R679. [Google Scholar] [CrossRef][Green Version]
- Goodyear, R.J.; Legan, P.K.; Wright, M.B.; Marcotti, W.; Oganesian, A.; Coats, S.A.; Booth, C.J.; Kros, C.J.; Seifert, R.A.; Bowen-Pope, D.F.; et al. A receptor-like inositol lipid phosphatase is required for the maturation of developing cochlear hair bundles. J. Neurosci. 2003, 23, 9208–9219. [Google Scholar] [CrossRef]
- McVie-Wylie, A.J.; Lamson, D.R.; Chen, Y.T. Molecular cloning of a novel member of the GLUT family of transporters, SLC2a10 (GLUT10), localized on chromosome 20q13.1: A candidate gene for NIDDM susceptibility. Genomics 2001, 72, 113–117. [Google Scholar] [CrossRef]
- Pausch, H.; Venhoranta, H.; Wurmser, C.; Hakala, K.; Iso-Touru, T.; Sironen, A.; Vingborg, R.K.; Lohi, H.; Söderquist, L.; Fries, R.; et al. A frameshift mutation in ARMC3 is associated with a tail stump sperm defect in Swedish Red (Bos taurus) cattle. BMC Genet. 2016, 17, 49. [Google Scholar] [CrossRef][Green Version]
- Touré, A.; Martinez, G.; Kherraf, Z.-E.; Cazin, C.; Beurois, J.; Arnoult, C.; Ray, P.F.; Coutton, C. The genetic architecture of morphological abnormalities of the sperm tail. Hum. Genet. 2021, 140, 21–42. [Google Scholar] [CrossRef] [PubMed]
- Lehti, M.S.; Sironen, A. Formation and function of sperm tail structures in association with sperm motility defects. Biol. Reprod. 2017, 97, 522–536. [Google Scholar] [CrossRef] [PubMed]
- Rather, H.A.; Kumaresan, A.; Nag, P.; Kumar, V.; Nayak, S.; Batra, V.; Ganaie, B.A.; Baithalu, R.K.; Mohanty, T.K.; Datta, T.K. Spermatozoa produced during winter are superior in terms of phenotypic characteristics and oviduct explants binding ability in the water buffalo (Bubalus bubalis). Reprod. Domest. Anim. 2020, 55, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- Gmel, A.I.; Burger, D.; Neuditschko, M. A novel QTL and a candidate gene are associated with the progressive motility of Franches-Montagnes stallion spermatozoa after thaw. Genes 2021, 12, 1501. [Google Scholar] [CrossRef] [PubMed]
- Van Oort, R.J.; Garbino, A.; Wang, W.; Dixit, S.S.; Landstrom, A.P.; Gaur, N.; De Almeida, A.C.; Skapura, D.G.; Rudy, Y.; Burns, A.R.; et al. Disrupted junctional membrane complexes and hyperactive ryanodine receptors after acute junctophilin knockdown in mice. Circulation 2011, 123, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Darszon, A.; Nishigaki, T.; Beltran, C.; Treviño, C.L. Calcium channels in the development, maturation, and function of spermatozoa. Physiol. Rev. 2011, 91, 1305–1355. [Google Scholar] [CrossRef] [PubMed]
- Perni, S.; Beam, K.G. Voltage-induced Ca2+ release is supported by junctophilins 1, 2 and 3, and not by junctophilin 4: Calcium signaling and excitation–contraction in cardiac, skeletal and smooth muscle. J. Gen. Physiol. 2021, 154, e2021ecc22. [Google Scholar] [CrossRef]
- Piggott, C.A.; Jin, Y. Junctophilins: Key membrane tethers in muscles and neurons. Front. Mol. Neurosci. 2021, 14, 709390. [Google Scholar] [CrossRef]
