Screening of Reference Genes for Quantitative Real-Time PCR Analysis in Tissues and during Testis Development, and Application to Analyze the Expression of kifc1 in Hemibarbus labeo (Teleostei, Cypriniformes, Cyprinidae)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Identification of Testis Development
2.3. RNA Extraction and Reverse Transcription
2.4. Primer Design, Validation, and PCR Amplification Efficiency
2.5. Quantitative Real-Time PCR
2.6. Data Analysis
3. Results
3.1. Primer Specificity and Amplification Efficiency
3.2. Cycle Threshold (Ct) Values of Candidate Reference Genes in Tissues and during Testis Development
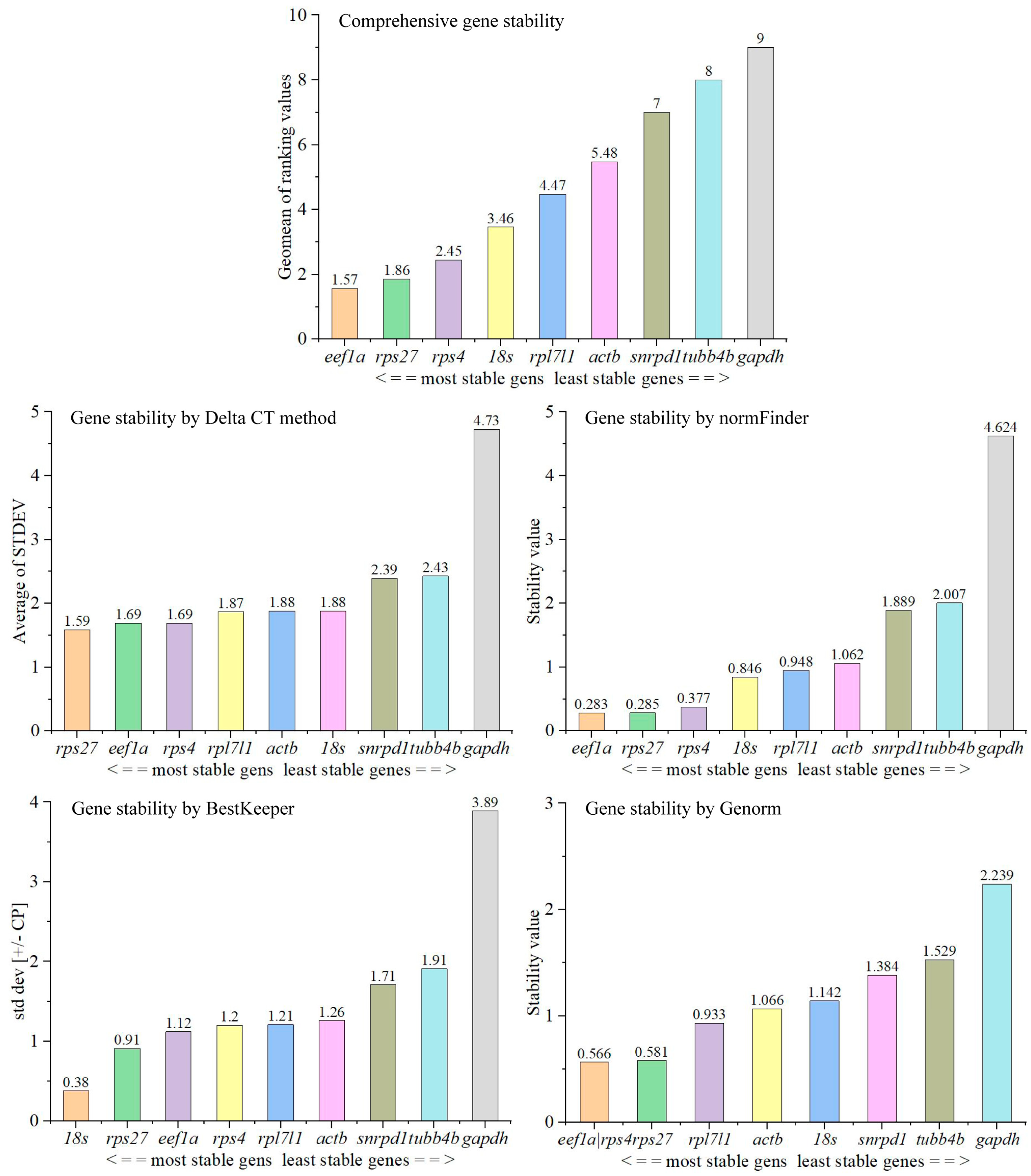
3.3. Stability of Expression of Candidate Reference Genes in Tissues in Adult Male Fish
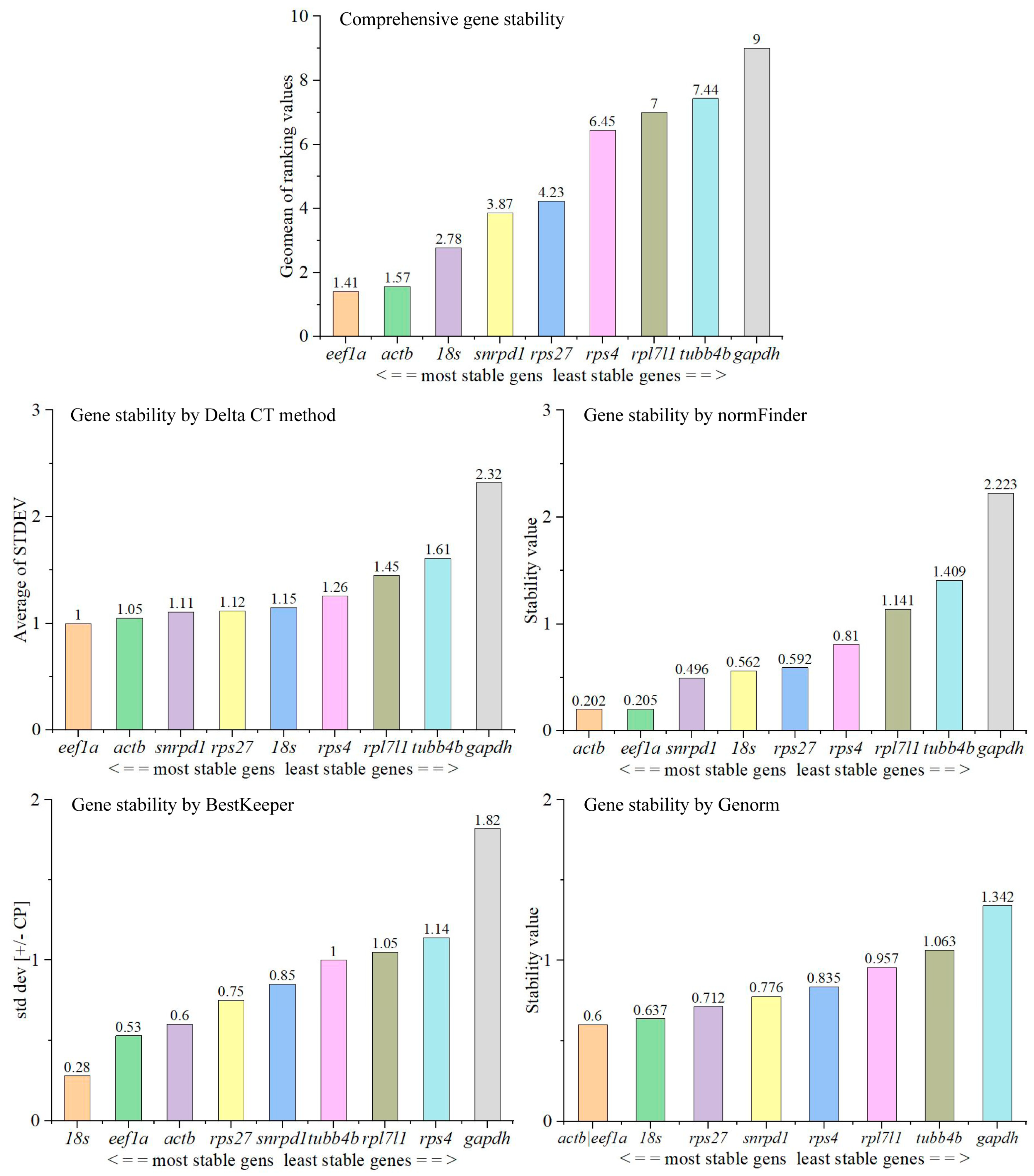
3.4. Stability of Expression of Candidate Reference Genes during Testes Development
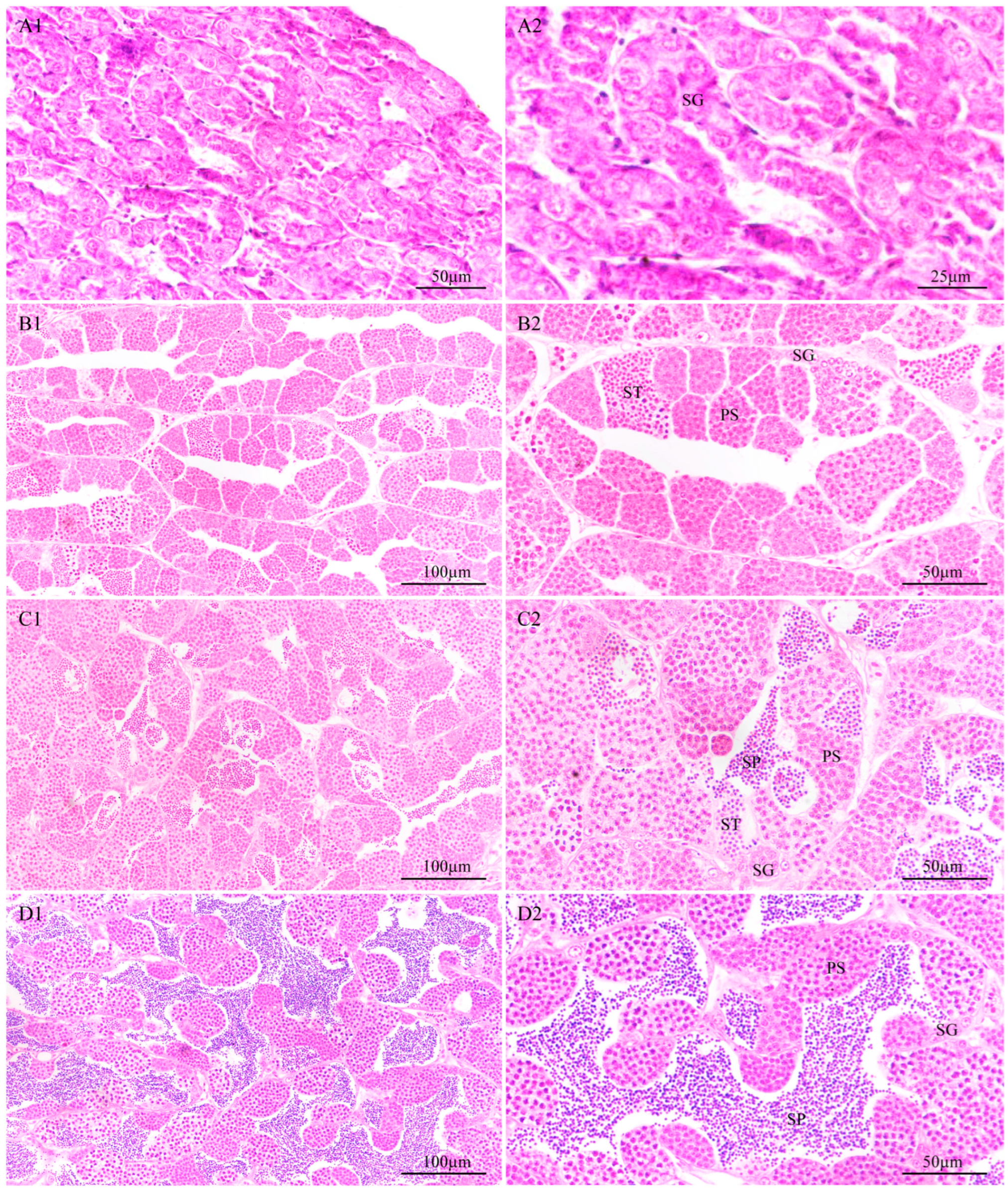
3.4.1. Identification of the Developmental Stage of the Testes
3.4.2. Selection of Reference Genes for qPCR Analysis during Testicular Development
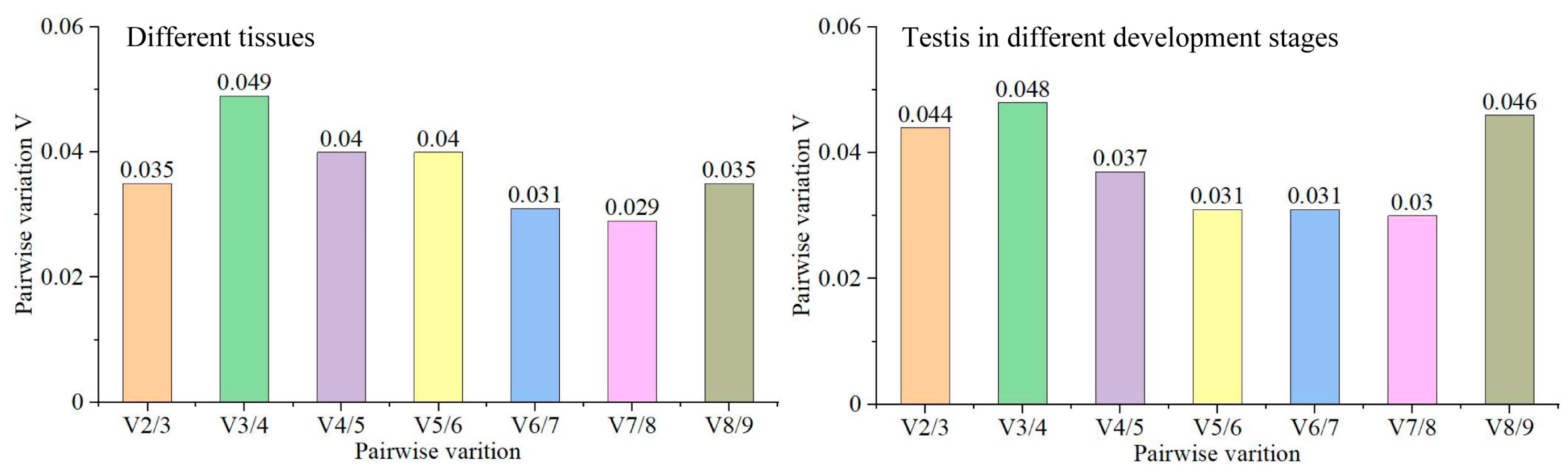
3.5. Optimal Number of Reference Genes Required for Quantification in Tissues and during Testis Development
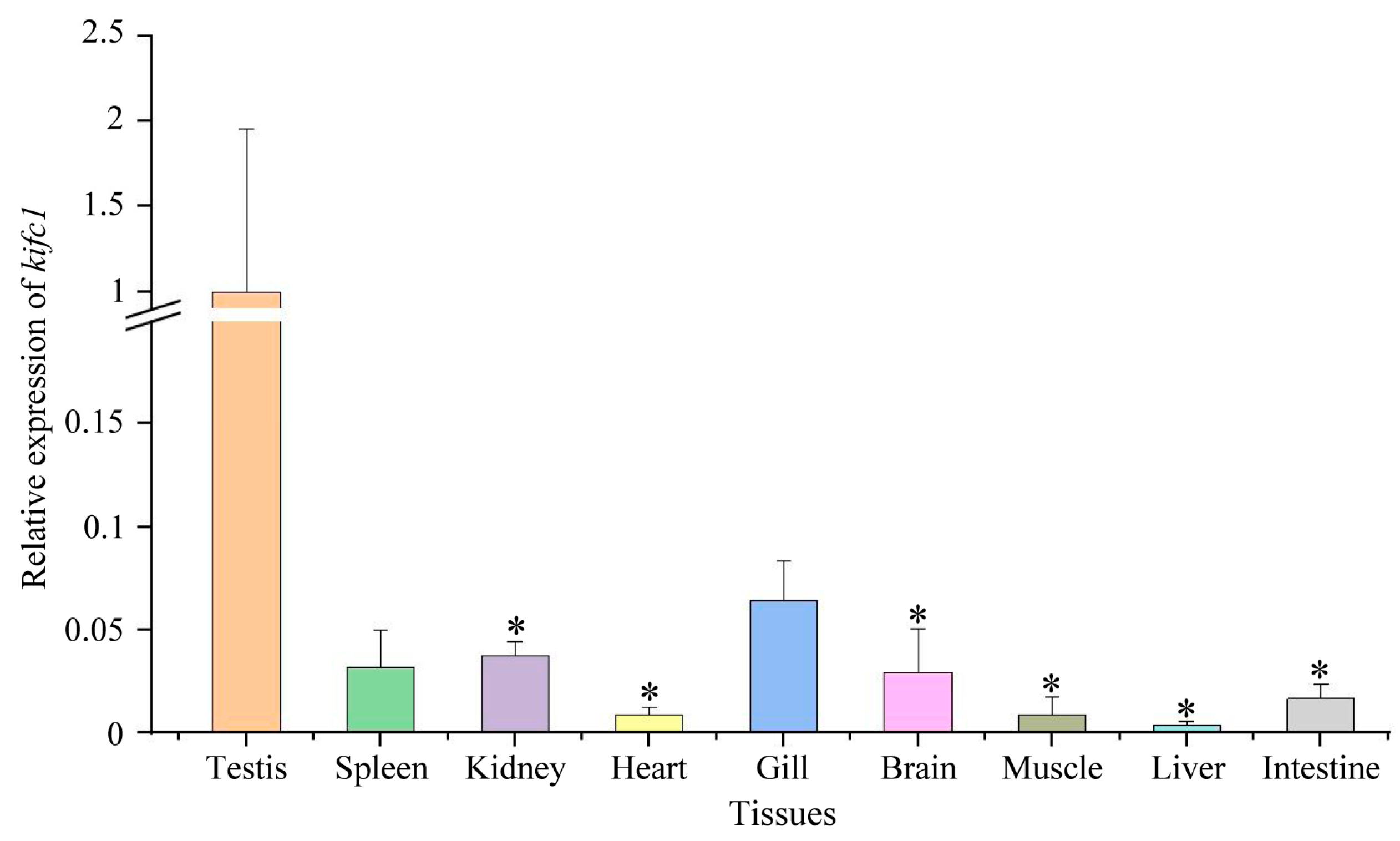
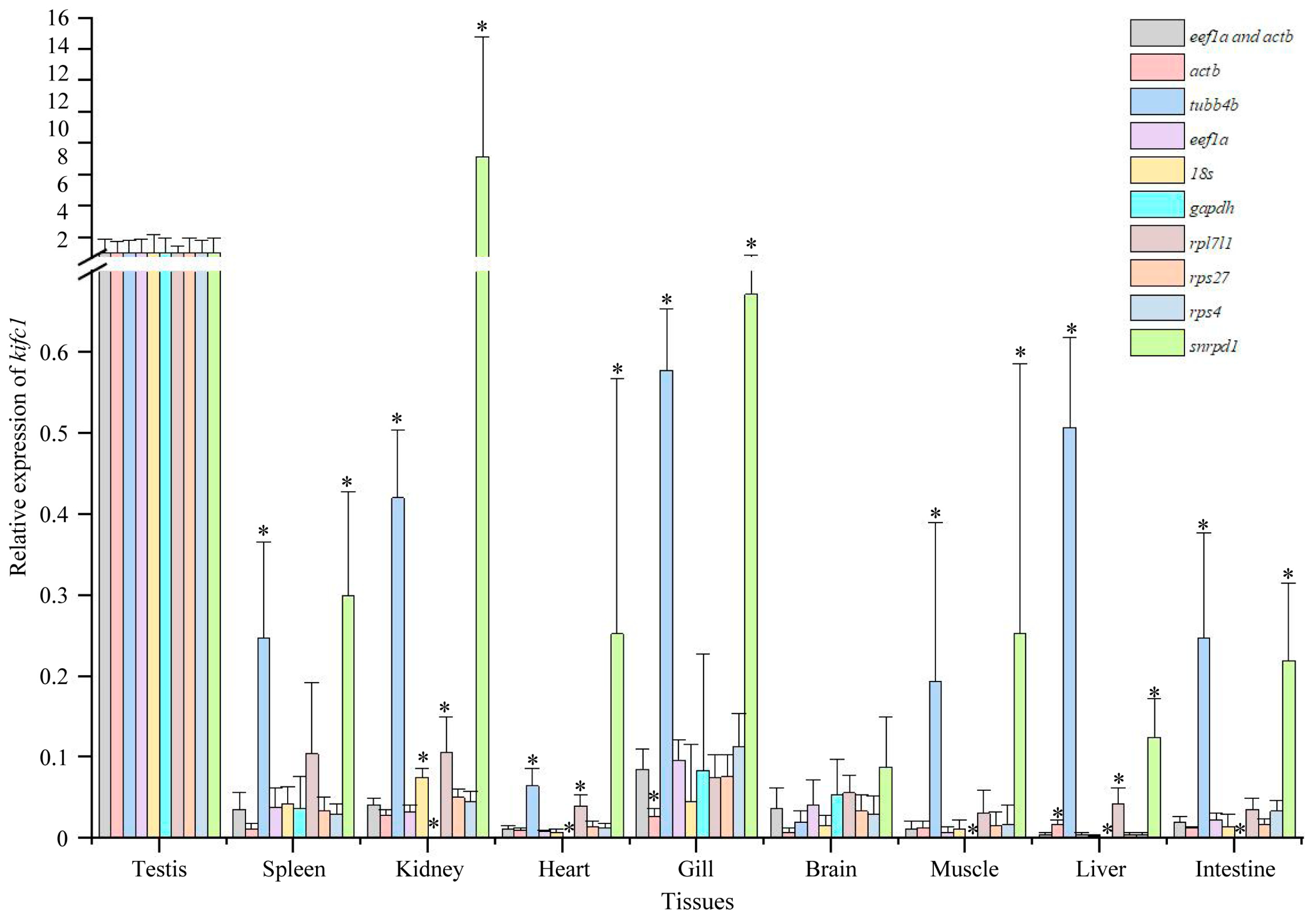
3.6. Expression of kifc1 in Different Tissues
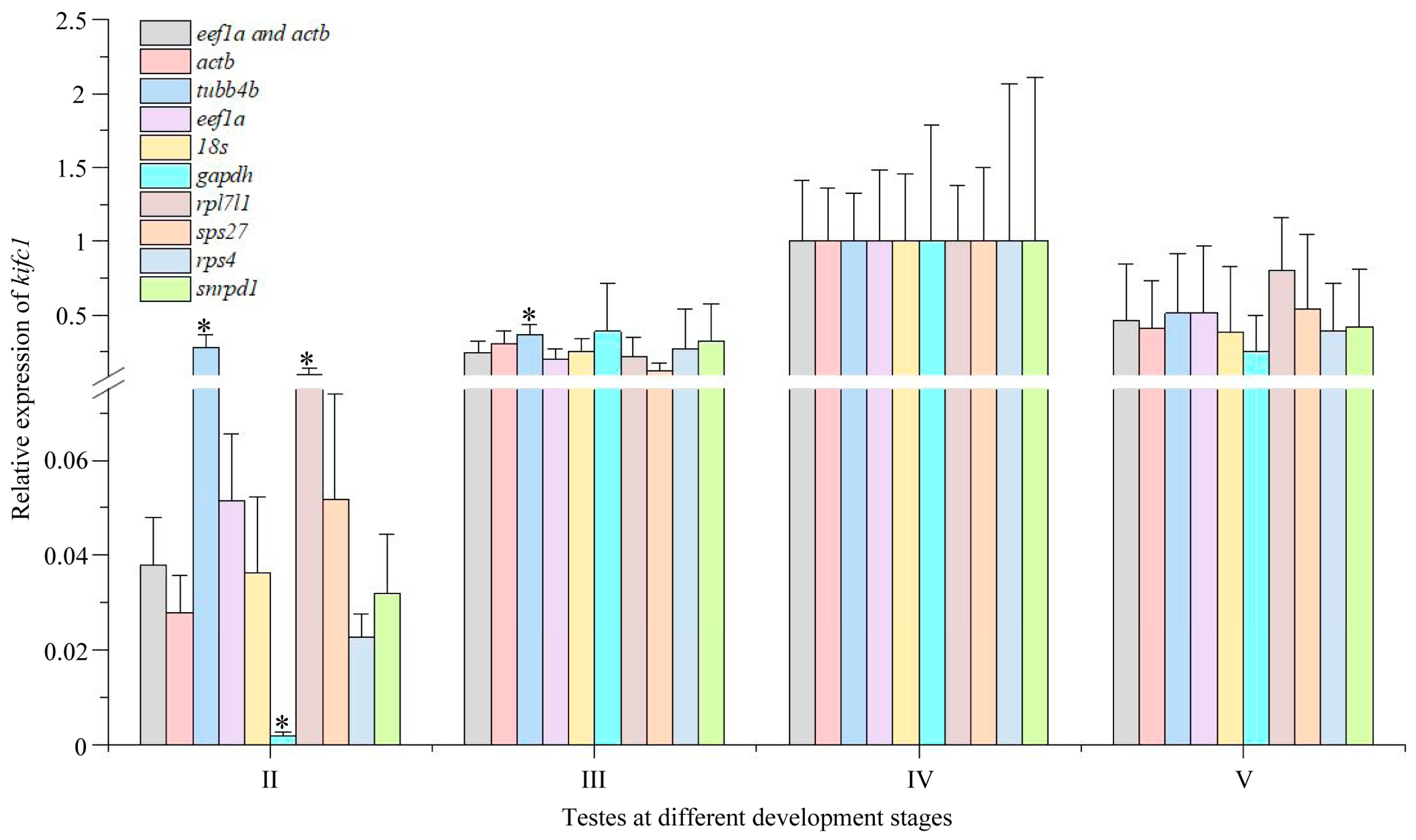
3.7. Expression of kifc1 during Testis Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bustin, S.A. Developments in real-time PCR research and molecular diagnostics. Expert. Rev. Mol. Diagn. 2010, 10, 713–715. [Google Scholar] [CrossRef] [PubMed]
