Mitochondrial Protein-Coding Gene Expression in the Lizard Sphenomorphus incognitus (Squamata:Scincidae) Responding to Different Temperature Stresses
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Lizard Capture and Acclimatization
2.2. Experimental Design
2.3. DNA Extraction, PCR, and Sequencing
2.4. Mitochondrial Genome Localization and Sequence Analysis
2.5. RNA Extraction and cDNA Synthesis
2.6. RT-qPCR Primer Design and Screening
2.7. Relative mRNA Quantification
2.8. Data Analysis
3. Results
3.1. General Features of the Sphenomorphus incognitus Mitogenome
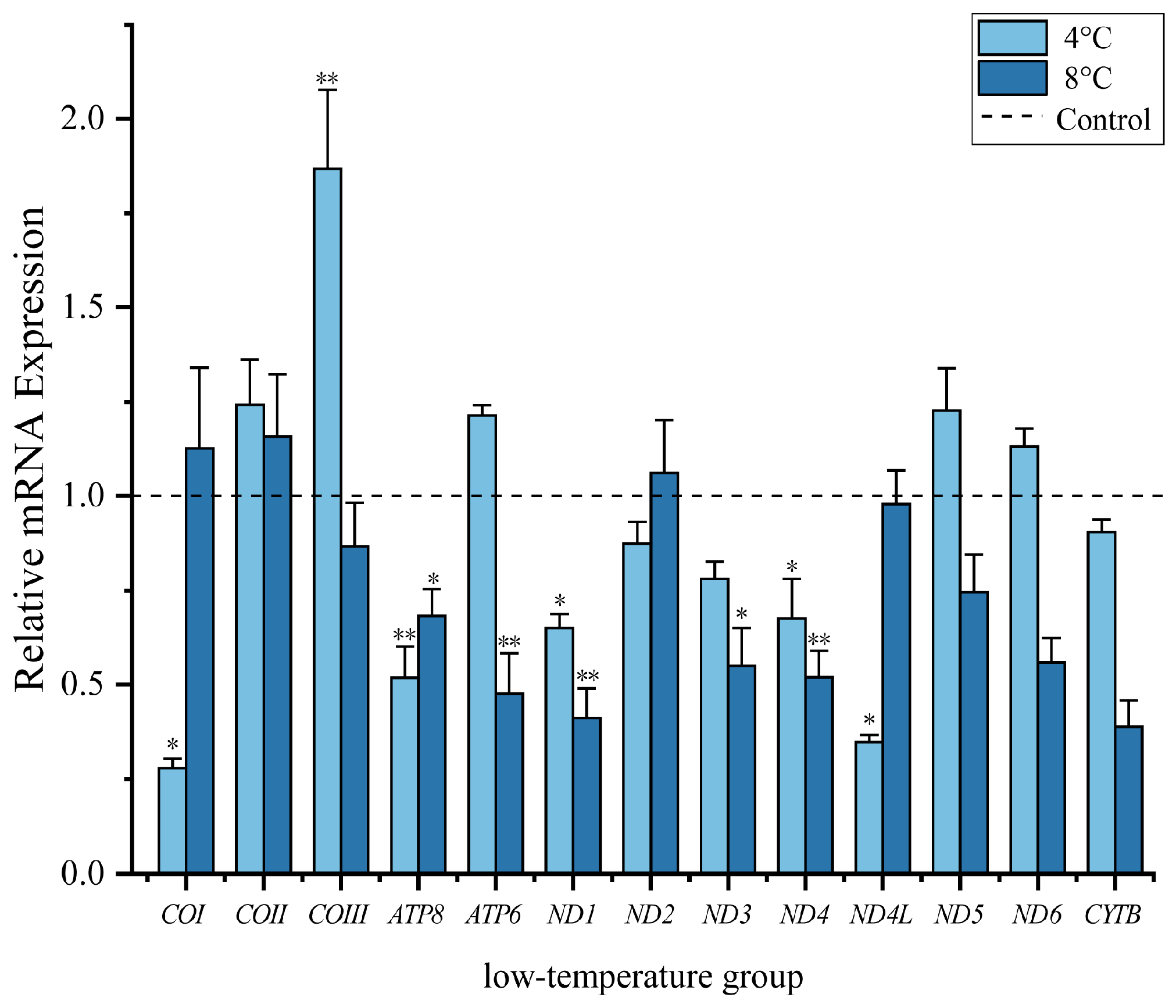
3.2. Quantification of Mitochondrial Protein-Coding Genes at Low Temperatures
3.3. Quantification of Mitochondrial Protein-Coding Genes at High Temperatures
4. Discussion
4.1. Analysis of Mitochondrial Transcript Levels at Low Temperature
4.2. Analysis of Mitochondrial Transcript Levels at High Temperature
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cosendey, B.N.; Duarte Rocha, C.F.; Menezes, V.A. Climate change, lizard populations, and species vulnerability/persistence: Trends in ecological and predictive climate studies. Environ. Dev. Sustain. 2023, 25, 8929–8950. [Google Scholar] [CrossRef]
- Lee, H.; Calvin, K.; Dasgupta, D.; Krinner, G.; Mukherji, A.; Thorne, P.; Trisos, C.; Romero, J.; Aldunce, P.; Barrett, K. Climate Change 2023: Synthesis Report; Contribution of working groups I, II and III to the sixth assessment report of the intergovernmental panel on climate change; IPCC: Geneva, Switzerland, 2023; pp. 33–115. [Google Scholar]
- Alexander, L.V.; Zhang, X.; Peterson, T.C.; Caesar, J.; Gleason, B.; Klein Tank, A.; Haylock, M.; Collins, D.; Trewin, B.; Rahimzadeh, F. Global observed changes in daily climate extremes of temperature and precipitation. J. Geophys. Res. Atmos. 2006, 111, 1–22. [Google Scholar] [CrossRef]
- Donihue, C.M.; Kowaleski, A.M.; Losos, J.B.; Algar, A.C.; Baeckens, S.; Buchkowski, R.W.; Fabre, A.C.; Frank, H.K.; Geneva, A.J.; Reynolds, R.G. Hurricane effects on neotropical lizards span geographic and phylogenetic scales. Proc. Natl. Acad. Sci. USA 2020, 117, 10429–10434. [Google Scholar] [CrossRef] [PubMed]
- Grant, P.R. Evolution, climate change, and extreme events. Science 2017, 357, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Burraco, P.; Orizaola, G.; Monaghan, P.; Metcalfe, N.B. Climate change and ageing in ectotherms. Global Chang. Biol. 2020, 26, 5371–5381. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, J.W.; Scott, D.E.; Ryan, T.J.; Buhlmann, K.A.; Tuberville, T.D.; Metts, B.S.; Greene, J.L.; Mills, T.; Leiden, Y.; Poppy, S.; et al. The Global Decline of Reptiles, Deja Vu Amphibians. Bioscience 2000, 50, 653–666. [Google Scholar] [CrossRef]
