Genome-Wide Association Study of Reproductive Traits in Large White Pigs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Traits
2.2. Genotypes and Quality Control
2.3. Statistical Analysis
2.4. Identification of Candidate Genes and Analysis of Functional Enrichment
3. Results and Discussion
3.1. Genetic Architecture and LD Decay
3.2. Heritability
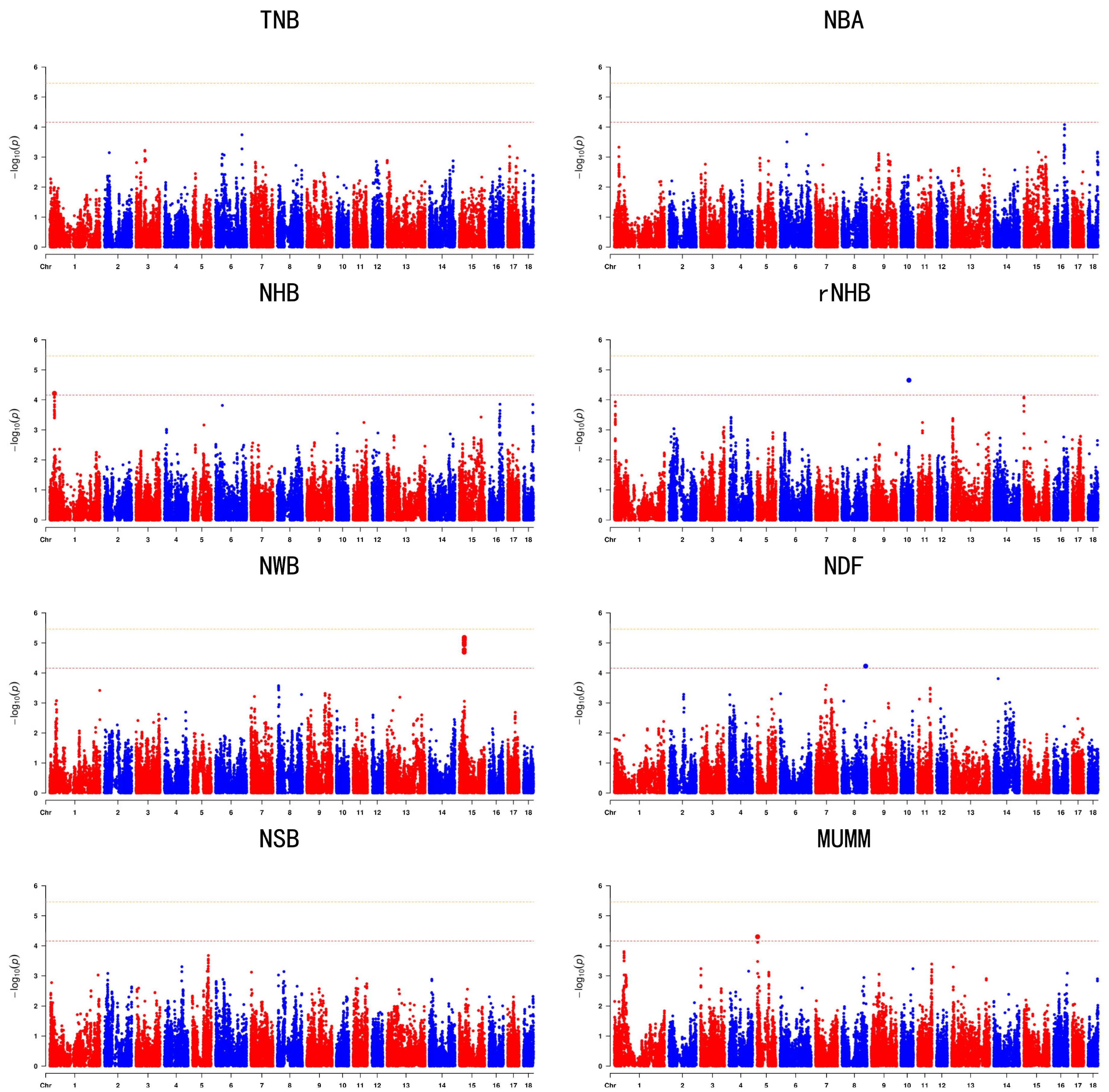
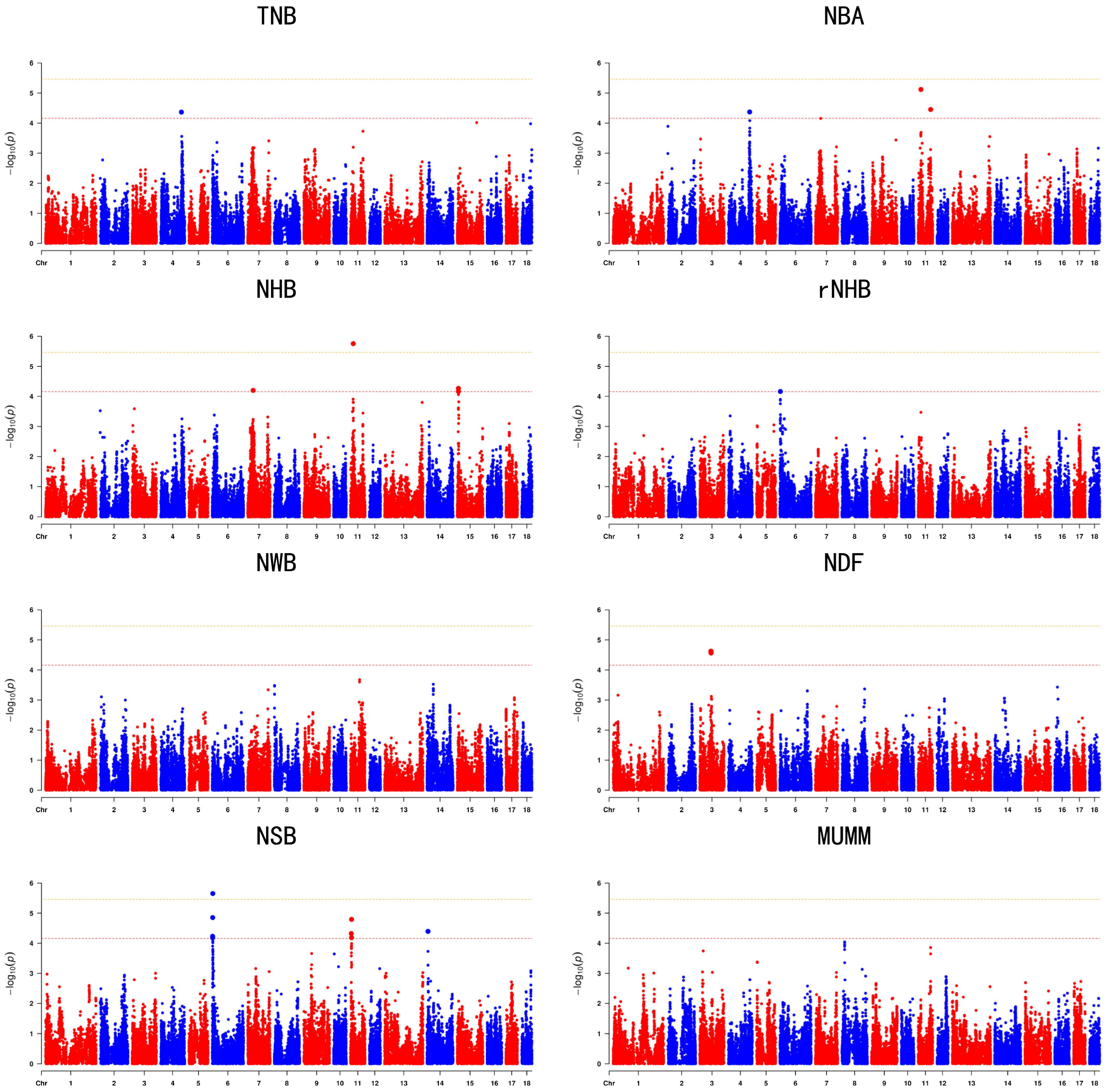
3.3. Genome’Wide Association Studies
3.4. Candidate Genes for Litter Traits
3.5. The Functional Enrichment Analysis for Litter Traits
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sell-Kubiak, E. Selection for litter size and litter birthweight in Large White pigs: Maximum, mean and variability of reproduction traits. Animal 2021, 15, 100352. [Google Scholar] [CrossRef]
- Knecht, D.; Środoń, S.; Duziński, K. The impact of season, parity and breed on selected reproductive performance parameters of sows. Arch. Anim. Breed. 2015, 58, 49–56. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, J.; Olasege, B.S.; Ma, P.; Qiu, X.; Gao, H.; Wang, C.; Wang, Y.; Zhang, Q.; Yang, H.; et al. Estimation of genetic parameters for reproductive traits in connectedness groups of Duroc, Landrace and Yorkshire pigs in China. J. Anim. Breed. Genet. 2020, 137, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Schiavo, G.; Bovo, S.; Tinarelli, S.; Gallo, M.; Dall’Olio, S.; Fontanesi, L. Genome-wide association analyses for coat colour patterns in the autochthonous Nero Siciliano pig breed. Livest. Sci. 2020, 236, 104015. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Gong, H.; Cui, L.; Zhang, W.; Ma, J.; Chen, C.; Ai, H.; Xiao, S.; Huang, L.; et al. Genetic correlation of fatty acid composition with growth, carcass, fat deposition and meat quality traits based on GWAS data in six pig populations. Meat Sci. 2018, 150, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, Z.; Chen, R.; Huang, Z.; Han, X.; Qiao, R.; Wang, K.; Yang, F.; Li, X.-J.; Li, X.-L. Identification of Genomic Regions and Candidate Genes for Litter Traits in French Large White Pigs Using Genome-Wide Association Studies. Animals 2022, 12, 1584. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.-S.; Xu, J.-Y.; He, W.-M.; Yang, T.