Analysis of Differential Gene Expression under Acute Lead or Mercury Exposure in Larval Zebrafish Using RNA-Seq
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Toxic Lead Exposure on Fish
2.3. The Measurement of Lead Concentrations
2.4. RNA Extraction
2.5. Illumina Sequencing
2.6. Bioinformatic Analysis
2.7. Validation Analysis via Quantitative Real-Time PCR (qRT-PCR)
2.8. Comparison of Lead and Mercury Exposure Transcriptomic Data in Zebrafish Larvae
2.9. Statistical Analysis
3. Results
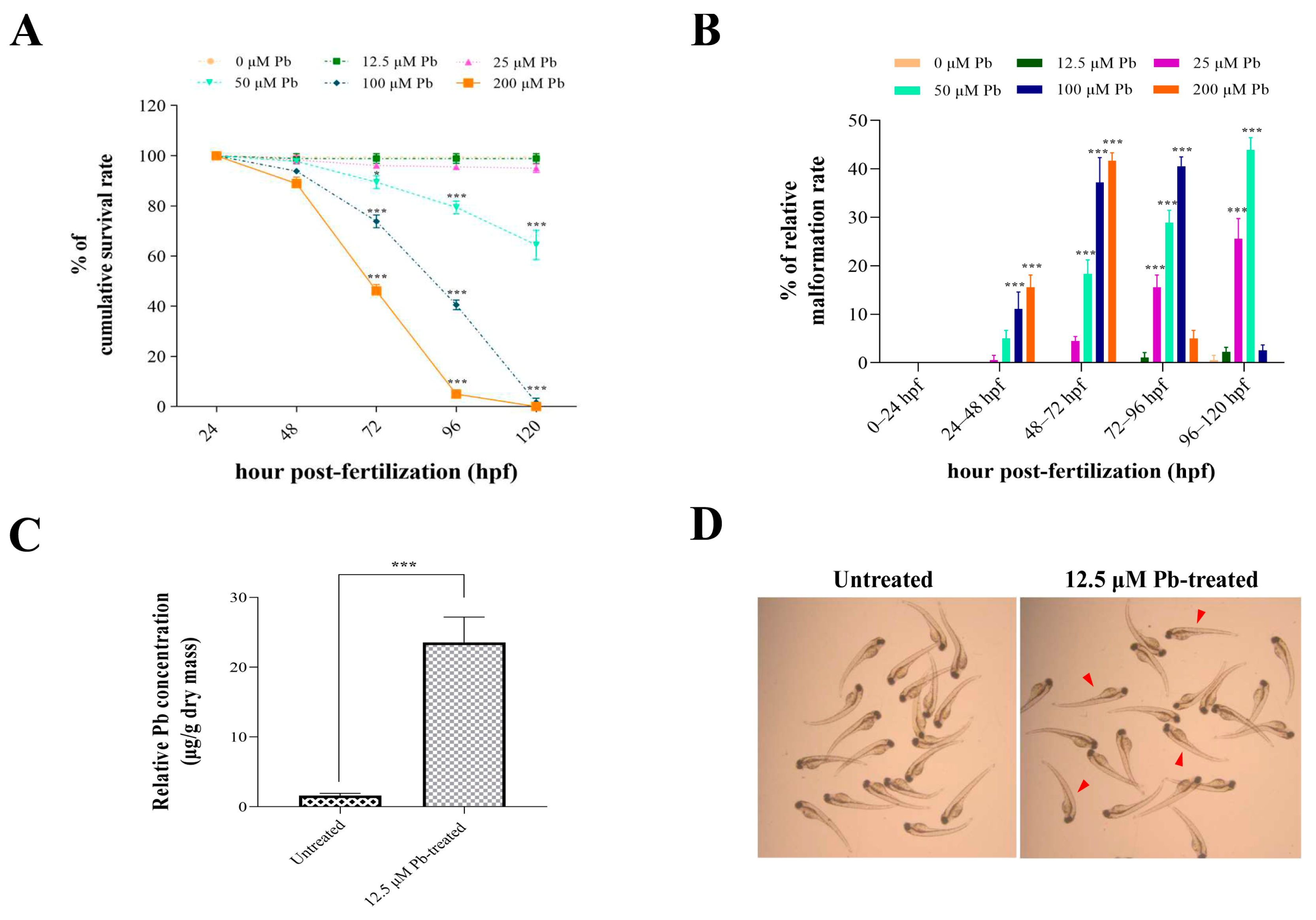
3.1. Effects of Lead Exposure on Zebrafish Development and Lead Bioaccumulation in Larval Zebrafish
3.2. Lead-Regulated Gene Expression
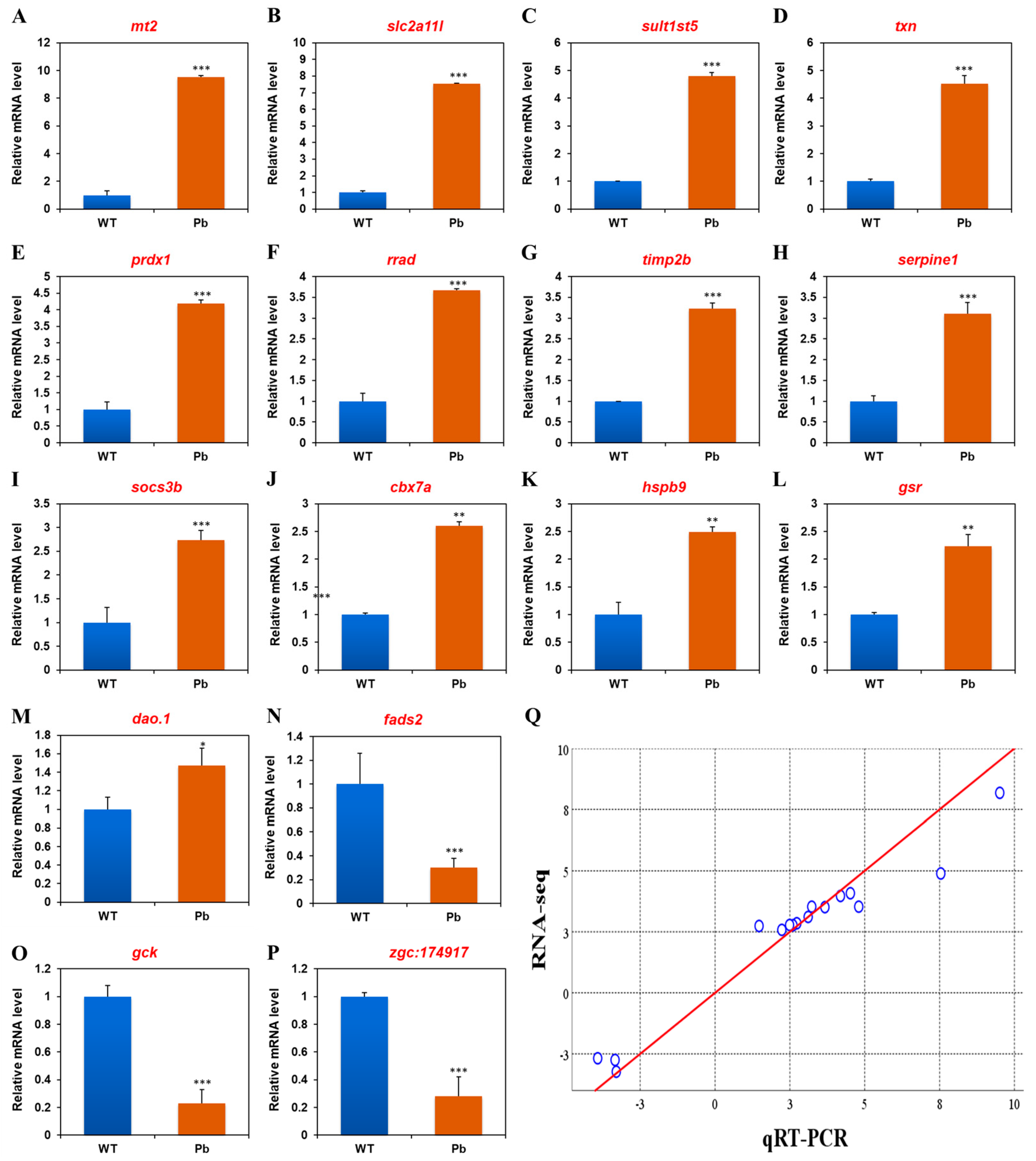
3.3. The Confirmation of RNA-Seq Data with qRT-PCR
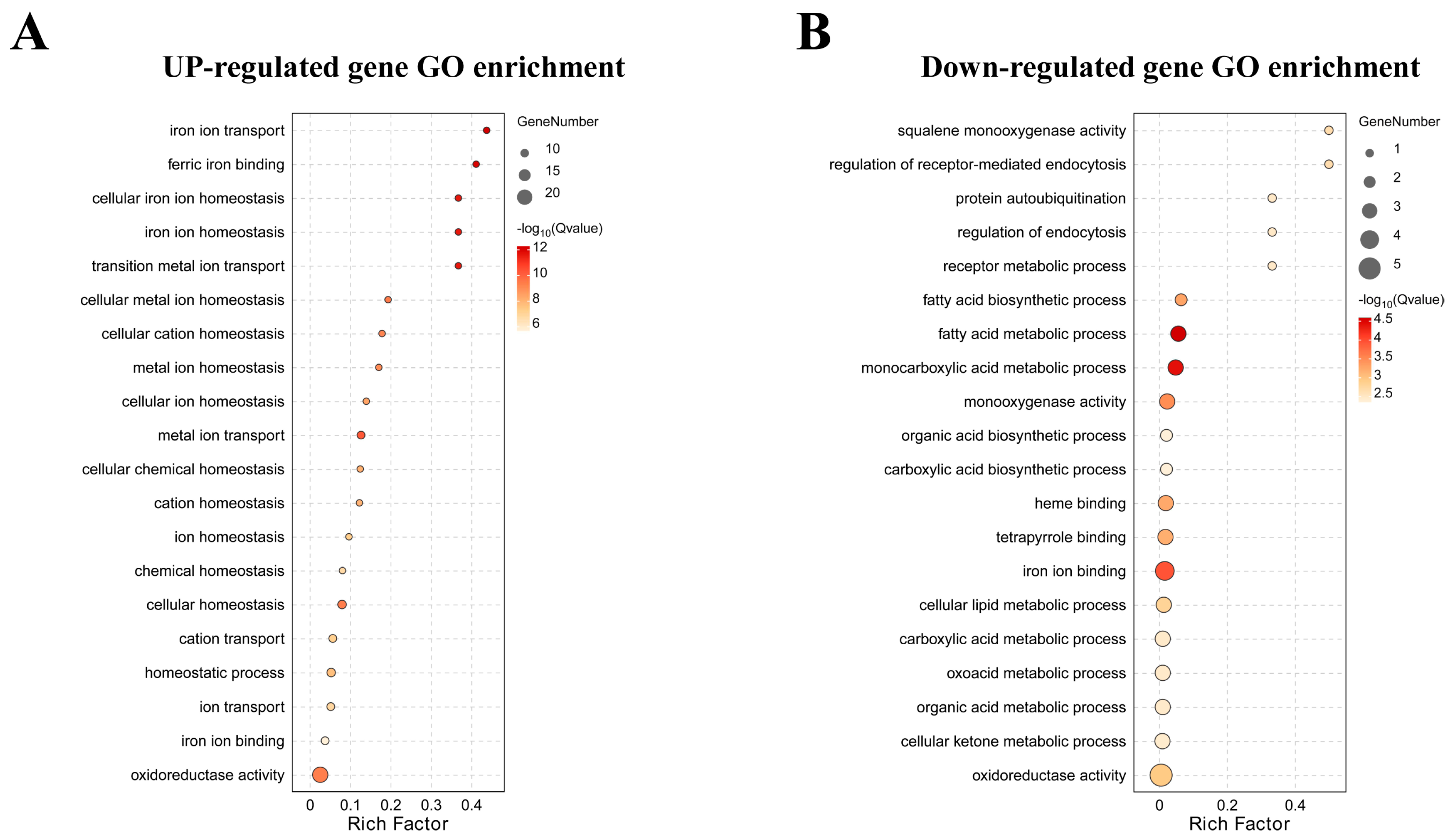
3.4. Functional Classification of Lead-Regulated DEGs
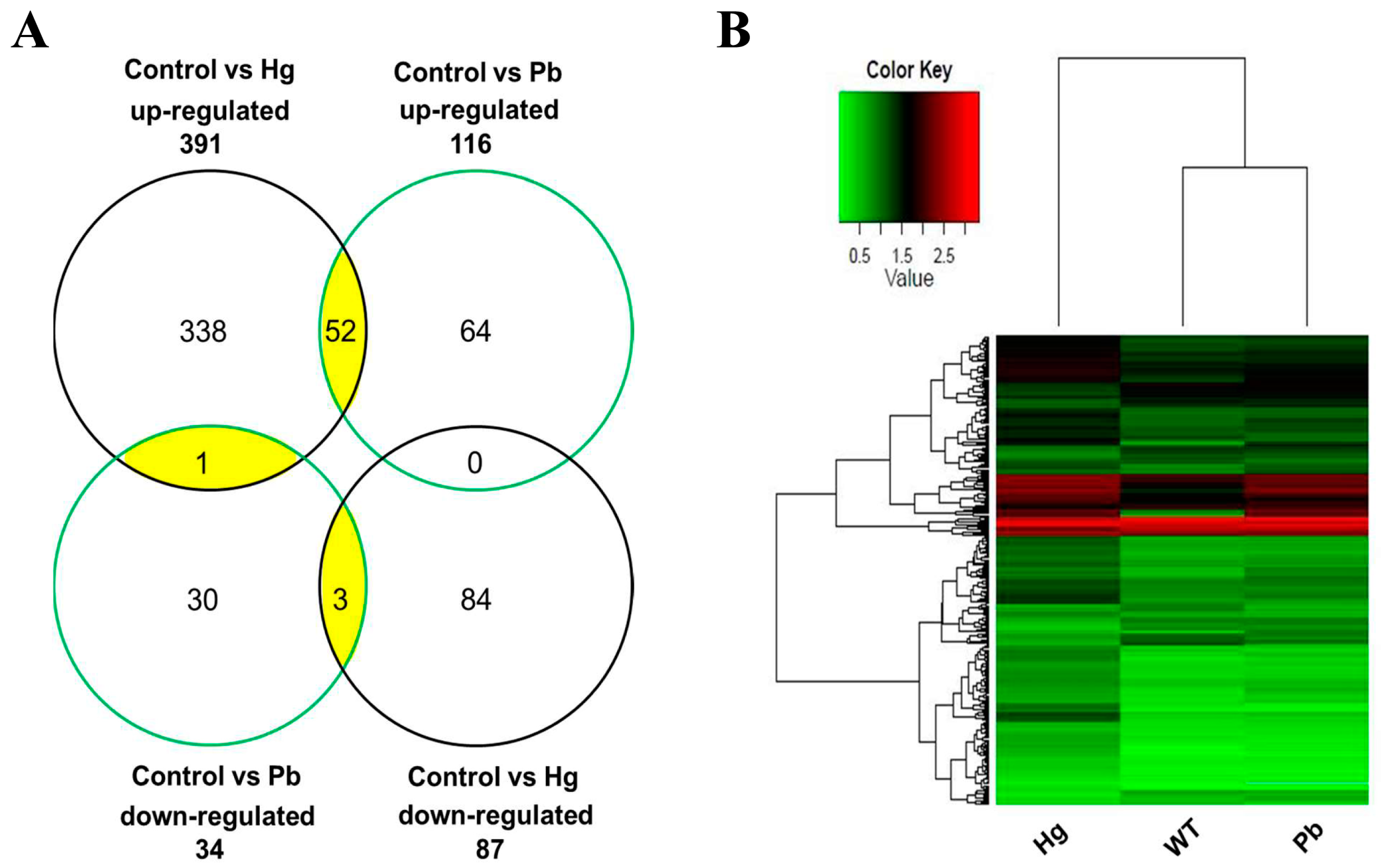
3.5. Genes Regulated by Mercury and Lead Exposure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mohmand, J.; Eqani, S.A.M.A.S.; Fasola, M.; Alamdar, A.; Mustafa, I.; Ali, N.; Liu, L.; Peng, S.; Shen, H. Human exposure to toxic metals via contaminated dust: Bio-accumulation trends and their potential risk estimation. Chemosphere 2015, 132, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Canaparo, R.; Foglietta, F.; Limongi, T.; Serpe, L. Biomedical applications of reactive oxygen species generation by metal nanoparticles. Materials 2020, 14, 53. [Google Scholar] [CrossRef] [PubMed]
- Shahjahan, M.; Taslima, K.; Rahman, M.S.; Al-Emran, M.D.; Alam, S.I.; Faggio, C. Effects of heavy metals on fish physiology—A review. Chemosphere 2022, 300, 134519. [Google Scholar] [CrossRef]
- Hou, L.P.; Yang, Y.; Shu, H.; Ying, G.G.; Zhao, J.L.; Chen, Y.B.; Chen, Y.H.; Fang, G.Z.; Liu, J.S. Changes in histopathology, enzyme activities, and the expression of relevant genes in zebrafish (Danio rerio) following long-term exposure to environmental levels of thallium. Bull. Environ. Contam. Toxicol. 