Integrative Meta-Analysis: Unveiling Genetic Factors in Meat Sheep Growth and Muscular Development through QTL and Transcriptome Studies
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
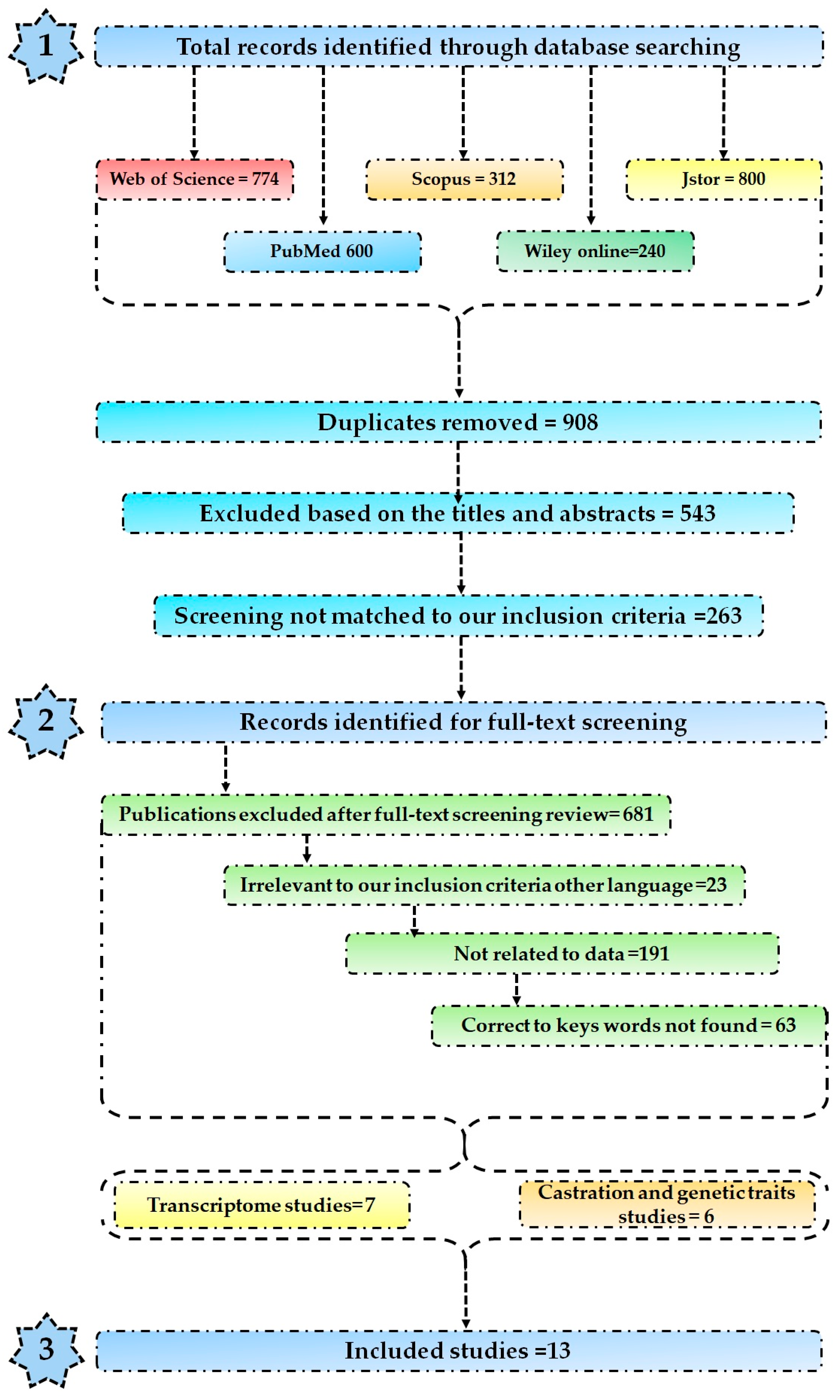
2.1. Selection Criteria
2.2. RNA Sequence Dataset
2.3. Differential Expression Analysis
2.4. Performance Measurement and Sampling
2.4.1. Collecting Data
2.4.2. Fisher’s Z Transformation
2.4.3. Heritability Estimates
2.4.4. Assessment of Heterogeneity
2.4.5. Differential Expression Gene Analysis
2.5. The RRA Methods
2.6. Gene Classification
2.7. Candidate Genes
2.8. Consistent Gene Selection
2.9. GO and KEGG Analyses Were Performed on Differentially Expressed mRNA
2.10. Protein–Protein Interaction (PPI) and Subcellular Localization of Differentially Expressed mRNAs
2.11. QTL Enrichment Analysis
3. Results and Discussion
3.1. Effects of Castration of Male Sheep on Performance, Carcass Characteristics, and Meat Quality
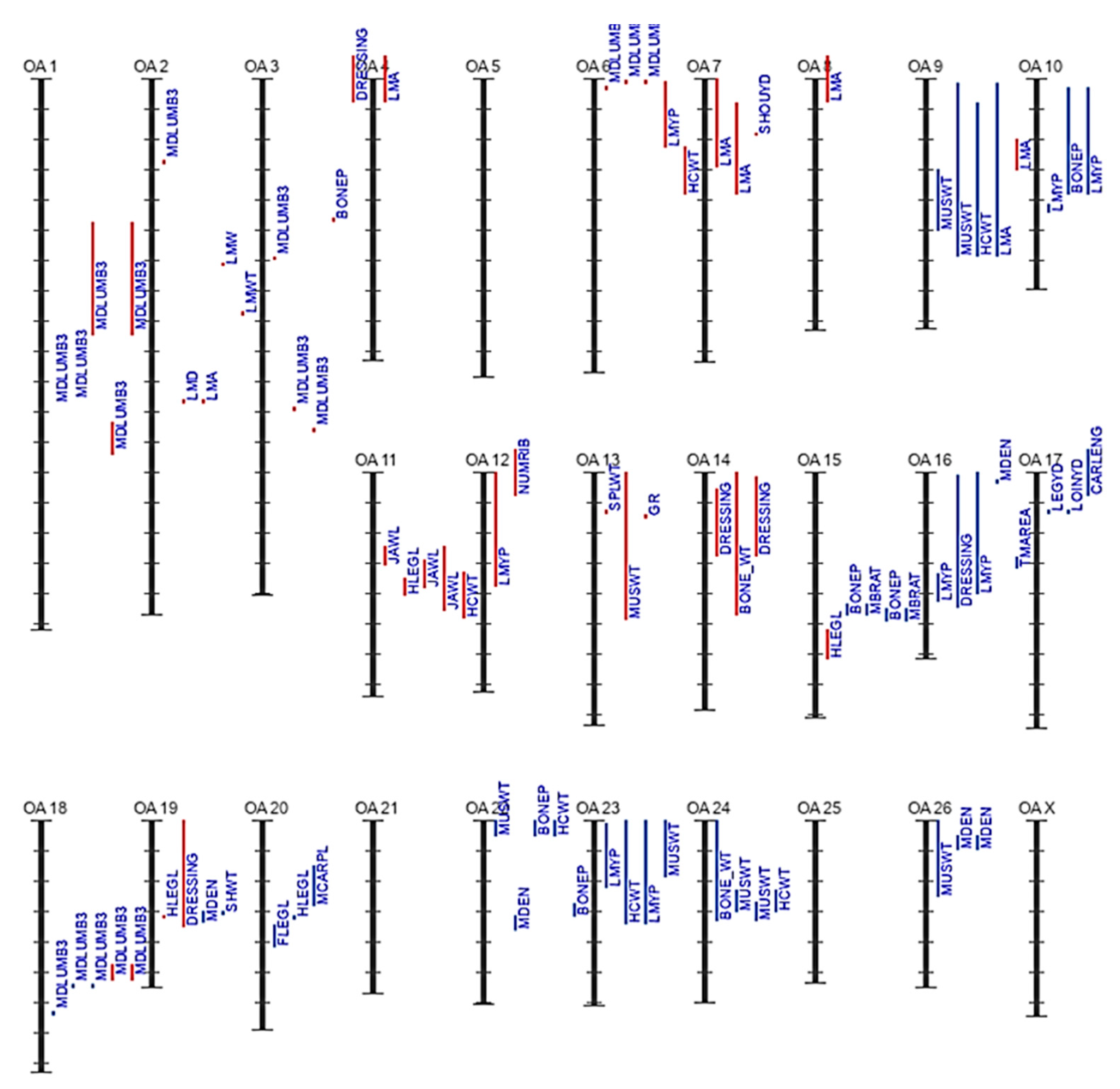
3.2. Genetic Background of Meat Production and Quality in Sheep
Top Sheep QTLs Associated with Meat Production and Quality in the 2022–2023 Database
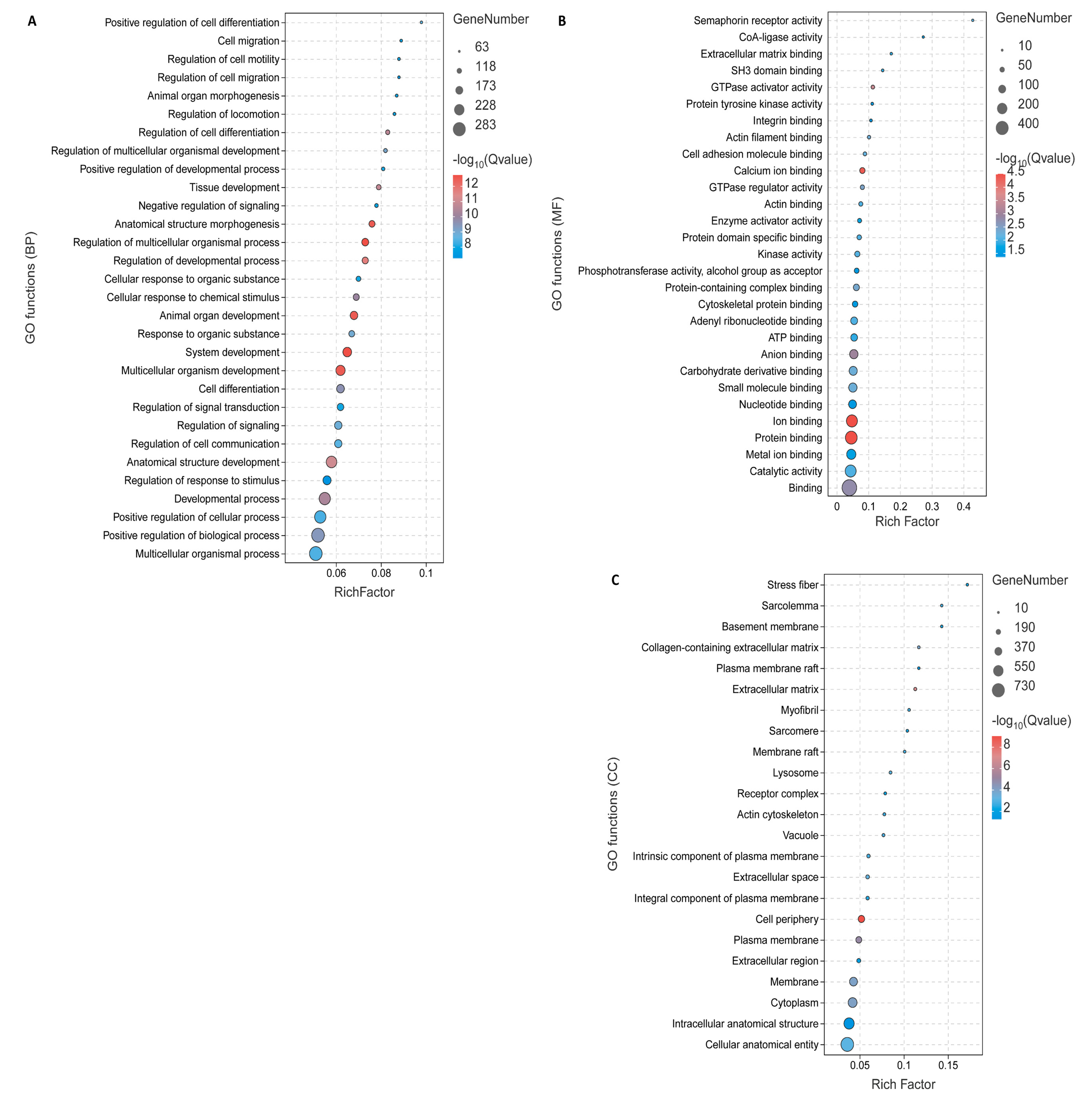
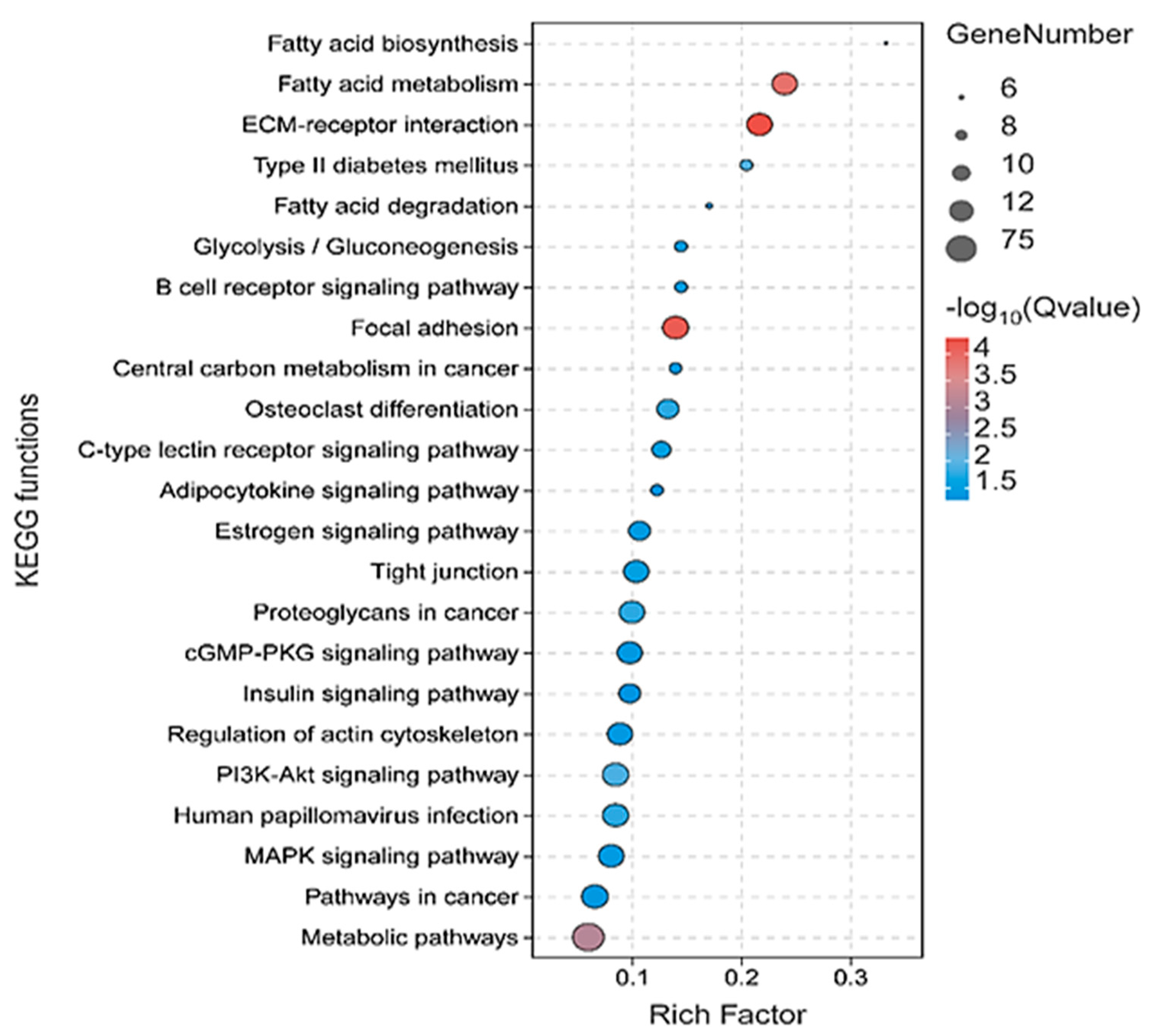
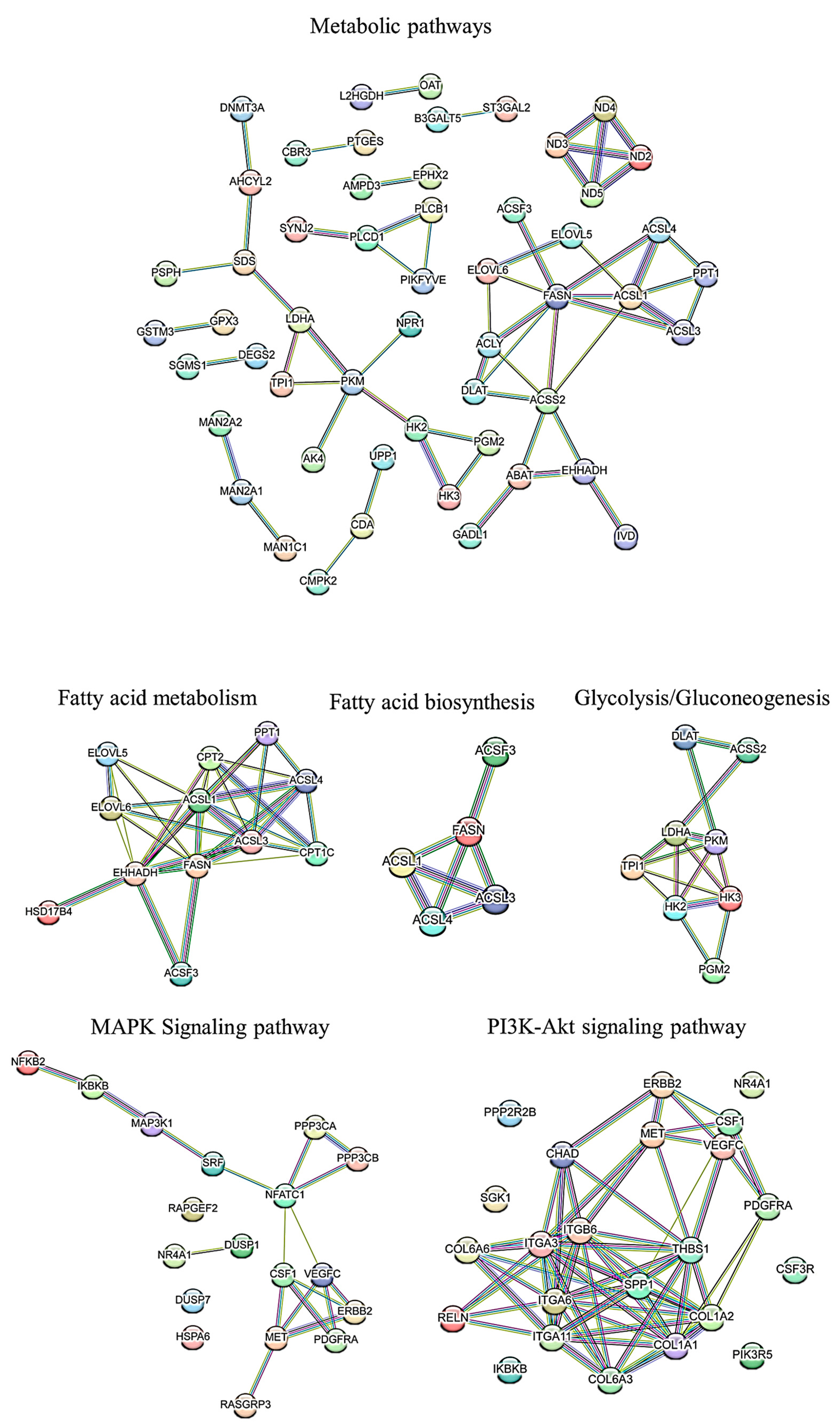
3.3. Functional Enrichment Analysis
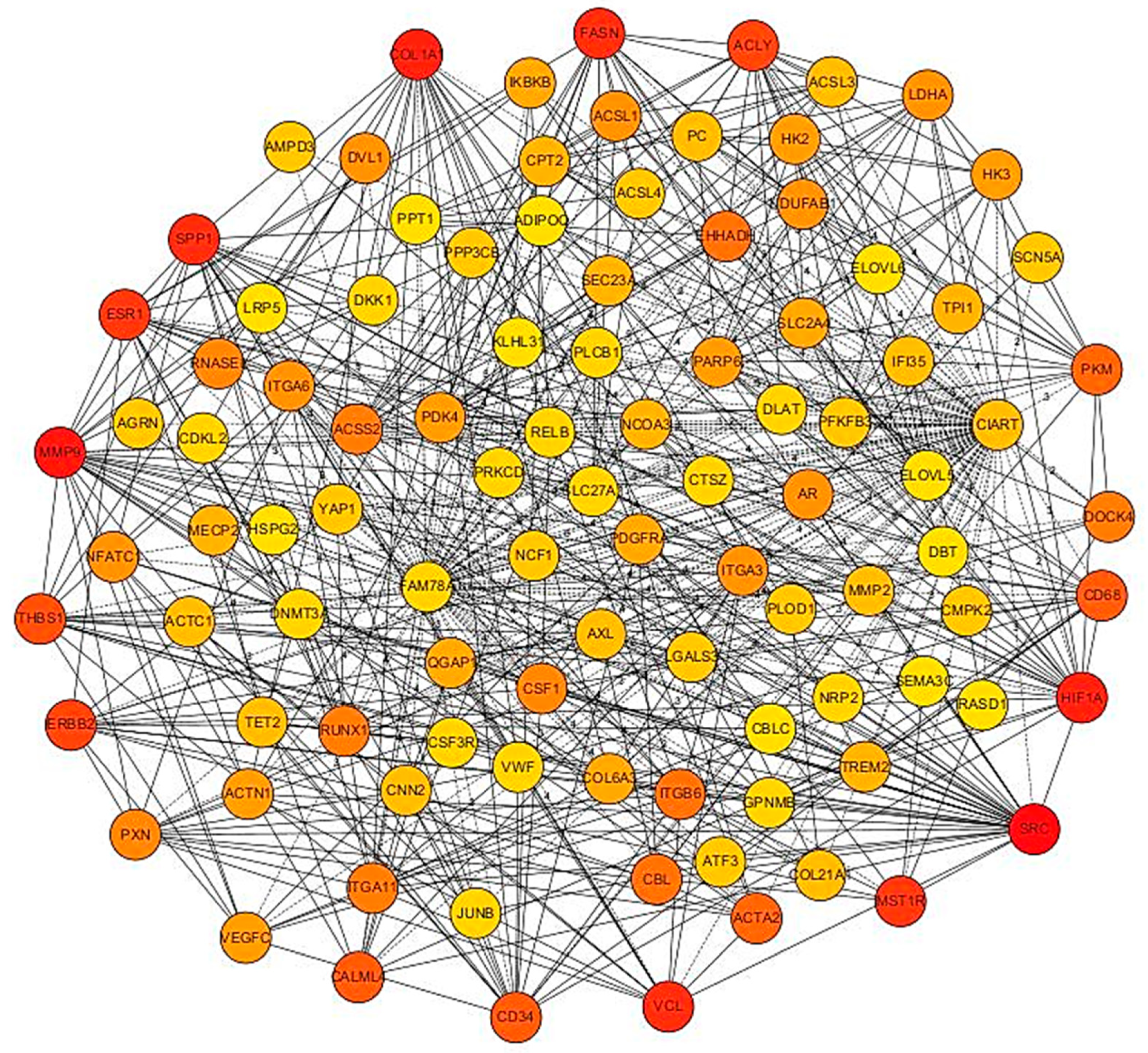
3.4. PPI Network Analysis
3.5. PPI Network of Top 100 Genes Associated with Growth Muscle
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abouheif, M.; Al-Owaimer, A.; Kraidees, M.; Metwally, H.; Shafey, T. Effect of restricted feeding and realimentation on feed performance and carcass characteristics of growing lambs. Rev. Bras. Zootec. 2013, 42, 95–101. [Google Scholar] [CrossRef]