-L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Genetic Investigation of ANthropometric Traits (GIANT) Consortium; Yang, J.; DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium; Ferreira, T.; Morris, A.P.; Medland, S.E.; Madden, P.A.F.; Heath, A.C.; Martin, N.G.; Montgomery, G.W.; et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012, 44, 369–375. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39 (Suppl. S2), W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Tan, C.; Hu, X.; Wang, A.; Wu, Z. Genetic parameters for reproductive traits at different parities in Large White pigs. J. Anim. Sci. 2018, 96, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.H.; Lee, D.H.; See, M.T. Estimation of Genetic Parameters for Reproductive Traits between First and Later Parities in Pig. Asian-Australas. J. Anim. Sci. 2005, 19, 7–12. [Google Scholar] [CrossRef]
- Yang, Y.; Gan, M.; Yang, X.; Zhu, P.; Luo, Y.; Liu, B.; Zhu, K.; Cheng, W.; Chen, L.; Zhao, Y.; et al. Estimation of genetic parameters of pig reproductive traits. Front. Vet. Sci. 2023, 10, 1172287. [Google Scholar] [CrossRef] [PubMed]
- Knol, E.F.; Nielsen, B.; Knap, P.W. Genomic selection in commercial pig breeding. Anim. Front. 2016, 6, 15–22. [Google Scholar] [CrossRef]
- Cameron, N.; Enser, M. Fatty Acid Composition of Lipid in Longissimus Dorsi Muscle of Duroc and British Landrace Pigs and Its Relationship with Eating Quality. Meat Sci. 1991, 29, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Roehe, R.; Kennedy, B.W. Effect of Selection for Maternal and Direct Genetic Effects on Genetic Improvement of Litter Size in Swine. J. Anim. Sci. 1993, 71, 2891–2904. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zou, S.; Li, Z.; Wang, Y.; Chen, T.; Song, P.; Chen, J.; He, X.; Xu, P.; Liang, M.; Luo, K.; et al. Association Study Between Polymorphisms of PRMT6, PEX10, SOX5, and Nonobstructive Azoospermia in the Han Chinese Population. Biol. Reprod. 2014, 90, 96. [Google Scholar] [CrossRef] [PubMed]
- Hering, D.; Olenski, K.; Kaminski, S. Genome-Wide Association Study for Sperm Concentration in Holstein-Friesian Bulls. Reprod. Domest. Anim. 2014, 49, 1008–1014. [Google Scholar] [CrossRef] [PubMed]