2017, 99, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Massányi, P.; Massányi, M.; Madeddu, R.; Stawarz, R.; Lukáč, N. Effects of cadmium, lead, and mercury on the structure and function of reproductive organs. Toxics 2020, 8, 94. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, N.; Syakir Ishak, M.I.; Bhawani, S.A.; Umar, K. Various natural and anthropogenic factors responsible for water quality degradation: A review. Water 2021, 13, 2660. [Google Scholar] [CrossRef]
- Renu, K.; Chakraborty, R.; Myakala, H.; Koti, R.; Famurewa, A.C.; Madhyastha, H.; Vellingiri, B.; George, A.; Gopalakrishnan, A.V. Molecular mechanism of heavy metals (lead, chromium, arsenic, mercury, nickel and cadmium)-induced hepatotoxicity–A review. Chemosphere 2021, 271, 129735. [Google Scholar] [CrossRef]
- Rana, M.N.; Tangpong, J.; Rahman, M.M. Toxicodynamics of lead, cadmium, mercury and arsenic-induced kidney toxicity and treatment strategy: A mini review. Toxicol. Rep. 2018, 5, 704–713. [Google Scholar] [CrossRef]
- de Paula Arrifano, G.; Crespo-Lopez, M.E.; Lopes-Araújo, A.; Santos-Sacramento, L.; Barthelemy, J.L.; de Nazaré, C.G.L.; Freitas, L.G.R.; Augusto-Oliveira, M. Neurotoxicity and the global worst pollutants: Astroglial involvement in arsenic, lead, and mercury intoxication. Neurochem. Res. 2023, 48, 1047–1065. [Google Scholar] [CrossRef]
- Mukherjee, A.G.; Wanjari, U.R.; Renu, K.; Vellingiri, B.; Gopalakrishnan, A.V. Heavy metal and metalloid-induced reproductive toxicity. Environ. Toxicol. Phar. 2022, 92, 103859. [Google Scholar] [CrossRef]
- Liang, C.; Zhang, Z.; Cao, Y.; Wang, J.; Shen, L.; Jiang, T.; Li, D.; Zou, W.; Zong, K.; Liang, D.; et al. Exposure to multiple toxic metals and polycystic ovary syndrome risk: Endocrine disrupting effect from As, Pb and Ba. Sci. Total Environ. 2022, 849, 157780. [Google Scholar] [CrossRef] [PubMed]