- Henchion, M.; McCarthy, M.; Resconi, V.C.; Troy, D. Meat consumption: Trends and quality matters. Meat Sci. 2014, 98, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Francisco, A.; Dentinho, M.; Alves, S.; Portugal, P.; Fernandes, F.; Sengo, S.; Jerónimo, E.; Oliveira, M.; Costa, P.; Sequeira, A. Growth performance, carcass and meat quality of lambs supplemented with increasing levels of a tanniferous bush (Cistus ladanifer L.) and vegetable oils. Meat Sci. 2015, 100, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.A.; Hammoud, M.; Dabour, N.A.; Hafez, E.; Sharaby, M.A. BMPR-1B, BMP-15 and GDF-9 genes structure and their relationship with litter size in six sheep breeds reared in Egypt. BMC Res. Notes 2020, 13, 215. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, D.; Fogarty, N.; Mortimer, S. Genetic related effects on sheep meat quality. Small Rumin. Res. 2011, 101, 160–172. [Google Scholar] [CrossRef]
- Mekuriaw, S.; Haile, A. Genetic parameter estimates for growth and reproductive trait of sheep for genetic improvement and designing breeding program in Ethiopia: A review. Open Access Libr. J. 2014, 1, 1–10. [Google Scholar] [CrossRef]
- Roudbar, M.A.; Mohammadabadi, M.; Mehrgardi, A.A.; Abdollahi-Arpanahi, R. Estimates of variance components due to parent-of-origin effects for body weight in Iran-Black sheep. Small Rumin. Res. 2017, 149, 1–5. [Google Scholar] [CrossRef]
- Saleh, A.A.; Rashad, A.M.; Hassanine, N.N.; Sharaby, M.A.; Sallam, S.M. History of the Goat and Modern versus Old Strategies to enhance the genetic performance. In Goat Science—From Keeping to Precision Production; IntechOpen: London, UK, 2023. [Google Scholar]