- Grassmann, F.; Harsch, S.; Brandl, C.; Kiel, C.; Nürnberg, P.; Toliat, M.R.; Fleckenstein, M.; Pfau, M.; Schmitz-Valckenberg, S.; Holz, F.G.; et al. Assessment of Novel Genome-Wide Significant Gene Loci and Lesion Growth in Geographic Atrophy Secondary to Age-Related Macular Degeneration. JAMA Ophthalmol 2019, 137, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Pei, L.; Lai, F.; Xiao, H.; Li, Z.; Zeng, R.; Chen, L.; Chen, W.; Liu, H.; Li, Y.; et al. Genome-wide analysis study of gestational diabetes mellitus and related pathogenic factors in a Chinese Han population. BMC Pregnancy Childbirth 2023, 23, 856. [Google Scholar] [CrossRef] [PubMed]
- Rajovski, S.; Vucic, N.; Karanovic, J.; Matijasevic, S.; Savic-Pavicevic, D.; Dobrijevic, Z.; Brajuskovic, G. Association of PRMT6, PEX10 and SOX5 genetic variants with idiopathic male infertility: Evidence from North Macedonian population and an updated meta-analysis. Genetika 2023, 55, 355–372. [Google Scholar] [CrossRef]
- Thompson, S.D.; Sudman, M.; Ramos, P.S.; Marion, M.C.; Ryan, M.; Tsoras, M.; Weiler, T.; Wagner, M.; Keddache, M.; Haas, J.P.; et al. The susceptibility loci juvenile idiopathic arthritis shares with other autoimmune diseases extend to PTPN2, COG6, and ANGPT1. Arthritis Rheum. 2010, 62, 3265–3276. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, H.; Wu, G. Investigating the genetic association of HCP5, SPATA2, TNIP1, TNFAIP3 and COG6 with psoriasis in Chinese population. Int. J. Immunogenet. 2014, 41, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Mesner, L.D.; Calabrese, G.M.; Al-Barghouthi, B.; Gatti, D.M.; Sundberg, J.P.; Churchill, G.A.; Godfrey, D.A.; Ackert-Bicknell, C.L.; Farber, C.R. Mouse genome-wide association and systems genetics identifies Lhfp as a regulator of bone mass. PLOS Genet. 2019, 15, e1008123. [Google Scholar] [CrossRef] [PubMed]
- De León, C.; Martínez, R.; Rocha, J.; Darghan, A. Research Article Selection of genomic regions and genes associated with adaptation and fertility traits in two Colombian creole cattle breeds. Evolution 2021, 20. [Google Scholar] [CrossRef]
- Hill-Burns, E.M.; Wissemann, W.T.; Hamza, T.H.; A Factor, S.; Zabetian, C.P.; Payami, H. Identification of a novel Parkinson’s disease locus via stratified genome-wide association study. BMC Genom. 2014, 15, 118. [Google Scholar] [CrossRef]