- Valasek, M.A.; Repa, J.J. The power of real-time PCR. Adv. Physiol. Educ. 2005, 29, 151–159. [Google Scholar] [CrossRef]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef]
- Hu, Q.; Guo, W.; Gao, Y.; Tang, R.; Li, D. Reference gene selection for real-time RT-PCR normalization in rice field eel (Monopterus albus) during gonad development. Fish. Physiol. Biochem. 2014, 40, 1721–1730. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Wang, K.; Chen, D.; Wang, J.; He, Y.; Long, B.; Yang, L.; Yang, Q.; Geng, Y.; Huang, X.; et al. Evaluation and selection of appropriate reference genes for real-time quantitative PCR analysis of gene expression in Nile Tilapia (Oreochromis niloticus) during vaccination and infection. Int. J. Mol. Sci. 2015, 16, 9998–10015. [Google Scholar] [CrossRef]
- Mahanty, A.; Purohit, G.K.; Mohanty, S.; Nayak, N.R.; Mohanty, B.P. Suitable reference gene for quantitative real-time PCR analysis of gene expression in gonadal tissues of minnow Puntius sophore under high-temperature stress. BMC Genom. 2017, 18, 617. [Google Scholar] [CrossRef]
- Li, Y.Y.; Chen, X.; Yang, J.X.; Chen, Q.; Song, T.Y.; Ge, J.Q. Evaluation of housekeeping genes as references for quantitative real-time PCR analysis of European eel, Anguilla anguilla. J. Fish Biol. 2023, 102, 141–154. [Google Scholar] [CrossRef] [PubMed]
- De Kretser, D.M.; Loveland, K.L.; Meinhardt, A.; Simorangkir, D.; Wreford, N. Spermatogenesis. Hum. Reprod. 1998, 13, 1–8. [Google Scholar] [CrossRef]
- Fu, S.Y.; Jiang, J.H.; Yang, W.X.; Zhu, J.Q. A histological study of testis development and ultrastructural features of spermatogenesis in cultured Acrossocheilus fasciatus. Tissue Cell 2016, 48, 49–62. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, D.; Guo, C.; Wang, Y.; Xu, S. The gonadal development and changes of serum steroid levels ofcultured Pampus argenteus. J. Fish. China 2017, 41, 198–211. [Google Scholar]