- Haverinen, E.; Fernandez, M.F.; Mustieles, V.; Tolonen, H. Metabolic syndrome and endocrine disrupting chemicals: An overview of exposure and health effects. Int. J. Environ. Res. Public Health 2021, 18, 13047. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Lu, L.; Jin, C.; Wang, S.; Zhou, J.; Ni, Y.; Fu, Z.; Jin, Y. Effects of short term lead exposure on gut microbiota and hepatic metabolism in adult zebrafish. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2018, 209, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bakar, N.A.; Ibrahim, W.N.W.; Zulkiflli, A.R.; Hodin, N.A.S.; Kim, T.Y.; Ling, Y.S.; Ajat, M.M.M.; Shaari, K.; Shohaimi, S.; Nasruddin, N.S.; et al. Embryonic mercury exposure in zebrafish: Alteration of metabolites and gene expression, related to visual and behavioral impairments. Ecotoxicol. Environ. Saf. 2023, 256, 114862. [Google Scholar] [CrossRef] [PubMed]
- Khatib, I.; Horyn, O.; Bodnar, O.; Lushchak, O.; Rychter, P.; Falfushynska, H. Molecular and biochemical evidence of the toxic effects of terbuthylazine and malathion in zebrafish. Animals 2023, 13, 1029. [Google Scholar] [CrossRef]
- Long, Y.; Song, G.; Yan, J.; He, X.; Li, Q.; Cui, Z. Transcriptomic characterization of cold acclimation in larval zebrafish. BMC Genom. 2013, 14, 612. [Google Scholar] [CrossRef]
- Taslima, K.; Al-Emran, M.; Rahman, M.S.; Hasan, J.; Ferdous, Z.; Rohani, M.F.; Shahjahan, M. Impacts of heavy metals on early development, growth and reproduction of fish—A review. Toxicol. Rep. 2022, 9, 858–868. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Long, Y.; Li, Q.; Zhong, S.; Wang, Y.; Cui, Z. Molecular characterization and functions of zebrafish ABCC2 in cellular efflux of heavy metals. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2011, 153, 381–391. [Google Scholar] [CrossRef]
- Lu, X.; Xiang, Y.; Yang, G.; Zhang, L.; Wang, H.; Zhong, S. Transcriptomic characterization of zebrafish larvae in response to mercury exposure. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2017, 192, 40–49. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Rresearch 2015, 4, 1521. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Long, Y.; Peng, J.; Li, Q.; Cui, Z. Transcriptomic characterization of the dorsal lobes after hepatectomy of the ventral lobe in zebrafish. BMC Genom. 2015, 16, 979. [Google Scholar] [CrossRef]