- Tammineni, E.R.; Figueroa, L.; Kraeva, N.; Manno, C.; Ibarra, C.A.; Klip, A.; Riazi, S.; Rios, E. Fragmentation and roles of junctophilin1 in muscle of patients with cytosolic leak of stored calcium: Calcium signaling and excitation–contraction in cardiac, skeletal and smooth muscle. J. Gen. Physiol. 2022, 154, e2021ecc32. [Google Scholar] [CrossRef]
- Ter Keurs, H.E.; Boyden, P.A. Calcium and arrhythmogenesis. Physiol. Rev. 2007, 87, 457–506. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Castelao, B.; Castano, J.G. Synphilin-1 inhibits alpha-synuclein degradation by the proteasome. Cell. Mol. Life Sci. 2011, 68, 2643–2654. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, S.E.; Dudiki, T.; Vijayaraghavan, S.; Carr, D.W. Loss of R2D2 proteins ROPN1 and ROPN1L causes defects in murine sperm motility, phosphorylation, and fibrous sheath integrity. Biol. Reprod. 2013, 88, 41. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-L.; Ni, B.; Wang, X.-W.; Huo, W.-Q.; Zhang, J.; Tian, Z.-Q.; Huang, Z.-M.; Tian, Y.; Tang, J.; Zheng, Y.-H.; et al. FSCB phosphorylation in mouse spermatozoa capacitation. BMB Rep. 2011, 44, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, M.; Yu, R.; Liu, B.; Tian, Z.; Liu, S. FSCB phosphorylation regulates mouse spermatozoa capacitation through suppressing SUMOylation of ROPN1/ROPN1L. Am. J. Transl. Res. 2016, 8, 2776–2782. Available online: https://e-century.us/files/ajtr/8/6/ajtr0020298.pdf (accessed on 12 December 2023). [PubMed]
- Kawashima, A.; Osman, B.A.; Takashima, M.; Kikuchi, A.; Kohchi, S.; Satoh, E.; Tamba, M.; Matsuda, M.; Okamura, N. CABS1 is a novel calcium-binding protein specifically expressed in elongate spermatids of mice. Biol. Reprod. 2009, 80, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, S.; Lai, Z.; Zhou, Z.; Wu, F.; Huang, Y.; Lan, X.; Lei, C.; Chen, H.; Dang, R. Analysis of long non-coding RNA and mRNA expression profiling in immature and mature bovine (Bos taurus) testes. Front. Genet. 2019, 10, 646. [Google Scholar] [CrossRef]
- Meiners, S.; Heyken, D.; Weller, A.; Ludwig, A.; Stangl, K.; Kloetzel, P.-M.; Krüger, E. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of mammalian proteasomes. J. Biol. Chem. 2003, 278, 21517–21525. [Google Scholar] [CrossRef]
- Yokota, N.; Kataoka, Y.; Hashii, N.; Kawasaki, N.; Sawada, H. Sperm-specific C-terminal processing of the proteasome PSMA1/α6 subunit. Biochem. Biophys. Res. Commun. 2011, 410, 809–815. [Google Scholar] [CrossRef]
- Kurta, K.; Jeuthe, H.; Naboulsi, R.; de Koning, D.J.; Palaiokostas, C. Seasonal and age-related changes in sperm quality of farmed arctic charr (Salvelinus alpinus). BMC Genom. 2023, 24, 519. [Google Scholar] [CrossRef]
| Disorders | BTA 1 | SNP Localization, Mb | Candidate Genes |
|---|---|---|---|
| Absence of acrosomes | 4 | 82.4–82.9 | POU6F2, VPS41, AMPH |