- Schulz, R.W.; de França, L.R.; Lareyre, J.J.; Le Gac, F.; Chiarini-Garcia, H.; Nobrega, R.H.; Miura, T. Spermatogenesis in fish. Gen. Comp. Endocrinol. 2010, 165, 390–411. [Google Scholar] [CrossRef] [PubMed]
- Uribe, M.C.; Grier, H.J.; Mejía-Roa, V. Comparative testicular structure and spermatogenesis in bony fishes. Spermatogenesis 2015, 4, e983400. [Google Scholar] [CrossRef] [PubMed]
- Maclean II, J.A.; Wilkinson, M.F. Gene regulation in spermatogenesis. Curr. Top. Dev. Biol. 2005, 71, 131–197. [Google Scholar]
- White-Cooper, H. Molecular mechanisms of gene regulation during Drosophila spermatogenesis. Reproduction 2010, 139, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Chen, W.; Cheng, Z.; Wu, S.; He, J.; Han, L.; He, Z.; Qin, W. Novel gene regulation in normal and abnormal spermatogenesis. Cells 2021, 10, 666. [Google Scholar] [CrossRef]
- Zhang, D.D.; Gao, X.M.; Zhao, Y.Q.; Hou, C.C.; Zhu, J.Q. The C-terminal kinesin motor KIFC1 may participate in nuclear reshaping and flagellum formation during spermiogenesis of Larimichthys crocea. Fish Physiol. Biochem. 2017, 43, 1351–1371. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Gao, X.; Wang, J.; Du, C.; Hou, C.; Xie, Q.; Lou, B.; Liu, F.; Zhu, J. KIFC1 functions in nuclear reshaping and midpiece formation during the spermatogenesis of small yellow croaker Larimichthys polyactis. Anim. Reprod. Sci. 2021, 226, 106702. [Google Scholar] [CrossRef]