- McCurley, A.T.; Callard, G.V. Characterization of housekeeping genes in zebrafish: Male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol. Biol. 2008, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jezierska, B.; Ługowska, K.; Witeska, M. The effects of heavy metals on embryonic development of fish (a review). Fish Physiol. Biochem. 2009, 35, 625–640. [Google Scholar] [CrossRef]
- Copeland, D.L.; Duff, R.J.; Liu, Q.; Prokop, J.; Londraville, R.L. Leptin in teleost fishes: An argument for comparative study. Front. Physiol. 2011, 2, 26. [Google Scholar] [CrossRef]
- Blanco, A.M.; Soengas, J.L. Leptin signalling in teleost fish with emphasis in food intake regulation. Mol. Cell. Endocrinol. 2021, 526, 111209. [Google Scholar] [CrossRef]
- Pandit, R.; Beerens, S.; Adan, R.A. Role of leptin in energy expenditure: The hypothalamic perspective. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2017, 312, R938–R947. [Google Scholar] [CrossRef]
- Friedman, J.M. Leptin and the endocrine control of energy balance. Nat. Metab. 2019, 1, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, O.; Patinote, A.; Nguyen, T.; Bobe, J. Multiple vitellogenins in zebrafish (Danio rerio): Quantitative inventory of genes, transcripts and proteins, and relation to egg quality. Fish Physiol. Biochem. 2018, 44, 1509–1525. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.J.; Shao, J.Z.; Xiang, L.X. Identification and characterization of suppressor of cytokine signaling 3 (SOCS-3) homologues in teleost fish. Mol. Immunol. 2007, 44, 1042–1051. [Google Scholar] [CrossRef]
- Sobah, M.L.; Liongue, C.; Ward, A.C. SOCS proteins in immunity, inflammatory diseases, and immune-related cancer. Front. Med. 2021, 8, 727987. [Google Scholar] [CrossRef]
- Zhang, J.; Bao, Z.; Guo, J.; Su, X.; Zou, Y.; Guo, H. Comparative transcriptome analysis of the hepatopancreas from Macrobrachium rosenbergii exposed to the heavy metal copper. Animals 2024, 14, 1117. [Google Scholar] [CrossRef]