- Sunday, J.M.; Bates, A.E.; Kearney, M.R.; Colwell, R.K.; Dulvy, N.K.; Longino, J.T.; Huey, R.B. Thermal-safety margins and the necessity of thermoregulatory behavior across latitude and elevation. Proc. Natl. Acad. Sci. USA 2014, 111, 5610–5615. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Sammy, M.J.; Ballinger, S.W. An evolutionary, or “Mitocentric” perspective on cellular function and disease. Redox Biol. 2020, 36, 101568. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, I. Mitochondrial adaptations to variable environments and their role in animals’ stress tolerance. Integr. Comp. Biol. 2018, 58, 519–531. [Google Scholar] [CrossRef]
- Camus, M.F.; Wolff, J.N.; Sgro, C.M.; Dowling, D.K. Experimental support that natural selection has shaped the latitudinal distribution of mitochondrial Haplotypes in Australian Drosophila melanogaster. Mol. Biol. Evol. 2017, 34, 2600–2612. [Google Scholar] [CrossRef]
- Chung, D.J.; Schulte, P.M. Mitochondria and the thermal limits of ectotherms. J. Exp. Biol. 2020, 223, jeb227801. [Google Scholar] [CrossRef] [PubMed]
- Moraes, C.T. What regulates mitochondrial DNA copy number in animal cells? Trends Genet. 2001, 17, 199–205. [Google Scholar] [CrossRef]
- Scheibye-Knudsen, M.; Fang, E.F.; Croteau, D.L.; Wilson, D.M.; Bohr, V.A. Protecting the mitochondrial powerhouse. Trends Cell Biol. 2015, 25, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.; Sterling, J.F.; Roberts, S.A.; Bhagwat, A.S.; Resnick, M.A.; Gordenin, D.A. Base damage within single-strand DNA underlies in vivo hypermutability induced by a ubiquitous environmental agent. PLoS Genet. 2012, 8, e1003149. [Google Scholar] [CrossRef] [PubMed]
- Rongvaux, A.; Jackson, R.; Harman, C.C.; Li, T.; West, A.P.; De Zoete, M.R.; Wu, Y.; Yordy, B.; Lakhani, S.A.; Kuan, C.-Y. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 2014, 159, 1563–1577. [Google Scholar] [CrossRef] [PubMed]
- West, P. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 2015, 520, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Sainz, A.G.; Shadel, G.S. Mitochondrial DNA: Cellular genotoxic stress sentinel. Trends Biochem. Sci. 2021, 46, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Das, J. The role of mitochondrial respiration in physiological and evolutionary adaptation. Bioessays 2006, 28, 890–901. [Google Scholar] [CrossRef]
- Sun, Y.B.; Shen, Y.Y.; Irwin, D.M.; Zhang, Y.P. Evaluating the roles of energetic functional constraints on teleost mitochondrial-encoded protein evolution. Mol. Biol. Evol. 2011, 28, 39–44. [Google Scholar] [CrossRef]
- Uetz, P.; Koo, M.; Aguilar, R.; Brings, E.; Catenazzi, A.; Chang, A.; Chaitanya, R.; Freed, P.; Gross, J.; Hammermann, M. A quarter century of reptile and amphibian databases. Herpetol. Rev. 2021, 52, 246–255. [Google Scholar]
- Cheng, Z.Q.; Liu, J.L.; Wu, Y.K.; Chen, S.L.; Guo, H.X.; Ding, G.Y. A new record of lizards from Zhejiang and Jiangxi Provinces—Sphenomorphus incognitus. Sichuan J. Zool. 2017, 36, 479–480. [Google Scholar]