- Wang, K.; Wu, P.; Yang, Q.; Chen, D.; Zhou, J.; Jiang, A.; Ma, J.; Tang, Q.; Xiao, W.; Jiang, Y.; et al. Detection of Selection Signatures in Chinese Landrace and Yorkshire Pigs Based on Genotyping-by-Sequencing Data. Front. Genet. 2018, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Sabik, O.L.; Calabrese, G.M.; Taleghani, E.; Ackert-Bicknell, C.L.; Farber, C.R. Identification of a Core Module for Bone Mineral Density through the Integration of a Co-expression Network and GWAS Data. Cell Rep. 2020, 32, 108145. [Google Scholar] [CrossRef] [PubMed]
- McLendon, J.M.; Zhang, X.; Stein, C.; Abouassaly, G.; Witmer, N.; Boudreau, R.L. TXLNB is a Novel Regulator of Cardiac Proteostasis. FASEB J. 2018, 32, 864.11. [Google Scholar] [CrossRef]
- Blaj, I.; Tetens, J.; Bennewitz, J.; Thaller, G.; Falker-Gieske, C. Integrating imputed structural variants and tandem repeats in a GWAS for growth and carcass traits in F2 pig crosses. In Proceedings of the 12th World Congress on Genetics Applied to Livestock Production (WCGALP); Veerkamp, R.F., de Haas, Y., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2022; p. 390. [Google Scholar]
- Dementieva, N.V.; Dysin, A.P.; Shcherbakov, Y.S.; Nikitkina, E.V.; Musidray, A.A.; Petrova, A.V.; Mitrofanova, O.V.; Plemyashov, K.V.; Azovtseva, A.I.; Griffin, D.K.; et al. Risk of Sperm Disorders and Impaired Fertility in Frozen–Thawed Bull Semen: A Genome-Wide Association Study. Animals 2024, 14, 251. [Google Scholar] [CrossRef]
- Ran, X.; Pan, H.; Huang, S.; Liu, C.; Niu, X.; Li, S.; Wang, J. Copy number variations of MTHFSD gene across pig breeds and its association with litter size traits in Chinese indigenous Xiang pig. J. Anim. Physiol. Anim. Nutr. 2018, 102, 1320–1327. [Google Scholar] [CrossRef] [PubMed]
- Balogh, E.E.; Gábor, G.; Bodó, S.; Rózsa, L.; Rátky, J.; Zsolnai, A.; Anton, I. Effect of single-nucleotide polymorphisms on specific reproduction parameters in Hungarian Large White sows. Acta Vet. Hung. 2019, 67, 256–273. [Google Scholar] [CrossRef] [PubMed]