- Ruttkay-Nedecky, B.; Nejdl, L.; Gumulec, J.; Zitka, O.; Masarik, M.; Eckschlager, T.; Stiborova, M.; Adam, V.; Kizek, R. The role of metallothionein in oxidative stress. Int. J. Mol. Sci. 2013, 14, 6044–6066. [Google Scholar] [CrossRef]
- Kim, J.H.; Oh, C.W.; Kang, J.C. Antioxidant responses, neurotoxicity, and metallothionein gene expression in juvenile Korean rockfish Sebastes schlegelii under dietary lead exposure. J. Aquat. Anim. Health 2017, 29, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yue, X.; Zhang, D.; Zhang, P.; Abdallah, A.; Yin, Y.; Cai, Y.; Li, Y. Study of bioaccumulation, hematological parameters, and antioxidant responses of Carassius auratus gibelio exposed to dietary lead and Bacillus subtilis. Biol. Trace Elem. Res. 2019, 189, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zheng, Y.G.; Feng, Y.H.; Li, M.Y.; Wang, G.Q.; Ma, Y.F. Toxic effects of waterborne lead (Pb) on bioaccumulation, serum biochemistry, oxidative stress and heat shock protein-related genes expression in Channa argus. Chemosphere 2020, 261, 127714. [Google Scholar] [CrossRef]
- Balali-Mood, M.; Naseri, K.; Tahergorabi, Z.; Khazdair, M.R.; Sadeghi, M. Toxic mechanisms of five heavy metals: Mercury, lead, chromium, cadmium, and arsenic. Front. Pharmacol. 2021, 12, 643972. [Google Scholar] [CrossRef]
- Quinlivan, V.H.; Farber, S.A. Lipid uptake, metabolism, and transport in the larval zebrafish. Front. Endocrinol. 2017, 8, 319. [Google Scholar] [CrossRef] [PubMed]
- Anzenbacher, P.; Anzenbacherova, E. Cytochromes P450 and metabolism of xenobiotics. Cell. Mol. Life Sci. 2001, 58, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Rubino, F.M. Toxicity of glutathione-binding metals: A review of targets and mechanisms. Toxics 2015, 3, 20–62. [Google Scholar] [CrossRef] [PubMed]
- Lalmanach, G.; Saidi, A.; Bigot, P.; Chazeirat, T.; Lecaille, F.; Wartenberg, M. Regulation of the proteolytic activity of cysteine cathepsins by oxidants. Int. J. Mol. Sci. 2020, 21, 1944. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Yan, J.; Song, G.; Li, X.; Li, X.; Li, Q.; Cui, Z. Transcriptional events co-regulated by hypoxia and cold stresses in zebrafish larvae. BMC Genom. 2015, 16, 385. [Google Scholar] [CrossRef]