- Luu, B.E.; Wijenayake, S.; Zhang, J.; Tessier, S.N.; Quintero-Galvis, J.F.; Gaitán-Espitia, J.D.; Nespolo, R.F.; Storey, K.B. Strategies of biochemical adaptation for hibernation in a south American marsupial, Dromiciops gliroides: 3. Activation of pro-survival response pathways. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2018, 224, 26–31. [Google Scholar] [CrossRef]
- Speers-Roesch, B.; Norin, T.; Driedzic, W.R. The benefit of being still: Energy savings during winter dormancy in fish come from inactivity and the cold, not from metabolic rate depression. Proc. R. Soc. B Biol. Sci. 2018, 285, 20181593. [Google Scholar] [CrossRef]
- Stroud, J.T.; Mothes, C.C.; Beckles, W.; Heathcote, R.J.P.; Donihue, C.M.; Losos, J.B. An extreme cold event leads to community-wide convergence in lower temperature tolerance in a lizard community. Biol. Lett. 2020, 16, 20200625. [Google Scholar] [CrossRef] [PubMed]
- He, J.Y.; Zhan, L.M.; Meng, S.Q.; Wang, Z.; Gao, L.L.; Wang, W.J.; Storey, K.B.; Zhang, Y.P.; Yu, D.N. Differential Mitochondrial Genome Expression of Three Sympatric Lizards in Response to Low-Temperature Stress. Animals 2024, 14, 1158. [Google Scholar] [CrossRef] [PubMed]
- Baker, E.; Peris, D.; Moriarty, R.; Li, X.; Fay, J.; Hittinger, C. Mitochondrial DNA and temperature tolerance in lager yeasts. Sci. Adv. 2019, 5, eaav1869. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.B. Differential Gene Expression under Environmental Stress in the Freeze Tolerant Wood Frog, Rana sylvatica. Ph.D. Thesis, Carleton University, Ottawa, ON, Canada, 1999. [Google Scholar]
- Wang, J.Y.; Zhang, L.H.; Hong, Y.H.; Cai, L.N.; Storey, K.B.; Zhang, J.Y.; Zhang, S.S.; Yu, D.N. How does mitochondrial protein-coding gene expression in Fejervarya kawamurai (Anura: Dicroglossidae) respond to extreme temperatures? Animals 2023, 13, 3015. [Google Scholar] [CrossRef]
- Bury, S.; Cichoń, M.; Bauchinger, U.; Sadowska, E.T. High oxidative stress despite low energy metabolism and vice versa: Insights through temperature acclimation in an ectotherm. J. Therm. Biol 2018, 78, 36–41. [Google Scholar] [CrossRef]
- Mutiibwa, D.; Vavrus, S.J.; McAfee, S.A.; Albright, T.P. Recent spatiotemporal patterns in temperature extremes across conterminous United States. J. Geophys. Res. Atmos. 2015, 120, 7378–7392. [Google Scholar] [CrossRef]
- Abdelghffar, E.A.; ALmohammadi, A.G.; Malik, S.; Khalphallah, A.; Soliman, M.M. Changes in clinicomorphometrical findings, lipid profiles, hepatorenal indices and oxidant/antioxidant status as thermoregulatory adaptive mechanisms in poikilothermic Dabb lizard (Uromastyx aegyptia). Sci. Rep. 2023, 13, 3409. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, I.M. Ectotherm mitochondrial economy and responses to global warming. Acta Physiol. 2023, 237, e13950. [Google Scholar] [CrossRef] [PubMed]