- Saleh, A.A.; Easa, A.A.; El-Hedainy, D.K.; Rashad, A.M. Prediction of some milk production traits using udder and teat measurements with a spotlight on their genetic background in Friesian cows. Sci. Rep. 2023, 13, 16193. [Google Scholar] [CrossRef]
- Arabpoor Raghabadi, Z.; Mohammadabadi, M.; Khezri, A. The expression pattern of p32 gene in femur, humeral muscle, back muscle and back fat tissues of Kermani lambs. Agric. Biotechnol. J. 2021, 13, 183–200. [Google Scholar]
- Akanno, E.; Schenkel, F.; Quinton, V.; Friendship, R.; Robinson, J. Meta-analysis of genetic parameter estimates for reproduction, growth and carcass traits of pigs in the tropics. Livest. Sci. 2013, 152, 101–113. [Google Scholar] [CrossRef]
- Bakhtiarizadeh, M.R.; Salehi, A.; Alamouti, A.A.; Abdollahi-Arpanahi, R.; Salami, S.A. Deep transcriptome analysis using RNA-Seq suggests novel insights into molecular aspects of fat-tail metabolism in sheep. Sci. Rep. 2019, 9, 9203. [Google Scholar] [CrossRef] [PubMed]
- Medrado, B.D.; Pedrosa, V.B.; Pinto, L.F.B. Meta-analysis of genetic parameters for economic traits in sheep. Livest. Sci. 2021, 247, 104477. [Google Scholar] [CrossRef]
- Ahmad, N.; Zhang, K.; Ma, J.; Yuan, M.; Zhao, S.; Wang, M.; Deng, L.; Ren, L.; Gangurde, S.S.; Pan, J. Transcriptional networks orchestrating red and pink testa color in peanut. BMC Plant Biol. 2023, 23, 44. [Google Scholar] [CrossRef] [PubMed]
- Raziq, A.; Zhang, K.; Sun, W.; Ahmad, N.; Zhao, H.; Raza, M.A.; Ahmed, S.; Din, A.M.U.; Zhao, S.; Pan, J. Transcriptome profiling of MYB-overexpressed transgenic lines provides crucial molecular insights into anthocyanin and remodel the biosynthesis regulatory network in Nicotiana tabacum. Ind. Crops Prod. 2024, 213, 118374. [Google Scholar] [CrossRef]