- Pagnamenta, A.T.; Bacchelli, E.; de Jonge, M.V.; Mirza, G.; Scerri, T.S.; Minopoli, F.; Chiocchetti, A.; Ludwig, K.U.; Hoffmann, P.; Paracchini, S.; et al. Characterization of a Family with Rare Deletions in CNTNAP5 and DOCK4 Suggests Novel Risk Loci for Autism and Dyslexia. Biol. Psychiatry 2010, 68, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.; Lam, M.; Zai, C.; Tay, J.; Karlsson, N.; Deshpande, S.N.; Thelma, B.K.; Ozaki, N.; Inada, T.; Sim, K.; et al. Genome wide study of tardive dyskinesia in schizophrenia. Transl. Psychiatry 2021, 11, 351. [Google Scholar] [CrossRef]
- Cantero, M.d.R.; Cantiello, H.F. Polycystin-2 (TRPP2): Ion channel properties and regulation. Gene 2022, 827, 146313. [Google Scholar] [CrossRef] [PubMed]
- Carrozzo, R.; Verrigni, D.; Rasmussen, M.; de Coo, R.; Amartino, H.; Bianchi, M.; Buhas, D.; Mesli, S.; Naess, K.; Born, A.P.; et al. Succinate-CoA ligase deficiency due to mutations in SUCLA2 and SUCLG1: Phenotype and genotype correlations in 71 patients. J. Inherit. Metab. Dis. 2016, 39, 243–252. [Google Scholar] [CrossRef]
- Zuo, Y.; Zhang, L.; Tang, W.; Tang, W. Identification of prognosis-related alternative splicing events in kidney renal clear cell carcinoma. J. Cell. Mol. Med. 2019, 23, 7762–7772. [Google Scholar] [CrossRef]


| Parity | Trait | N | Mean | SD | Minimum | Maximum | Heritability |
|---|---|---|---|---|---|---|---|
| 1 | TNB | 2036 | 17.56 | 3.59 | 6 | 25 | 0.04 |
| NBA | 1955 | 14.72 | 3.66 | 5 | 23 | 0.02 | |
| NHB | 1958 | 13.61 | 3.50 | 4 | 21 | 0.04 | |
| rNHB | 1978 | 77.27 | 16.60 | 15 | 100 | 0.02 | |
| NWB | 1991 | 0.89 | 1.34 | 0 | 5 | 0.01 | |
| NDF | 2022 | 0.02 | 0.15 | 0 | 1 | 0.01 | |
| NSB | 1937 | 1.88 | 1.66 | 0 | 6 | 0.01 | |
| MUMM | 1914 | 0.66 | 1.07 | 0 | 4 | 0.01 | |
| 2 | TNB | 1332 | 18.53 | 3.34 | 6 | 25 | 0.04 |
| NBA | 1306 | 15.59 | 3.64 | 5 | 23 | 0.06 | |
| NHB | 1306 | 14.14 | 3.27 | 4 | 21 | 0.05 | |
| rNHB | 1308 | 76.64 | 14.47 | 15 | 100 | 0.01 | |
| NWB | 1298 | 1.28 | 1.45 | 0 | 5 | 0.01 | |
| NDF | 1328 | 0.02 | 0.15 | 0 | 1 | 0.01 | |
| NSB | 1258 | 1.98 | 1.69 | 0 | 6 | 0.01 | |
| MUMM | 1292 | 0.61 | 1.02 | 0 | 4 | 0.01 |
| Trait | SSC | rs | Position | Allele | Maf | p | Candidate Gene |
|---|---|---|---|---|---|---|---|
| NHB | 1 | rs55618047 | 25284574 | C/T | 0.315315 | 6.05448 × 10−5 | TXLNB |
| rNHB | 10 | rs81294332 | 44308657 | A/G | 0.180471 | 2.21049 × 10−5 | SLC39A12 |