- Conradie, S.R.; Woodborne, S.M.; Cunningham, S.J.; McKechnie, A.E. Chronic, sublethal effects of high temperatures will cause severe declines in southern African arid-zone birds during the 21st century. Proc. Natl. Acad. Sci. USA 2019, 116, 14065–14070. [Google Scholar] [CrossRef] [PubMed]
- Uller, T.; Isaksson, C.; Olsson, M. Immune challenge reduces reproductive output and growth in a lizard. Funct. Ecol. 2006, 20, 873–879. [Google Scholar] [CrossRef]
- Chang, J.; Pan, Y.F.; Liu, W.T.; Xie, Y.; Hao, W.Y.; Xu, P.; Wang, Y.H. Acute temperature adaptation mechanisms in the native reptile species Eremias argus. Sci. Total Environ. 2022, 818, 151773. [Google Scholar] [CrossRef]
- Kumazawa, Y.; Endo, H. Mitochondrial genome of the Komodo dragon: Efficient sequencing method with reptile-oriented primers and novel gene rearrangements. DNA Res. 2004, 11, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [PubMed]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; DePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 1–31. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Xiang, C.Y.; Gao, F.L.; Jakovlić, I.; Lei, H.P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.T.; Guan, J.Y.; Dai, X.Y.; Wu, G.J.; Zhang, L.P.; Storey, K.B.; Zhang, J.Y.; Zheng, R.Q.; Yu, D.N. Mitochondrial gene expression in different organs of Hoplobatrachus rugulosus from China and Thailand under low-temperature stress. BMC Zool. 2022, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.N.; Zhang, L.H.; Lin, Y.J.; Wang, J.Y.; Storey, K.B.; Zhang, J.Y.; Yu, D.N. Two-fold ND5 genes, three-fold control regions, incRNA, and the “missing” ATP8 found in the mitogenomes of polypedates megacephalus (Rhacophridae: Polypedates). Animals 2023, 13, 2857. [Google Scholar] [CrossRef] [PubMed]
- Biederman, J.; Yee, J.; Cortes, P. Validation of internal control genes for gene expression analysis in diabetic glomerulosclerosis. Kidney Int. 2004, 66, 2308–2314. [Google Scholar] [CrossRef] [PubMed]
- Formosa, L.E.; Dibley, M.G.; Stroud, D.A.; Ryan, M.T. Building a complex complex: Assembly of mitochondrial respiratory chain complex I. Semin. Cell Dev. Biol. 2018, 76, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Guppy, M.; Fuery, C.; Flanigan, J. Biochemical principles of metabolic depression. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1994, 109, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.D.; Guan, J.Y.; Zhang, Z.Y.; Cao, Y.R.; Cai, Y.Y.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. Insight into the phylogenetic relationships among three subfamilies within Heptageniidae (Insecta: Ephemeroptera) along with low-temperature selection pressure analyses using mitogenomes. Insects 2021, 12, 656. [Google Scholar] [CrossRef]