- Borenstein, M.; Hedges, L.V.; Higgins, J.P.; Rothstein, H.R. A basic introduction to fixed-effect and random-effects models for meta-analysis. Res. Synth. Methods 2010, 1, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, R.; Hu, X.; Li, N. Application of genomic technologies to the improvement of meat quality of farm animals. Meat Sci. 2007, 77, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Kroll, L.; Grooms, D.; Siegford, J.; Schweihofer, J.; Metz, K.; Rust, S. Effects of tail docking on health and performance of beef cattle in confined, slatted-floor feedlots. J. Anim. Sci. 2014, 92, 4108–4114. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, P.; Escribano, M.; Mesías, F.; De Ledesma, A.R.; Pulido, F. Sheep farms in the Spanish rangelands (dehesas): Typologies according to livestock management and economic indicators. Small Rumin. Res. 2008, 74, 52–63. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, M.; Wang, J.; Wang, S.; Wang, X.; Yang, J.; Gao, L.; Gan, S. Comparative transcriptome analysis of key genes and pathways activated in response to fat deposition in two sheep breeds with distinct tail phenotype. Front. Genet. 2021, 12, 639030. [Google Scholar] [CrossRef]
- Cheng, S.; Wang, X.; Wang, Q.; Yang, L.; Shi, J.; Zhang, Q. Comparative analysis of Longissimus dorsi tissue from two sheep groups identifies differentially expressed genes related to growth, development and meat quality. Genomics 2020, 112, 3322–3330. [Google Scholar] [CrossRef]
- Sun, L.; Bai, M.; Xiang, L.; Zhang, G.; Ma, W.; Jiang, H. Comparative transcriptome profiling of longissimus muscle tissues from Qianhua Mutton Merino and Small Tail Han sheep. Sci. Rep. 2016, 6, 33586. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, Q.; Huang, X.; Qu, C.; Ma, S.; Mao, Y.; Yang, J.; Li, Y.; Li, Y.; Tan, C. Mutation screening in genes known to be responsible for Retinitis Pigmentosa in 98 Small Han Chinese Families. Sci. Rep. 2017, 7, 1948. [Google Scholar] [CrossRef]