- Zhou, X.; Wu, X.; Chu, M.; Liang, C.; Ding, X.; Pei, J.; Xiong, L.; Bao, P.; Guo, X.; Yan, P. Validation of suitable reference genes for gene expression studies on yak testis development. Animals 2020, 10, 182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.B.; Shen, X.; Li, X.J.; Tian, Y.B.; Ouyang, H.J.; Huang, Y.M. Reference gene selection for expression studies in the reproductive axis tissues of Magang geese at different reproductive stages under light treatment. Sci. Rep. 2021, 11, 7573. [Google Scholar] [CrossRef]
- Li, J.; Luo, X.; Li, J.; Jin, G.; Xia, D.; Qi, Z.; Liu, Z.; Liu, Y.; Zhao, L. Evaluation of nutritive quality and nutrient components in the muscle of Hemibarbus labeo Pallas. J. Shenyang Agric. Univ. 2011, 42, 59–64. [Google Scholar]
- Xu, W.; Li, C.T.; Gao, D.C.; Geng, L.W. Scale and growth characteristics of Hemibarbus labeo in the Wusulijiang River. Chin. J. Zool. 2008, 43, 108–112. [Google Scholar]
- Huang, J.S.; Wang, Y.; Chen, W.; Yang, Y. Effects of animal and plant protein percentage on growth performance and nutrient excretion of Hemibarbus labeo. Feed Ind. 2014, 35, 125–128. [Google Scholar]
- Yu, J.; Guo, L.; Zhang, S.H.; Zhu, Q.Y.; Chen, R.Y.; Wong, B.H.; Ding, G.H.; Chen, J. Transcriptomic analysis of intermuscular bone development in barbel steed (Hemibarbus labeo). Comp. Biochem. Phys. D 2022, 44, 101030. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jiang, W.; Xu, Y.W.; Chen, R.Y.; Xu, Q. Sequence analysis of hepcidin in barbel steed (Hemibarbus labeo): QSHLS motif confers hepcidin iron-regulatory activity but limits its antibacterial activity. Dev. Comp. Immunol. 2021, 114, 103845. [Google Scholar] [CrossRef]
- Lan, Z.; Fan, M.; Huang, X.; Zhao, J. Population diversity and phylogeography of Hemibarbus labeo and Hemibarbus medius in South China. Acta Ecol. Sin. 2016, 36, 6091–6102. [Google Scholar]