- Kovacs, G.; Montalbetti, N.; Franz, M.C.; Graeter, S.; Simonin, A.; Hediger, M.A. Human TRPV5 and TRPV6: Key players in cadmium and zinc toxicity. Cell Calcium 2013, 54, 276–286. [Google Scholar] [CrossRef]
- Ahmed, I.; Zakiya, A.; Fazio, F. Effects of aquatic heavy metal intoxication on the level of hematocrit and hemoglobin in fishes: A review. Front. Environ. Sci. 2022, 10, 919204. [Google Scholar] [CrossRef]

| Group Name | Control | Control% | Lead | Lead% |
|---|---|---|---|---|
| Total reads | 77,597,408 | 100.00% | 59,858,446 | 100.00% |
| Total bases pairs | 7,682,143,392 | 100.00% | 5,925,986,154 | 100.00% |
| Processed reads | 67,329,394 | 86.77% | 51,432,756 | 85.92% |
| Processed bases pairs | 6,665,610,006 | 86.77% | 5,091,842,844 | 85.92% |
| Low-quality reads | 6,769,040 | 8.72% | 5,304,506 | 8.86% |
| Adapter polluted reads | 3,498,974 | 4.51% | 3,121,184 | 5.21% |
| Total mapped reads | 58,407,461 | 86.75% | 44,685,271 | 86.88% |
| Unique mapping | 49,365,912 | 73.32% | 37,937,786 | 73.76% |
| Total unmapped reads | 8,921,933 | 13.25% | 6,747,485 | 13.12% |
| Gene Symbol | Gene Name | Fold Change (Pb) | |
|---|---|---|---|
| RNA-Seq | qRT-PCR | ||
| mt2 | metallothionein 2 | 8.18 | 9.51 ± 0.13 |
| slc2a11l | solute carrier family 2 (facilitated glucose transporter), member 11-like | 4.89 | 7.54 ± 0.03 |
| sult1st5 | sulfotransferase family 1, cytosolic sulfotransferase 5 | 3.53 | 4.80 ± 0.12 |
| txn | thioredoxin | 4.08 | 4.52 ± 0.30 |
| prdx1 | peroxiredoxin 1 | 3.97 | 4.19 ± 0.11 |
| rrad | Ras-related associated with diabetes | 3.51 | 3.67 ± 0.03 |
| timp2b | TIMP metallopeptidase inhibitor 2b | 3.53 | 3.23 ± 0.13 |
| serpine1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 3.11 | 3.11 ± 0.26 |
| socs3b | suppressor of cytokine signaling 3b | 2.85 | 2.73 ± 0.21 |
| cbx7a | chromobox homolog 7a | 2.77 | 2.60 ± 0.08 |
| hspb9 | heat shock protein, alpha-crystallin-related, 9 | 2.78 | 2.49 ± 0.09 |