| NWB | 15 | rs336401964 | 27459405 | T/C | 0.229584 | 2.02398 × 10−5 | CNTNAP5 |
| NWB | 15 | rs333569636 | 27509302 | T/C | 0.229439 | 2.02405 × 10−5 | CNTNAP5 |
| NWB | 15 | rs81304168 | 27531806 | A/G | 0.218686 | 9.90197 × 10−6 | CNTNAP5 |
| NWB | 15 | rs335973133 | 27581413 | C/T | 0.221593 | 1.69344 × 10−5 | CNTNAP5 |
| NWB | 15 | rs333665093 | 27738575 | A/G | 0.220866 | 8.04687 × 10−6 | CNTNAP5 |
| NWB | 15 | rs343368776 | 27746996 | C/G | 0.221011 | 9.97989 × 10−6 | CNTNAP5 |
| NWB | 15 | rs320619304 | 27755517 | A/G | 0.220866 | 8.04687 × 10−6 | CNTNAP5 |
| NWB | 15 | rs81305599 | 27775994 | A/G | 0.220866 | 8.04687 × 10−6 | CNTNAP5 |
| NWB | 15 | rs337432591 | 27785758 | A/G | 0.221593 | 1.14719 × 10−5 | CNTNAP5 |
| NWB | 15 | rs327976032 | 27883239 | A/T | 0.220575 | 6.59095 × 10−6 | CNTNAP5 |
| NWB | 15 | rs332278643 | 27892641 | T/G | 0.220575 | 6.57329 × 10−6 | CNTNAP5 |
| NWB | 15 | rs81478872 | 28021915 | T/G | 0.220285 | 6.82459 × 10−6 | CNTNAP5 |
| NWB | 15 | rs3470958220 | 28183106 | T/C | 0.219849 | 7.14878 × 10−6 | CNTNAP5 |
| NWB | 15 | rs327135140 | 28279474 | G/T | 0.219994 | 7.4039 × 10−6 | CNTNAP5 |
| NWB | 15 | rs81452141 | 28293171 | A/C | 0.219994 | 7.4039 × 10−6 | CNTNAP5 |
| NWB | 15 | rs322582049 | 28347013 | A/G | 0.219994 | 7.4039 × 10−6 | CNTNAP5 |
| NWB | 15 | rs338760245 | 28395070 | T/C | 0.219704 | 6.6822 × 10−6 | CNTNAP5 |
| NDF | 8 | rs81310054 | 131016420 | A/C | 0.104911 | 5.92047 × 10−5 | PKD2 |
| MUMM | 5 | rs321202796 | 4183089 | T/C | 0.121331 | 5.00831 × 10−5 | KIAA0930 |
| Trait | SSC | rs | Position | Allele | MAF | p | Genes |
|---|---|---|---|---|---|---|---|
| TNB | 4 | rs344560170 | 113119104 | C/T | 0.387242 | 4.308 × 10−5 | PRMT6 |
| NBA | 4 | rs3474737609 | 116388595 | A/T | 0.0956117 | 4.258 × 10−5 | OLFM3 |
| NBA | 11 | rs341909772 | 14909631 | T/C | 0.447835 | 7.613 × 10−6 | COG6 |
| NBA | 11 | rs81431697 | 67830653 | G/A | 0.372857 | 3.524 × 10−5 | DOCK9 |
| NHB | 7 | rs341878379 | 29614695 | A/C | 0.406423 | 6.299 × 10−5 | COL21A1 |
| NHB | 11 | rs341909772 | 14909631 | T/C | 0.447835 | 1.759 × 10−6 | COG6 |
| NHB | 15 | rs1108768276 | 6547063 | C/A | 0.409183 | 6.827 × 10−5 | ENSSSCG00000053606 |
| NHB | 15 | rs320524315 | 6384510 | C/T | 0.343795 | 5.407 × 10−5 | ENSSSCG00000053606 |