- Sun, Y.; Wang, Q.; Ren, J.; Yang, S. Identification and phylogenetic analysis of the mitochondrial genome of Hemibarbus labeo BML (Cypriniformes: Cyprinidae). Mitochondrial DNA B 2020, 5, 2655–2657. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]
- Jiang, B.; Li, Q.; Zhang, Z.; Huang, Y.; Wu, Y.; Li, X.; Huang, M.; Huang, Y.; Jian, J. Selection and evaluation of stable reference genes for quantitative real-time PCR in the head kidney leukocyte of Oreochromis niloticus. Aquacult. Rep. 2023, 31, 101660. [Google Scholar] [CrossRef]
- Hirokawa, N.; Tanaka, Y. Kinesin superfamily proteins (KIFs): Various functions and their relevance for important phenomena in life and diseases. Exp. Cell Res. 2015, 334, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Cross, R.A.; McAinsh, A. Prime movers: The mechanochemistry of mitotic kinesins. Nat. Rev. Mol. Cell Biol. 2014, 15, 257–271. [Google Scholar] [CrossRef]
- She, Z.Y.; Yang, W.X. Molecular mechanisms of kinesin-14 motors in spindle assembly and chromosome segregation. J. Cell Sci. 2017, 130, 2097–2110. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.D.; Wang, D.H.; Yang, W.X. Kinesins in spermatogenesis. Biol. Reprod. 2017, 96, 267–276. [Google Scholar] [CrossRef]
- Yang, W.X.; Sperry, A.O. C-terminal kinesin motor KIFC1 participates in acrosome biogenesis and vesicle transport. Biol. Reprod. 2003, 69, 1719–1729. [Google Scholar] [CrossRef]
- Yang, W.X.; Jefferson, H.; Sperry, A.O. The molecular motor KIFC1 associates with a complex containing nucleoporin NUP62 that is regulated during development and by the small GTPase RAN. Biol. Reprod. 2006, 74, 684–690. [Google Scholar] [CrossRef]