- Hong, Y.H.; Huang, H.M.; Wu, L.; Storey, K.B.; Zhang, J.Y.; Zhang, Y.P.; Yu, D.N. Characterization of two mitogenomes of Hyla sanchiangensis (Anura: Hylidae), with phylogenetic relationships and selection pressure analyses of Hylidae. Animals 2023, 13, 1593. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Tong, Y.; Ayivi, S.P.G.; Storey, K.B.; Zhang, J.Y.; Yu, D.N. The complete mitochondrial genomes of three Sphenomorphinae species (Squamata: Scincidae) and the selective pressure analysis on mitochondrial genomes of limbless Isopachys gyldenstolpei. Animals 2022, 12, 2015. [Google Scholar] [CrossRef]
- Lemieux, H.; Blier, P.U. Exploring thermal sensitivities and adaptations of oxidative phosphorylation pathways. Metabolites 2022, 12, 360. [Google Scholar] [CrossRef]
- Price, E.R.; Sirsat, T.S.; Sirsat, S.K.; Kang, G.; Keereetaweep, J.; Aziz, M.; Chapman, K.D.; Dzialowski, E.M. Thermal acclimation in American alligators: Effects of temperature regime on growth rate, mitochondrial function, and membrane composition. J. Therm. Biol. 2017, 68, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.D. Uncoupling to survive? The role of mitochondrial inefficiency in ageing. Exp. Gerontol. 2000, 35, 811–820. [Google Scholar] [CrossRef]
- Dilberger, B.; Baumanns, S.; Schmitt, F.; Schmiedl, T.; Hardt, M.; Wenzel, U.; Eckert, G.P. Mitochondrial oxidative stress impairs energy metabolism and reduces stress resistance and longevity of C. elegans. Oxid. Med. Cell. Longev. 2019, 2019, 6840540. [Google Scholar] [CrossRef] [PubMed]
- Allan, M.E.; Storey, K.B. Expression of NF-κB and downstream antioxidant genes in skeletal muscle of hibernating ground squirrels, Spermophilus tridecemlineatus. Cell Biochem. Funct. 2012, 30, 166–174. [Google Scholar] [CrossRef]
- Sun, B.; Williams, C.M.; Li, T.; Speakman, J.R.; Jin, Z.; Lu, H.; Luo, L.; Du, W. Higher metabolic plasticity in temperate compared to tropical lizards suggests increased resilience to climate change. Ecol. Monogr. 2022, 92, e01512. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Molecular physiology of freeze tolerance in vertebrates. Physiol. Rev. 2017, 97, 623–665. [Google Scholar] [CrossRef] [PubMed]
- Rogowitz, G.L. Evaluation of thermal acclimation and altitudinal variation of metabolism in a Neotropical lizard, Anolis gundlachi. Copeia 1996, 1996, 535–542. [Google Scholar] [CrossRef]
- Tsuji, J.S. Thermal acclimation of metabolism in Sceloporus lizards from different latitudes. Physiol. Zool. 1988, 61, 241–253. [Google Scholar] [CrossRef]
- Patterson, J. Emergence, basking behaviour, mean selected temperature and critical thermal minimum in high and low altitude subspecies of the tropical lizard Mabuya striata. Afr. J. Ecol. 1991, 29, 330–339. [Google Scholar] [CrossRef]
- Salin, K.; Auer, S.K.; Rey, B.; Selman, C.; Metcalfe, N.B. Variation in the link between oxygen consumption and ATP production, and its relevance for animal performance. Proc. R. Soc. B 2015, 282, 10–25. [Google Scholar] [CrossRef]
- Salin, K.; Auer, S.K.; Rudolf, A.M.; Anderson, G.J.; Cairns, A.G.; Mullen, W.; Hartley, R.C.; Selman, C.; Metcalfe, N.B. Individuals with higher metabolic rates have lower levels of reactive oxygen species in vivo. Biol. Lett. 2015, 11, 20150538. [Google Scholar] [CrossRef] [PubMed]