- Liu, N.; He, J.; Yu, W.; Liu, K.; Cheng, M.; Liu, J.; He, Y.; Zhao, J.; Qu, X. Transcriptome analysis of skeletal muscle at prenatal stages in Polled Dorset versus Small-tailed Han sheep. Genet. Mol. Res. 2015, 14, 1085–1095. [Google Scholar] [CrossRef]
- Mukherjee, S.; Mukherjee, A.; Jasrotia, R.S.; Jaiswal, S.; Iquebal, M.; Longkumer, I.; Mech, M.; Vüpru, K.; Khate, K.; Rajkhowa, C. Muscle transcriptome signature and gene regulatory network analysis in two divergent lines of a hilly bovine species Mithun (Bos frontalis). Genomics 2020, 112, 252–262. [Google Scholar] [CrossRef]
- Cui, R.; Kang, X.; Liu, Y.; Liu, X.; Chan, S.; Wang, Y.; Li, Z.; Ling, Y.; Feng, D.; Li, M.; et al. Integrated analysis of the whole transcriptome of skeletal muscle reveals the ceRNA regulatory network related to the formation of muscle fibers in Tan sheep. Front. Genet. 2022, 13, 991606. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Siddaraju, N.K.; Manjunatha, S.; Sudarshan, S.; Fairoze, M.N.; Kumar, A.; Chhabra, P.; Kaur, M.; Sreesujatha, R.; Ahlawat, S. Muscle transcriptome provides the first insight into the dynamics of gene expression with progression of age in sheep. Sci. Rep. 2021, 11, 22360. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Borenstein, M.; Rothstein, H. Comprehensive Meta-Analysis; Biostat: Englewood Cliffs, NJ, USA, 1999. [Google Scholar]
- Borenstein, M.; Hedges, L.V.; Higgins, J.P.T.; Rothstein, H.R. Comprehensive Meta-Analysis, version 2.2.027; Computer Software; Biostat: Englewood, NJ, USA, 2006.