- Wang, W.; Zhu, J.Q.; Yu, H.M.; Tan, F.Q.; Yang, W.X. KIFC1-like motor protein associates with the cephalopod manchette and participates in sperm nuclear morphogenesis in Octopus tankahkeei. PLoS ONE 2010, 5, e15616. [Google Scholar] [CrossRef][Green Version]
- Lehti, M.S.; Sironen, A. Formation and function of the manchette and flagellum during spermatogenesis. Reproduction 2016, 151, R43–R54. [Google Scholar] [CrossRef]
- Ercolani, L.; Florence, B.; Denaro, M.; Alexander, M. Isolation and complete sequence of a functional human glyceraldehyde-3-phosphate dehydrogenase gene. J. Biol. Chem. 1988, 263, 15335–15341. [Google Scholar] [CrossRef]
- Tisdale, E.J. Glyceraldehyde-3-phosphate dehydrogenase is phosphorylated by protein kinase Ciota/lambda and plays a role in microtubule dynamics in the early secretory pathway. J. Biol. Chem. 2002, 277, 3334–3341. [Google Scholar] [CrossRef] [PubMed]
- Kanatsu-Shinohara, M.; Tanaka, T.; Ogonuki, N.; Ogura, A.; Morimoto, H.; Cheng, P.F.; Eisenman, R.N.; Trumpp, A.; Shinohara, T. Myc/Mycn-mediated glycolysis enhances mouse spermatogonial stem cell self-renewal. Genes Dev. 2016, 30, 2637–2648. [Google Scholar] [CrossRef] [PubMed]
- Helsel, A.R.; Oatley, M.J.; Oatley, J.M. Glycolysis-optimized conditions enhance maintenance of regenerative integrity in mouse spermatogonial stem cells during long-term culture. Stem Cell Rep. 2017, 8, 1430–1441. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhang, Z.; Chang, C.; Yang, Z.; Wang, P.; Fu, H.; Wei, X.; Chen, E.; Tan, S.; Huang, W.; et al. A bioenergetic shift is required for spermatogonial differentiation. Cell Discov. 2020, 6, 56. [Google Scholar] [CrossRef]