- Kotlyar, A.B.; Vinogradov, A.D. Slow active/inactive transition of the mitochondrial NADH-ubiquinone reductase. Biochim. Biophys. Acta (BBA)-Bioenerg. 1990, 1019, 151–158. [Google Scholar] [CrossRef]
- Dupuis, A.; Darrouzet, E.; Duborjal, H.; Pierrard, B.; Chevallet, M.; van Belzen, R.; Albracht, S.P.; Lunardi, J. Distal genes of the nuo operon of Rhodobacter capsulatus equivalent to the mitochondrial ND subunits are all essential for the biogenesis of the respiratory NADH-ubiquinone oxidoreductase. Mol. Microbiol. 1998, 28, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.Y.; Zhang, Z.Y.; Cao, Y.R.; Xu, X.D.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. The complete mitochondrial genome of Choroterpe (Euthralus) yixingensis (Ephemeroptera: Leptophlebiidae) and its mitochondrial protein-coding gene expression under imidacloprid stress. Gene 2021, 800, 145833. [Google Scholar] [CrossRef] [PubMed]
- Refsnider, J.M.; Vazquez, T.K.; Clifton, I.T.; Jayawardena, D.M.; Heckathorn, S.A. Cellular and whole-organism effects of prolonged versus acute heat stress in a montane, desert lizard. J. Exp. Zool. Part A 2021, 335, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Heckrotte, C. Temperature and light effects on the circadian rhythm and locomotory activity of the plains garter snake (Thamnophis radix hayendi). J. Interdiscip. Cycle Res. 1975, 6, 279–289. [Google Scholar] [CrossRef]
- Nie, Y.F.; Wang, Z.K.; Yu, S.M.; Liu, Y.P.; Zhang, L.Y.; Liu, R.; Zhou, Z.Q.; Zhu, W.T.; Diao, J.L. Combined effects of abamectin and temperature on the physiology and behavior of male lizards (Eremias argus): Clarifying adaptation and maladaptation. Sci. Total Environ. 2022, 837, 155794. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.W.; Lutz, T.J.; Fokidis, H.B.; Stahlschmidt, Z.R. Eating increases oxidative damage in a reptile. J. Exp. Biol. 2016, 219, 1969–1973. [Google Scholar] [CrossRef]
- Vaanholt, L.M.; Speakman, J.R.; Garland, T., Jr.; Lobley, G.E.; Visser, G.H. Protein synthesis and antioxidant capacity in aging mice: Effects of long-term voluntary exercise. Physiol. Biochem. Zool. 2008, 81, 148–157. [Google Scholar] [CrossRef]
- Chai, L.; Chen, A.; Luo, P.; Zhao, H.; Wang, H. Histopathological changes and lipid metabolism in the liver of Bufo gargarizans tadpoles exposed to Triclosan. Chemosphere 2017, 182, 255–266. [Google Scholar] [CrossRef]
- Wu, C.; Zhang, Y.; Chai, L.; Wang, H. Histological changes, lipid metabolism and oxidative stress in the liver of Bufo gargarizans exposed to cadmium concentrations. Chemosphere 2017, 179, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Pörtner, H. Climate variations and the physiological basis of temperature dependent biogeography: Systemic to molecular hierarchy of thermal tolerance in animals. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2002, 132, 739–761. [Google Scholar] [CrossRef] [PubMed]