- de Oliveira, H.R.; Ventura, H.T.; Costa, E.V.; Pereira, M.A.; Veroneze, R.; de Souza Duarte, M.; e Silva, F.F. Meta-analysis of genetic-parameter estimates for reproduction, growth and carcass traits in Nellore cattle by using a random-effects model. Anim. Prod. Sci. 2017, 58, 1575–1583. [Google Scholar] [CrossRef]
- Ravi, S. Book review: Methods for meta-analysis in medical research. Stat. Methods Med. Res. 2005, 14, 319–320. [Google Scholar] [CrossRef]
- Higgins, J.P.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef]
- Cochran, W.G. The combination of estimates from different experiments. Biometrics 1954, 10, 101–129. [Google Scholar] [CrossRef]
- Ma, X.; Liu, A.; Tian, S. A meta-analysis of mRNA expression profiling studies in sheep with different FecB genotypes. Anim. Genet. 2023, 54, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Salehian-Dehkordi, H.; Xu, Y.-X.; Xu, S.-S.; Li, X.; Luo, L.-Y.; Liu, Y.-J.; Wang, D.-F.; Cao, Y.-H.; Shen, M.; Gao, L. Genome-wide detection of copy number variations and their association with distinct phenotypes in the world’s sheep. Front. Genet. 2021, 12, 670582. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.F.; Bakhtiarizadeh, M.R.; Salehi, A. Meta-analysis of RNA-Seq datasets highlights novel genes/pathways involved in fat deposition in fat-tail of sheep. Front. Vet. Sci. 2023, 10, 1159921. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Ordóñes, S.; Domínguez-Díaz, D.; Salmerón-Zamora, J.J.; Villalobos-Villalobos, G.; Ortega-Gutiérrez, J.A. Producción y calidad del forraje de variedades de avena en función del sistema de siembra y de la etapa de madurez al corte. Rev. Fitotec. Mex. 2013, 36, 395–403. [Google Scholar] [CrossRef]
- Zhang, M.; Su, R.; Corazzin, M.; Hou, R.; Zhang, Y.; Sun, L.; Hu, G.; Dou, L.; Guo, Y.; Su, L. Lipid transformation during postmortem chilled aging in Mongolian sheep using lipidomics. Food Chem. 2023, 405, 134882. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, G.; Wang, J.; Ji, Z.; Liu, Z.; Pi, X.; Chen, C. Characterization and comparative analyses of muscle transcriptomes in Dorper and small-tailed Han sheep using RNA-Seq technique. PLoS ONE 2013, 8, e72686. [Google Scholar] [CrossRef]
- Sun, L.; Jiang, H. Research on meat quality of Qianhua Mutton Merino sheep and Small-tail Han sheep. Open Life Sci. 2022, 17, 1315–1323. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Ahlawat, S.; Sharma, R. Unique attributes of indigenous AnGR-A case study on Bandur sheep. Adv. Manag. Anim. Genet. Resour. 2022, 87–91. [Google Scholar]
- Shabbir, S.; Boruah, P.; Xie, L.; Kulyar, M.F.-E.-A.; Nawaz, M.; Yousuf, S.; Liu, T.; Jabeen, F.; Miao, X. Genome-wide transcriptome profiling uncovers differential miRNAs and lncRNAs in ovaries of Hu sheep at different developmental stages. Sci. Rep. 2021, 11, 5865. [Google Scholar] [CrossRef]
- Claffey, N.A.; Fahey, A.G.; Gkarane, V.; Moloney, A.P.; Monahan, F.J.; Diskin, M.G. Effect of breed and castration on production and carcass traits of male lambs following an intensive finishing period. Transl. Anim. Sci. 2018, 2, 407–418. [Google Scholar] [CrossRef]
- Gkarane, V.; Allen, P.; Gravador, R.S.; Diskin, M.G.; Claffey, N.A.; Fahey, A.G.; Brunton, N.P.; Farmer, L.J.; Moloney, A.P.; Monahan, F.J. Effect of castration and age at slaughter on sensory perception of lamb meat. Small Rumin. Res. 2017, 157, 65–74. [Google Scholar] [CrossRef]
- Gashu, M.; Urge, M.; Animut, G.; Tadesse, D. Feedlot Comparative Fattening, Nutrient Utilization and Profitability Evaluation of Intact and Castrated Washera Sheep. East Afr. J. Vet. Anim. Sci. 2017, 1, 111–118. [Google Scholar]
- Polidori, P.; Pucciarelli, S.; Cammertoni, N.; Polzonetti, V.; Vincenzetti, S. The effects of slaughter age on carcass and meat quality of Fabrianese lambs. Small Rumin. Res. 2017, 155, 12–15. [Google Scholar] [CrossRef]
- De Brito, G.F.; Holman, B.W.; McGrath, S.R.; Friend, M.A.; Van de Ven, R.; Hopkins, D.L. The effect of forage-types on the fatty acid profile, lipid and protein oxidation, and retail colour stability of muscles from White Dorper lambs. Meat Sci. 2017, 130, 81–90. [Google Scholar] [CrossRef]
- Amad, A.A.; Badeer, M.; Zentek, J. Effects of Feeding Systems and Castration on the Growth Performance and some Carcass Traits in Lambs of Dhamari sheep-Yemen. Arab. J. Agric. Sci. 2023, 6, 105–132. [Google Scholar] [CrossRef]
- Diogénes, L.; Medeiros, E.d.S.; Moura, J.F.; Edvan, R.; de Oliveira, J.; Silva Filho, E.; Silva, A.d.A.; Araujo, M.; Oliveira, R.; Bezerra, L. Cactus pear mucilage processing enhances corn silage’s quality, ruminal degradability, goat kids’ performance, carcass traits, and dietary economic profitability. Small Rumin. Res. 2023, 227, 107083. [Google Scholar] [CrossRef]
- Carrillo-Muro, O.; Rivera-Villegas, A.; Hernandez-Briano, P.; Lopez-Carlos, M.A.; Castro-Perez, B.I. Effect of Dietary Calcium Propionate Inclusion Period on the Growth Performance, Carcass Characteristics, and Meat Quality of Feedlot Ram Lambs. Agriculture 2023, 13, 1577. [Google Scholar] [CrossRef]
- Raja, K.N.; Sonika, A.; Sagar, S.D.; Mahesh, S.D.; Mishra, B.P. Advances in Management of Animal Genetic Resources; Hyderabad National Institute of Agricultural Extension Management (MANAGE): Telangana, India; ICAR-National Bureau of Animal Genetic Resources: Karnal, India, 2022; Volume 132001, pp. 1–187. [Google Scholar]
- Ali, A.S.; Atta, M.; Mohamed, M.B.; Fatnassi, M.; Al-Dosari, H.M.; Al-Shamari, H.S. Effect of feeding on different levels of crushed cactus in total mixed rations on fattening performance, carcass characteristics, and meat quality of Awassi sheep: Cactus feeding in sheep and meat quality. Lett. Anim. Biol. 2023, 3, 6–12. [Google Scholar] [CrossRef]