| rNHB | 6 | rs320041489 | 2728087 | T/C | 0.402645 | 6.856 × 10−5 | MTHFSD |
| NSB | 6 | rs333336780 | 2814834 | C/T | 0.274629 | 1.398 × 10−5 | IRF8 |
| NSB | 6 | rs334764615 | 2387727 | T/C | 0.321854 | 5.853 × 10−5 | MTHFSD |
| NSB | 6 | rs336729684 | 2454772 | T/C | 0.273612 | 6.613 × 10−5 | MTHFSD |
| NSB | 6 | rs342135123 | 3342934 | G/C | 0.380703 | 2.223 × 10−6 | GSE1 |
| NSB | 11 | rs329467410 | 5544200 | C/T | 0.106946 | 6.536 × 10−5 | PAN3 |
| NSB | 11 | rs345895836 | 5600114 | C/T | 0.0998256 | 1.604 × 10−5 | PAN3 |
| NSB | 11 | rs81430859 | 4296303 | C/T | 0.140221 | 4.744 × 10−5 | WASF3 |
| NSB | 14 | rs337919249 | 5225585 | C/T | 0.0893636 | 4.035 × 10−5 | GFRA2 |
| NDF | 3 | rs81304023 | 60157640 | T/C | 0.0838419 | 2.389 × 10−5 | SUCLG1 |
| NDF | 3 | rs81330647 | 60211109 | T/C | 0.0838419 | 2.389 × 10−5 | SUCLG1 |
| NDF | 3 | rs81327030 | 60216589 | T/C | 0.0838419 | 2.389 × 10−5 | SUCLG1 |
| NDF | 3 | rs332244679 | 60266762 | G/A | 0.0838419 | 2.389 × 10−5 | SUCLG1 |
| NDF | 3 | rs81299811 | 60914644 | C/G | 0.0841325 | 2.392 × 10−5 | SUCLG1 |
| NDF | 3 | rs336799801 | 61636339 | C/T | 0.0842778 | 2.743 × 10−5 | SUCLG1 |
| NDF | 3 | rs81244504 | 61851017 | C/T | 0.0844231 | 2.541 × 10−5 | SUCLG1 |
| NDF | 3 | rs81326050 | 61878711 | A/G | 0.0839872 | 2.391 × 10−5 | SUCLG1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, Y.; Tan, C.; He, X.; Wu, D.; Zhang, Y.; Song, C.; Wu, Z. Genome-Wide Association Study of Reproductive Traits in Large White Pigs. Animals 2024, 14, 2874. https://doi.org/10.3390/ani14192874
Hong Y, Tan C, He X, Wu D, Zhang Y, Song C, Wu Z. Genome-Wide Association Study of Reproductive Traits in Large White Pigs. Animals. 2024; 14(19):2874. https://doi.org/10.3390/ani14192874
Chicago/Turabian StyleHong, Yifeng, Cheng Tan, Xiaoyan He, Dan Wu, Yuxing Zhang, Changxu Song, and Zhenfang Wu. 2024. "Genome-Wide Association Study of Reproductive Traits in Large White Pigs" Animals 14, no. 19: 2874. https://doi.org/10.3390/ani14192874
APA StyleHong, Y., Tan, C., He, X., Wu, D., Zhang, Y., Song, C., & Wu, Z. (2024). Genome-Wide Association Study of Reproductive Traits in Large White Pigs. Animals, 14(19), 2874. https://doi.org/10.3390/ani14192874