- Pörtner, H.O.; Knust, R. Climate change affects marine fishes through the oxygen limitation of thermal tolerance. Science 2007, 315, 95–97. [Google Scholar] [CrossRef] [PubMed]
- Pörtner, H.-O.; Bock, C.; Mark, F.C. Oxygen-and capacity-limited thermal tolerance: Bridging ecology and physiology. J. Exp. Biol. 2017, 220, 2685–2696. [Google Scholar] [CrossRef]
- Frederich, M.; Pörtner, H.O. Oxygen limitation of thermal tolerance defined by cardiac and ventilatory performance in spider crab, Maja squinado. Am. J. Physiol. 2000, 279, R1531–R1538. [Google Scholar] [CrossRef]

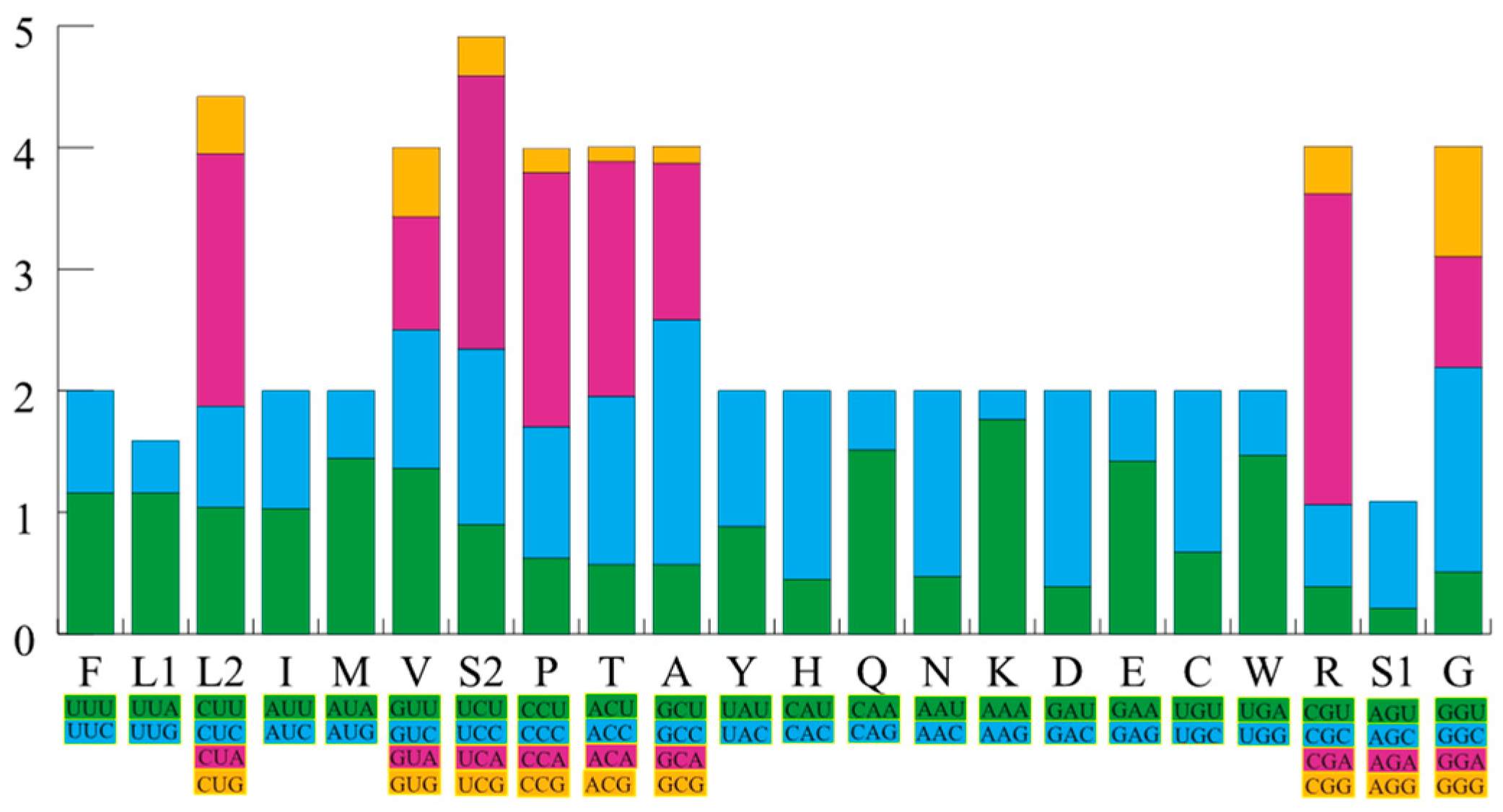

| Gene Name | Forward Primers (5′-3′) | Reverse Primers (5′-3′) |
|---|---|---|
| COI | GXXA-COI-J1 | GXXA-COI-N1 |
| GCATTGTCCTAGCCAATTCATC | AACATTACGCCGAAGTGGATT | |
| COII | GXXA-COII-J1 | GXXA-COII-N1 |
| ATCGCCCTTCCCTCACTTC | TCGTAGTCTGTGTATTCGTAGC | |
| COIII | GXXA-COIII-J1 | GXXA-COIII-N1 |
| CCACCAAGCACACGCATAC | GGTAATGTCTCGTCATCACTGT | |
| ATP8 | GXXA-ATP8-J1 | GXXA-ATP8-N1 |
| CTGAGCCACAATTCTTCTAATC | TAGGTTCATGGTCAGGTTCA | |
| ATP6 | GXXA-ATP6-J1 | GXXA-ATP6-N1 |
| TTGTGGCTGTCTACTGTCCTTA | CGGTTAGGTTGGCGGTTAGT | |
| ND1 | GXXA-ND1-J1 | GXXA-ND1-N1 |
| GACTACTATTCTGCTCGTGACC | AATGTTGGCGTATTCGGCTAG | |
| ND2 | GXXA-ND2-J1 | GXXA-ND2-N1 |
| ACCTGAGACATTACACAACTGA | TGTAGGACCTCTGGCAATCA | |
| ND3 | GXXA-ND3-J1 | GXXA-ND3-N1 |
| GCTTCCGTTCTCACTTCGTT | GGCTGCTCACTCGTATACTAAG | |
| ND4 | GXXA-ND4-J1 | GXXA-ND4-N1 |
| AACCTTCACCATCAACTCTTCC | TAGGAGCCAGCAGGATAGAAC | |
| ND4L | GXXA-ND4L-J2 | GXXA-ND4L-N2 |
| GGCATCCTCGGCTTATCAATAC | GGAGCATAGTTGTGGAGGGTAA | |
| ND5 | GXXA-ND5-J1 | GXXA-ND5-N1 |
| CAGCATACAGTCTACGGTTGAT | AAGGCTAGGCGGAGGATTG | |
| ND6 | GXXA-ND6-J1 | GXXA-ND6-N1 |
| CCCAAGAGTAAAGCAAAGAGAT | GTGTTGTTGTTGTTCGTGTT | |
| Cytb | GXXA-CYTB-J1 | GXXA-CYTB-N1 |
| TCTGCCGAGATGTTCAATATGG | GACGAAGGCTGTTGCTATTACT | |
| β-actin | GXXA-ACT-J1 | GXXA-ACT-N1 |
| GCCATGTACGTTGCCATCC | CCAGAGTCCATCACGATACCA |
| Gene/Region | Start Position | Stop Position | Spacer (+) Overlap (−) | Length (bp) | Start Codon | Stop Codon | Strand |
|---|---|---|---|---|---|---|---|
| tRNAPhe | 1 | 72 | 72 | H | |||
| 12S rRNA | 73 | 1010 | 938 | H | |||
| tRNAVal | 1011 | 1081 | 71 | H | |||
| 16S rRNA | 1082 | 2621 | 1540 | H | |||
| tRNALeu (UUR) | 2622 | 2697 | 76 | H | |||
| ND1 | 2698 | 3660 | +1 | 963 | ATG | TAA | H |
| tRNAIle | 3662 | 3734 | 73 | H | |||
| tRNAGln | 3735 | 3806 | −2 | 72 | L | ||
| tRNAMet | 3805 | 3874 | 70 | H | |||
| ND2 | 3875 | 4909 | 1035 | ATG | TAA | H | |
| tRNATrp | 4910 | 4981 | −1 | 72 | H | ||
| tRNAAla | 4981 | 5049 | 69 | L | |||
| tRNAAsn | 5050 | 5122 | +13 | 73 | L | ||
| tRNACys | 5136 | 5199 | 64 | L | |||
| tRNATyr | 5200 | 5266 | +1 | 67 | L | ||
| COI | 5268 | 6815 | −5 | 1548 | GTG | AGA | H |
| tRNASer (UCN) | 6811 | 6883 | +4 | 73 | L | ||