| Gene | GenBank Accession Number |
|---|---|
| glyceraldehyde-3-phosphate dehydrogenase (gapdh) | PP530450 |
| beta-actin (actb) | PP530451 |
| beta-tubulin (tubb4b) | PP530452 |
| elongation factor 1-alpha (eef1a) | PP530453 |
| 40S ribosomal protein S27 (rps27) | PP530455 |
| 40S ribosomal protein S4 (rps4) | PP530454 |
| 18s rRNA small subunit ribosomal RNA (18s) | PP527010 |
| small nuclear ribonucleoprotein D1 polypeptide (snrpd1) | PP530456 |
| ribosomal protein L7-like protein 1 (rpl7l1) | PP530457 |
| kinesin family member C1 (kifc1) | PP946169 |
| Primer Name | Sequence (5′ to 3′) | Dosage (µL) 1 | Amplification Size (bp) | PCR Amplification Efficiency | Correlation Coefficients |
|---|---|---|---|---|---|
| gapdhF | CCGTGCTGCTATCCAGTCCAAGA | 0.3 | 140 | 103.7% | 0.990 |
| gapdhR | TGCCGCCTTCTGCCTTAACCT | 0.3 | |||
| actbF | ATGGTATCGTGATGGACTCTGGTGAT | 0.6 | 162 | 101.4% | 0.999 |
| actbR | TGGTGGTGAAGCTGTAGCCTCTC | 0.6 | |||
| tubb4bF | GCCGTATGTCCATGAAGGAGGTG | 0.6 | 163 | 100.2% | 0.999 |
| tubb4bR | GCTGTGCTATTGCCGATGAAGGT | 0.6 | |||
| eef1aF | ACTGCCACACTGCTCACATTGC | 1.0 | 221 | 95.4% | 0.997 |
| eef1aR | AACAGCGACGGTCTGCCTCAT | 1.0 | |||
| rpl7l1F | CACTCATAGAGCAGCATCTTGGACAA | 0.3 | 125 | 97.9% | 0.993 |
| rpl7l1R | TGACAAATGGAACGGCAACAGGAA | 0.3 | |||
| rps27F | GGAGAGGAGGAGGCACAAGAAGAA | 0.5 | 146 | 94.8% | 0.994 |
| rps27R | GCACAGTTGAGCAACCGACACA | 0.5 | |||
| rps4F | CGAGGACCGAAGAAGCATCTGAAG | 0.5 | 232 | 102.1% | 0.999 |
| rps4R | CATCAGTGCGGACCTTGCCATC | 0.5 | |||
| 18sF | GGACACGGAAAGGATTGACAGATTGA | 1.0 | 120 | 99.4% | 0.999 |
| 18sR | CGGAGTCTCGTTCGTTATCGGAATG | 1.0 | |||
| snrpd1F | AACCGTCACCATTGAGCTGAAGAAT | 0.8 | 173 | 108.7% | 1.000 |