| gsr | glutathione reductase | 2.58 | 2.23 ± 0.22 |
| dao.1 | D-amino-acid oxidase, tandem duplicate 1 | 2.74 | 1.47 ± 0.19 |
| fads2 | fatty acid desaturase 2 | −2.75 | −3.34 ± 0.08 |
| gck | glucokinase | −2.68 | −3.92 ± 0.10 |
| zgc:174917 | uncharacterized gene | −3.23 | −3.30 ± 0.14 |
| KEGG Pathways for Up-Regulated Genes | KEGG Pathways for Down-Regulated Genes | ||
|---|---|---|---|
| Pathway Name | p-Value | Pathway Name | p-Value |
| Metabolism of xenobiotics by cytochrome P450 | 1.02 × 10−5 | Steroid biosynthesis | 0.00020935 |
| Glutathione metabolism | 3.15 × 10−5 | Sesquiterpenoid and triterpenoid biosynthesis | 0.00306591 |
| Drug metabolism–cytochrome P450 | 0.00019061 | Butirosin and neomycin biosynthesis | 0.00510528 |
| Complement and coagulation cascades | 0.00193363 | Streptomycin biosynthesis | 0.01120151 |
| Adipocytokine signaling pathway | 0.00221758 | Pancreatic secretion | 0.01418717 |
| Gene Symbol | Fold Change (Mercury) | Fold Change (Lead) | Biological Process | ||
|---|---|---|---|---|---|
| RNA-Seq | qRT-PCR | RNA-Seq | qRT-PCR | ||
| Regulated by specific exposure to mercury | |||||
| intl2 | 16.43 | 12.22 ± 0.06 | Signal transduction | ||
| per2 | 11.70 | 14.92 ± 0.21 | Response to hydrogen peroxide | ||
| cry5 | 9.31 | 10.17 ± 0.15 | DNA repair | ||
| cybb | 3.72 | 3.10 ± 0.16 | Oxidoreductase activity | ||
| hamp1 | 2.55 | 2.14 ± 0.30 | Cellular iron ion homeostasis | ||
| nyx | −5.06 | −5.56 ± 0.36 | Neurological system process | ||
| opn1sw1 | −3.74 | −4.32 ± 0.32 | Visual perception | ||
| Regulated by specific exposure to lead | |||||
| sult1st5 | 3.53 | 4.80 ± 0.12 | Xenobiotic metabolic process | ||
| rrad | 3.51 | 3.67 ± 0.03 | Small GTPase-mediated signal transduction | ||
| socs3b | 2.85 | 2.73 ± 0.21 | Intracellular signal transduction | ||
| hspb9 | 2.78 | 2.49 ± 0.09 | Response to stress | ||
| gck | −2.68 | −3.92 ± 0.10 | Glycolysis | ||
| Co-regulated by mercury and lead exposure | |||||
| mt2 | 28.01 | 24.04 ± 0.12 | 8.18 | 9.51 ± 0.13 | Metal ion binding |