- Hassan, C.S.; Alkass, J.; Baker, I. Effect of sex and slaughter weight on meat quality of black goat and meriz kids. Iraqi J. Agric. Sci. 2023, 54, 784–791. [Google Scholar] [CrossRef]
- Donaldson, C.L. Spine Characteristics in Sheep: Metrology, Relationship to Meat Yield and Their Genetic Parameters. Ph.D. Thesis, The University of Edinburgh, Edinburgh, UK, 2016. [Google Scholar]
- Yoshida, K.; Morishima, Y.; Ano, S.; Sakurai, H.; Kuramoto, K.; Tsunoda, Y.; Yazaki, K.; Nakajima, M.; Sherpa, M.T.; Matsuyama, M. ELOVL6 deficiency aggravates allergic airway inflammation through the ceramide-S1P pathway in mice. J. Allergy Clin. Immunol. 2023, 151, 1067–1080.e1069. [Google Scholar] [CrossRef] [PubMed]
- Kelly, K.L.; Reagan, W.J.; Sonnenberg, G.E.; Clasquin, M.; Hales, K.; Asano, S.; Amor, P.A.; Carvajal-Gonzalez, S.; Shirai, N.; Matthews, M.D. De novo lipogenesis is essential for platelet production in humans. Nat. Metab. 2020, 2, 1163–1178. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, X.; Cao, Y.; Xiao, C.; Liu, Y.; Jin, H.; Cao, Y. Effect of the ACAA1 gene on preadipocyte differentiation in sheep. Front. Genet. 2021, 12, 649140. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, X.; Yang, Z.; Zhang, Q.; Hao, W.; Pang, Y.; Zhang, D.; Liu, D. Transcriptional Regulation Associated with Subcutaneous Adipogenesis in Porcine ACSL1 Gene. Biomolecules 2023, 13, 1057. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Guo, T.; Yue, Y.; Lu, Z.; Liu, J.; Yuan, C.; Niu, C.; Yang, M.; Yang, B. Quantitative proteomic analysis identified differentially expressed proteins with tail/rump fat deposition in Chinese thin-and fat-tailed lambs. PLoS ONE 2021, 16, e0246279. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Yu, Y.; Zhang, L.; Liu, Y.; Zheng, K.; Wang, S.; Jin, H.; Liu, L.; Cao, Y. Transcript variants of long-chain acyl-CoA synthase 1 have distinct roles in sheep lipid metabolism. Front. Genet. 2022, 13, 1021103. [Google Scholar] [CrossRef]
- Qiao, L.; Zhao, B.; Liu, X.; Liu, J.; Yang, K.; Pan, Y.; Liu, W. TPT1 promotes the adipogenic differentiation of stromal vascular fractions via the PI3K/AKT pathway and FOXO1 in sheep. J. Appl. Anim. Res. 2023, 51, 388–396. [Google Scholar] [CrossRef]
- Li, X.; Yao, X.; Bao, Y.; Deng, K.; Deng, M.; Yang, F.; Sun, X.; You, P.; Cai, Q.; Wang, F. PPP2R2A promotes testosterone secretion in Hu sheep Leydig cells via activation of the AKT/mTOR signaling pathway. J. Mol. Endocrinol. 2023, 17, 70. [Google Scholar] [CrossRef] [PubMed]
- Dudek, M.; Swift, J.; Meng, Q.-J. The circadian clock and extracellular matrix homeostasis in aging and age-related diseases. Am. J. Physiol.-Cell Physiol. 2023, 325, C52–C59. [Google Scholar] [CrossRef] [PubMed]
- de Frutos, S.; Griera, M.; Hatem-Vaquero, M.; Campillo, S.; Gutiérrez-Calabres, E.; García-Ayuso, D.; Pardo, M.; Calleros, L.; Rodríguez-Puyol, M.; Rodríguez-Puyol, D. The integrin beta1 modulator Tirofiban prevents adipogenesis and obesity by the overexpression of integrin-linked kinase: A pre-clinical approach in vitro and in vivo. Cell Biosci. 2022, 12, 10. [Google Scholar] [CrossRef] [PubMed]
- Karagöz, Z.; Rijns, L.; Dankers, P.Y.; van Griensven, M.; Carlier, A. Towards understanding the messengers of extracellular space: Computational models of outside-in integrin reaction networks. Comput. Struct. Biotechnol. J. 2021, 19, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Tegeler, A.P.; Fiallo-Diez, J.; Michelotti, T.; Flores, L.; Benitez, O.J.; Johnson, B.J.; Strieder-Barboza, C. 32 Depot-Specific Transcriptome and Adipocyte Metabolism in Intramuscular and Subcutaneous Adipose Tissue in Beef Cattle. J. Anim. Sci. 2023, 101, 16–17. [Google Scholar] [CrossRef]
- Li, X.; Bai, Y.; Li, J.; Chen, Z.; Ma, Y.; Shi, B.; Han, X.; Luo, Y.; Hu, J.; Wang, J. Transcriptional analysis of microRNAs related to unsaturated fatty acid synthesis by interfering bovine adipocyte ACSL1 gene. Front. Genet. 2022, 13, 994806. [Google Scholar] [CrossRef] [PubMed]
- Stahl, A. SLC27 family of fatty acid transporters in GtoPdb v. 2023.1. IUPHAR/BPS Guide Pharmacol. 2023. [Google Scholar] [CrossRef]
- Oh, J.; Kim, C.-S.; Kim, M.; Jo, W.; Sung, Y.H.; Park, J. Type VI collagen and its cleavage product, endotrophin, cooperatively regulate the adipogenic and lipolytic capacity of adipocytes. Metabolism 2021, 114, 154430. [Google Scholar] [CrossRef]
- Shang, K.; Ma, N.; Che, J.; Li, H.; Hu, J.; Sun, H.; Cao, B. SLC27A2 mediates FAO in colorectal cancer through nongenic crosstalk regulation of the PPARs pathway. BMC Cancer 2023, 23, 335. [Google Scholar] [CrossRef]
- Chen, H.-J.; Yan, X.-Y.; Sun, A.; Zhang, L.; Zhang, J.; Yan, Y.-E. Adipose extracellular matrix deposition is an indicator of obesity and metabolic disorders. J. Nutr. Biochem. 2023, 111, 109159. [Google Scholar] [CrossRef]
| Fat (Breeds) 1 | Thin (Breeds) 2 | Number of Replicates | Upregulated Genes 3 | Downregulated Genes 4 | Type of Clean Reads 5 | Gender 6 | References |
|---|---|---|---|---|---|---|---|
| STH 7 | PD 8 × STH | 100 | 457 | 417 | 10,278,304,671 |  | [21] |
| QHMM 9 | Merino × STH | 60 | 405 | 555 | 47,441,397 |  | [22] |
| Lori-Bakhtiari | Zel × Lori-Bakhtiari | 6 | 80 | 184 | 150 bp |  | [12] |
| Wagyu | Hostein × Wagyu | 98 | 321 | 341 | 129,097,834 |  | [23] |
| PD versus | STH× PD versus | 21 | 33 | 32 | 120 bp |  | [24] |
| India Bandur | Bandur | 20 | 98 | 43 | 93,434,064 |  | [27] |
| Gayal | Mithun/Gayal | 4 | 173 | 124 | 241,738,022 |  | [25] |
 : Male and
: Male and  : Female; 7 STH, Small Tail Han; 8 PD: Polled Dorset; 9 QHMM, Qianhua Mutton Merino sheep.