| snrpd1R | GCAGGATGAAGTAGCGGATGTTGT | 0.8 | |||
| kifc1F | GCAGCGGGAAGACCTTTACTATGG | 0.6 | 112 | 106.1% | 0.999 |
| kifc1R | CCTTGCTCTCGGAGTGCTTTGG | 0.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Liu, S.; Lv, Y.; Dai, Q.; Zhu, L.; Hu, Z.; Lu, J.; Zhou, H.; Jin, J. Screening of Reference Genes for Quantitative Real-Time PCR Analysis in Tissues and during Testis Development, and Application to Analyze the Expression of kifc1 in Hemibarbus labeo (Teleostei, Cypriniformes, Cyprinidae). Animals 2024, 14, 2006. https://doi.org/10.3390/ani14132006
Gao X, Liu S, Lv Y, Dai Q, Zhu L, Hu Z, Lu J, Zhou H, Jin J. Screening of Reference Genes for Quantitative Real-Time PCR Analysis in Tissues and during Testis Development, and Application to Analyze the Expression of kifc1 in Hemibarbus labeo (Teleostei, Cypriniformes, Cyprinidae). Animals. 2024; 14(13):2006. https://doi.org/10.3390/ani14132006
Chicago/Turabian StyleGao, Xinming, Siqi Liu, Yaoping Lv, Qingmin Dai, Ling Zhu, Zehui Hu, Junkai Lu, Haidong Zhou, and Jing Jin. 2024. "Screening of Reference Genes for Quantitative Real-Time PCR Analysis in Tissues and during Testis Development, and Application to Analyze the Expression of kifc1 in Hemibarbus labeo (Teleostei, Cypriniformes, Cyprinidae)" Animals 14, no. 13: 2006. https://doi.org/10.3390/ani14132006
APA StyleGao, X., Liu, S., Lv, Y., Dai, Q., Zhu, L., Hu, Z., Lu, J., Zhou, H., & Jin, J. (2024). Screening of Reference Genes for Quantitative Real-Time PCR Analysis in Tissues and during Testis Development, and Application to Analyze the Expression of kifc1 in Hemibarbus labeo (Teleostei, Cypriniformes, Cyprinidae). Animals, 14(13), 2006. https://doi.org/10.3390/ani14132006