| 11 | 70.9–72.5 | TCF23, SPDYA, PPP1CB, ABHD1 | |
| Sperm head abnormalities | 2 | 47.8–48.4 | ORC4, EPC2, MBD5 |
| Swollen acrosomes | 10 | 28.3–28.5 | LPCAT4, CACNB2 |
| Wrinkled acrosomes | 4 | 72.0–76.6 | IGFBP3, MON2, NPY, STEAP1 |
| Damaged sperm tails and cell necks | 1 | 1.2–5.9 | GRIK1, SON, GART |
| 2 | 47.7–48.4 | EPC2, MBD5, ORC4 | |
| 5 | 9.6–10.3 | PTPRQ, ATF7IP | |
| 9 | 32.2–35.1 | MAN1A1, FRK | |
| 90.2–91.3 | RGS17, ESR1 | ||
| 10 | 14.3–14.4 | C10H15orf61, MAP2K5 | |
| 80.2–80.3 | ZFYVE26, RAD51B | ||
| 17 | 11.5–11.7 | TTC29, POU4F2 | |
| 26 | 33.2–38.6 | ACSL5, RAB11FIP2 | |
| 42.0–43.7 | SPADH2, GPR26, SPADH1, FGFR2, ATE1 |
| SNP | BTA 1 | SNP Position (bp) | p-Value | Alleles | SNP Location | Candidate Genes 2 |
|---|---|---|---|---|---|---|
| Total motility | ||||||
| ARS-BFGL-NGS-29995 | 15 | 38,746,059 | 1.40 × 10−5 | A/G | intron variant | PSMA1 |
| HAPMAP51688-BTA-115008 | 21 | 54,879,224 | 6.70 × 10−6 | A/G | intergenic variant | FSCB (126.2 Kb), ENSBTAG00000052854 (135.8 Kb) |
| Progressive motility | ||||||
| BOVINEHD0400031627 | 4 | 110,457,609 | 9.17 × 10−6 | T/G | intergenic variant | ENSBTAG00000053666 (145 Kb) |
| BTB-00549286 | 7 | 32,855,047 | 8.72 × 10−6 | G/A | intron variant | SNCAIP |
| ARS-BFGL-NGS-113271 | 14 | 39,616,301 | 1.22 × 10−6 | T/C | intergenic variant | ENSBTAG00000032944 (108.5 Kb), JPH1 (17 Kb) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dementieva, N.V.; Dysin, A.P.; Shcherbakov, Y.S.; Nikitkina, E.V.; Musidray, A.A.; Petrova, A.V.; Mitrofanova, O.V.; Plemyashov, K.V.; Azovtseva, A.I.; Griffin, D.K.; et al. Risk of Sperm Disorders and Impaired Fertility in Frozen–Thawed Bull Semen: A Genome-Wide Association Study. Animals 2024, 14, 251. https://doi.org/10.3390/ani14020251
Dementieva NV, Dysin AP, Shcherbakov YS, Nikitkina EV, Musidray AA, Petrova AV, Mitrofanova OV, Plemyashov KV, Azovtseva AI, Griffin DK, et al. Risk of Sperm Disorders and Impaired Fertility in Frozen–Thawed Bull Semen: A Genome-Wide Association Study. Animals. 2024; 14(2):251. https://doi.org/10.3390/ani14020251
Chicago/Turabian StyleDementieva, Natalia V., Artem P. Dysin, Yuri S. Shcherbakov, Elena V. Nikitkina, Artem A. Musidray, Anna V. Petrova, Olga V. Mitrofanova, Kirill V. Plemyashov, Anastasiia I. Azovtseva, Darren K. Griffin, and et al. 2024. "Risk of Sperm Disorders and Impaired Fertility in Frozen–Thawed Bull Semen: A Genome-Wide Association Study" Animals 14, no. 2: 251. https://doi.org/10.3390/ani14020251
APA StyleDementieva, N. V., Dysin, A. P., Shcherbakov, Y. S., Nikitkina, E. V., Musidray, A. A., Petrova, A. V., Mitrofanova, O. V., Plemyashov, K. V., Azovtseva, A. I., Griffin, D. K., & Romanov, M. N. (2024). Risk of Sperm Disorders and Impaired Fertility in Frozen–Thawed Bull Semen: A Genome-Wide Association Study. Animals, 14(2), 251. https://doi.org/10.3390/ani14020251