: Female; 7 STH, Small Tail Han; 8 PD: Polled Dorset; 9 QHMM, Qianhua Mutton Merino sheep.| Breed | Method of Castration | Age of Castration | Slaughter Age/Weight | Rams | Wethers | Response Variables | Country | Reference |
|---|---|---|---|---|---|---|---|---|
| MSB 1 × BB 2 | RRi 9 | 40 kg | 18 | 18 | IMF | Mexico | [41] | |
| Mongolian sheep | 3 m 10 | 3 m/40 kg | 3 | 4 | CW | China | [42] | |
| STS 3 × DP 4 | RRi | 11 m | 11 m/39 kg, 64 kg | 2 | 2 | China | [43] | |
| QHMM 5 × STH 6 | 1 y 11 | 1 y/55 kg, 45 kg | 30 | 30 | CW | China | [44] | |
| Bandur × local sheep | - | 2 m | 6 m/38 kg | 16 | - | CW | India | [45] |
| 40 h 12 | 8 m/49.86, 40.64 kg | 3 | - | China | [46] | |||
| Native sheep | surgical | 5 d 13 | 45 kg | 47.6 | 12 | DM, CP | Bangladesh | [47] |
| SB 7 × TXSB 8 | RRi | 48 h | 64 kg, 66 kg | 16.50 | 17.25 | DM | Ireland | |
| SB × TXSB | RRi | 50 h | 20.8 kg, 23.3 kg | 50 | 50 | IMF | Ireland | [48] |
| Merino | RRi | 11 weeks | 20.7 | 30 | - | CW | Australia | [47] |
| Washera sheep | RRi | 1 m 15 d | 1 y/53.3, 53.9 kg | 12 | 12 | CW, DP | Ethiopia | [49] |
| Fabrianese | - | 5 d | 23, 37.4, 41 | 14 | 14 | DM | Italy | [50] |
| White Dorper | - | 24 h | 34.4 | 6 | 6 | DM | Australia | [51] |
| Dhamari sheep | - | 14 d | 23.7 | 3 | 3 | Yemen | [52] | |
| Boer goat | - | - | 30.4 | 32 | - | DM | Brazil | [53] |
| Doper lambs | - | 40.17 | 45 | - | DM | Mexico | [54] | |
| Santa Ines × Dorper | - | - | 28 d/31 kg | 72 | 36 | DM, CP, NDF | Brazil | [55] |
| Awassi sheep | - | - | 4 m/43.5 kg | 12 | - | CW | Qatar | [56] |
| Black goat | RRi | 24 h | 20 d/25 kg | - | 24 | DG, REA | Iraq | [57] |
| No. | Traits | Number of QTLs | |
|---|---|---|---|
| 3 | Body dimensions | Tail fat deposition | 64 |
| 4 | Body length | 8 | |
| 5 | Chest width | 7 | |
| 6 | Longissimus muscle area | 7 | |
| 7 | Shin circumference | 7 | |
| 8 | Backfat thickness | 18 | |
| 9 | Heart girth | 18 | |
| 10 | Body weight at hatch | 17 | |
| 11 | Withers height | 17 | |
| 12 | Chest depth | 16 | |
| 13 | Weight, Production and Quality | Body weight | 279 |
| 14 | Average daily gain | 95 | |
| 15 | Weaning weight | 4464 | |
| 16 | Birth weight | 4185 | |
| 17 | Body weight at 6 months | 2232 | |
| 18 | Yearling weight | 1674 | |
| 19 | Body weight at 8 weeks | 1116 | |
| 20 | Body weight at 20 weeks | 1116 | |
| 21 | Body weight at 180 days | 837 | |
| 22 | Body weight at 9 months | 837 | |
| 23 | Litter size (first parity) | 708 | |
| 24 | Slaughter weight | 558 | |
| 25 | Adult weight | 558 | |
| 26 | Growth | 589 | |
| 27 | Fatness | 174 | |
| 28 | Production | 610 | |
| 29 | Meat and carcass | 563 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehman, S.U.; Zhen, Y.; Ding, L.; Saleh, A.A.; Zhang, Y.; Zhang, J.; He, F.; Husien, H.M.; Zhou, P.; Wang, M. Integrative Meta-Analysis: Unveiling Genetic Factors in Meat Sheep Growth and Muscular Development through QTL and Transcriptome Studies. Animals 2024, 14, 1679. https://doi.org/10.3390/ani14111679
Rehman SU, Zhen Y, Ding L, Saleh AA, Zhang Y, Zhang J, He F, Husien HM, Zhou P, Wang M. Integrative Meta-Analysis: Unveiling Genetic Factors in Meat Sheep Growth and Muscular Development through QTL and Transcriptome Studies. Animals. 2024; 14(11):1679. https://doi.org/10.3390/ani14111679
Chicago/Turabian StyleRehman, Shahab Ur, Yongkang Zhen, Luoyang Ding, Ahmed A. Saleh, Yifan Zhang, Jinying Zhang, Feiyang He, Hosameldeen Mohamed Husien, Ping Zhou, and Mengzhi Wang. 2024. "Integrative Meta-Analysis: Unveiling Genetic Factors in Meat Sheep Growth and Muscular Development through QTL and Transcriptome Studies" Animals 14, no. 11: 1679. https://doi.org/10.3390/ani14111679
APA StyleRehman, S. U., Zhen, Y., Ding, L., Saleh, A. A., Zhang, Y., Zhang, J., He, F., Husien, H. M., Zhou, P., & Wang, M. (2024). Integrative Meta-Analysis: Unveiling Genetic Factors in Meat Sheep Growth and Muscular Development through QTL and Transcriptome Studies. Animals, 14(11), 1679. https://doi.org/10.3390/ani14111679