| ctssb.1 | 15.00 | 12.36 ± 0.18 | 5.56 | 2.88 ± 0.02 | Proteolysis |
| prdx1 | 6.29 | 5.82 ± 0.24 | 3.97 | 4.19 ± 0.11 | Peroxisome, antioxidant activity |
| txn | 4.60 | 4.33 ± 0.06 | 4.08 | 4.52 ± 0.30 | Antioxidant activity |
| sqrdl | 4.53 | 5.21 ± 0.02 | 2.96 | 3.62 ± 0.08 | Oxidoreductase activity |
| tmprss13a | 4.05 | 3.61 ± 0.13 | 2.38 | 2.49 ± 0.06 | Proteolysis |
| socs3a | 4.03 | 6.57 ± 0.07 | 2.57 | 3.45 ± 0.19 | Protein ubiquitination, intracellular signal transduction |
| trpv6 | 3.74 | 2.64 ± 0.07 | 2.82 | 2.65 ± 0.00 | Calcium ion transmembrane transport |
| abcb6a | 3.21 | 2.54 ± 0.46 | 2.58 | 3.66 ± 0.38 | Transmembrane transport, ATP catabolic process |
| gsr | 3.19 | 3.38 ± 0.08 | 2.58 | 2.23 ± 0.22 | Glutathione metabolism, oxidoreductase activity |
| hbz | 2.55 | 2.14 ± 0.01 | 3.06 | 4.03 ± 0.04 | Oxygen transporter activity |
| fads2 | −2.70 | −2.85 ± 0.12 | −2.75 | −3.34 ± 0.08 | Fatty acid biosynthetic process |
| zgc:92590 | −9.13 | −22.55 ± 0.16 | −7.19 | −9.24 ± 0.09 | Protein digestion and absorption |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, X.; Zhang, L.; Lin, G.-M.; Lu, J.-G.; Cui, Z.-B. Analysis of Differential Gene Expression under Acute Lead or Mercury Exposure in Larval Zebrafish Using RNA-Seq. Animals 2024, 14, 2877. https://doi.org/10.3390/ani14192877
Lu X, Zhang L, Lin G-M, Lu J-G, Cui Z-B. Analysis of Differential Gene Expression under Acute Lead or Mercury Exposure in Larval Zebrafish Using RNA-Seq. Animals. 2024; 14(19):2877. https://doi.org/10.3390/ani14192877
Chicago/Turabian StyleLu, Xing, Lang Zhang, Gen-Mei Lin, Jian-Guo Lu, and Zong-Bin Cui. 2024. "Analysis of Differential Gene Expression under Acute Lead or Mercury Exposure in Larval Zebrafish Using RNA-Seq" Animals 14, no. 19: 2877. https://doi.org/10.3390/ani14192877
APA StyleLu, X., Zhang, L., Lin, G.-M., Lu, J.-G., & Cui, Z.-B. (2024). Analysis of Differential Gene Expression under Acute Lead or Mercury Exposure in Larval Zebrafish Using RNA-Seq. Animals, 14(19), 2877. https://doi.org/10.3390/ani14192877