| tRNAAsp | 6888 | 6955 | 68 | H | |||
| COII | 6956 | 7643 | 688 | ATG | T | H | |
| tRNALys | 7644 | 7708 | +2 | 65 | H | ||
| ATP8 | 7711 | 7878 | −10 | 168 | ATG | TAA | H |
| ATP6 | 7869 | 8552 | −1 | 684 | ATG | TAA | H |
| COIII | 8552 | 9335 | 784 | ATG | T | H | |
| tRNAGly | 9336 | 9405 | 70 | H | |||
| ND3 | 9406 | 9751 | 346 | ATG | TAA | H | |
| tRNAArg | 9752 | 9820 | 69 | H | |||
| ND4L | 9821 | 10,117 | −7 | 297 | ATG | TAA | H |
| ND4 | 10,111 | 11,491 | 1381 | ATG | T | H | |
| tRNAHis | 11,492 | 11,563 | 72 | H | |||
| tRNASer (AGY) | 11,564 | 11,629 | −1 | 66 | H | ||
| tRNALeu (CUN) | 11,629 | 11,701 | +1 | 73 | H | ||
| ND5 | 11,703 | 13,529 | −5 | 1827 | ATG | TAA | H |
| ND6 | 13,525 | 14,046 | 522 | ATG | AGG | L | |
| tRNAGlu | 14,047 | 14,115 | +2 | 69 | L | ||
| Cyt b | 14,118 | 15,260 | +6 | 1143 | ATG | TAA | H |
| tRNAThr | 15,267 | 15,335 | 69 | H | |||
| tRNAPro | 15,336 | 15,404 | 69 | L | |||
| D-loop | 15,405 | 16,759 | Complete | H |
| Region | Strand | Length (bp) | A (%) | T (%) | C (%) | G (%) | A + T (%) | C + G (%) | AT Skew | GC Skew |
|---|---|---|---|---|---|---|---|---|---|---|
| Whole genome | 16,759 | 30.9 | 25.8 | 28.2 | 15.0 | 56.7 | 43.2 | 0.090 | −0.307 | |
| PCGs | + | 10,860 | 28.9 | 27.5 | 29.4 | 14.2 | 56.4 | 43.6 | 0.025 | −0.347 |
| − | 522 | 9.6 | 40.8 | 11.7 | 37.9 | 50.4 | 49.6 | −0.620 | 0.529 | |
| tRNAs | + | 986 | 31.5 | 25.4 | 23.3 | 19.8 | 56.9 | 43.1 | 0.109 | −0.082 |
| − | 556 | 25.9 | 29.7 | 16.0 | 28.4 | 55.6 | 44.4 | −0.068 | 0.279 | |
| rRNAs | + | 2478 | 35.6 | 20.9 | 25.3 | 18.3 | 56.5 | 43.6 | 0.260 | −0.161 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhan, L.; He, J.; Meng, S.; Guo, Z.; Chen, Y.; Storey, K.B.; Zhang, J.; Yu, D. Mitochondrial Protein-Coding Gene Expression in the Lizard Sphenomorphus incognitus (Squamata:Scincidae) Responding to Different Temperature Stresses. Animals 2024, 14, 1671. https://doi.org/10.3390/ani14111671
Zhan L, He J, Meng S, Guo Z, Chen Y, Storey KB, Zhang J, Yu D. Mitochondrial Protein-Coding Gene Expression in the Lizard Sphenomorphus incognitus (Squamata:Scincidae) Responding to Different Temperature Stresses. Animals. 2024; 14(11):1671. https://doi.org/10.3390/ani14111671
Chicago/Turabian StyleZhan, Lemei, Jingyi He, Siqi Meng, Zhiqiang Guo, Yuxin Chen, Kenneth B. Storey, Jiayong Zhang, and Danna Yu. 2024. "Mitochondrial Protein-Coding Gene Expression in the Lizard Sphenomorphus incognitus (Squamata:Scincidae) Responding to Different Temperature Stresses" Animals 14, no. 11: 1671. https://doi.org/10.3390/ani14111671
APA StyleZhan, L., He, J., Meng, S., Guo, Z., Chen, Y., Storey, K. B., Zhang, J., & Yu, D. (2024). Mitochondrial Protein-Coding Gene Expression in the Lizard Sphenomorphus incognitus (Squamata:Scincidae) Responding to Different Temperature Stresses. Animals, 14(11), 1671. https://doi.org/10.3390/ani14111671





