A Machine Learning Framework Based on Extreme Gradient Boosting to Predict the Occurrence and Development of Infectious Diseases in Laying Hen Farms, Taking H9N2 as an Example
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. H9N2 Cases and Data Collection
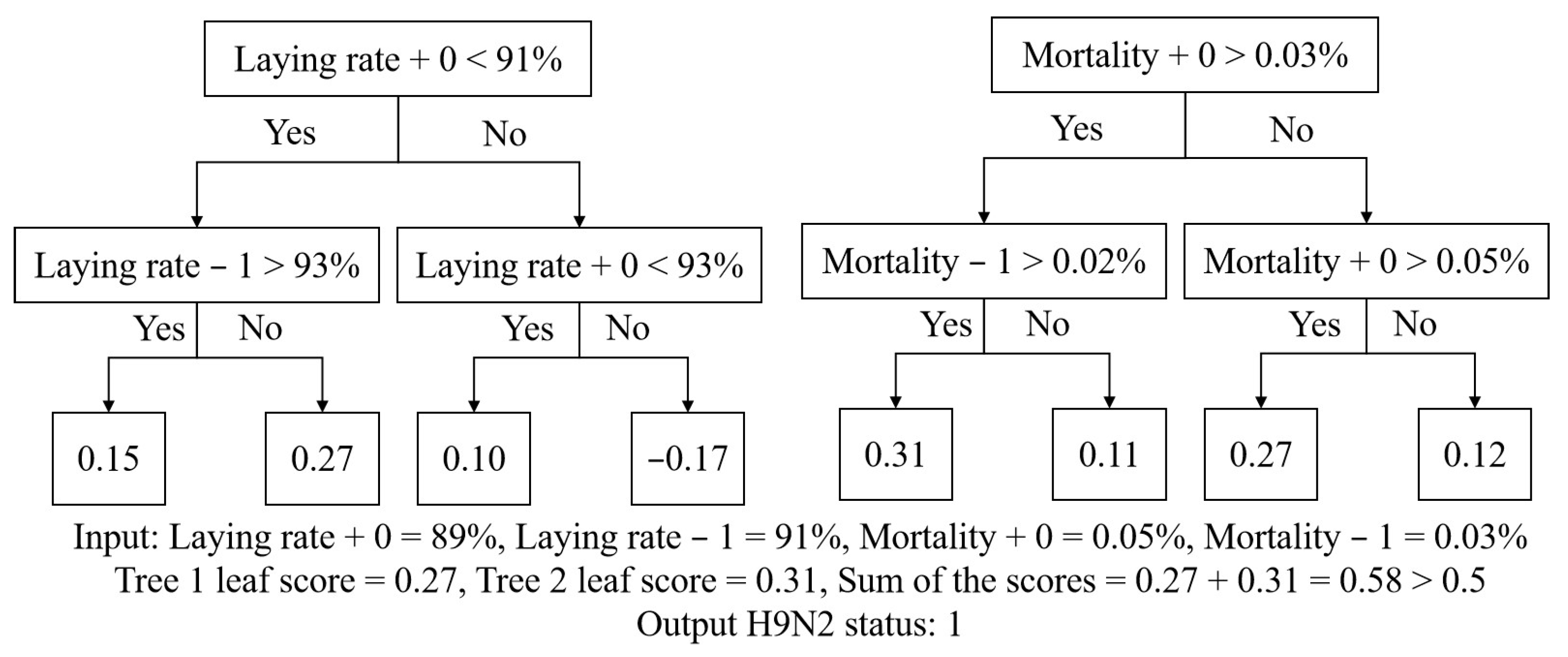
2.2. Xgboost Algorithm
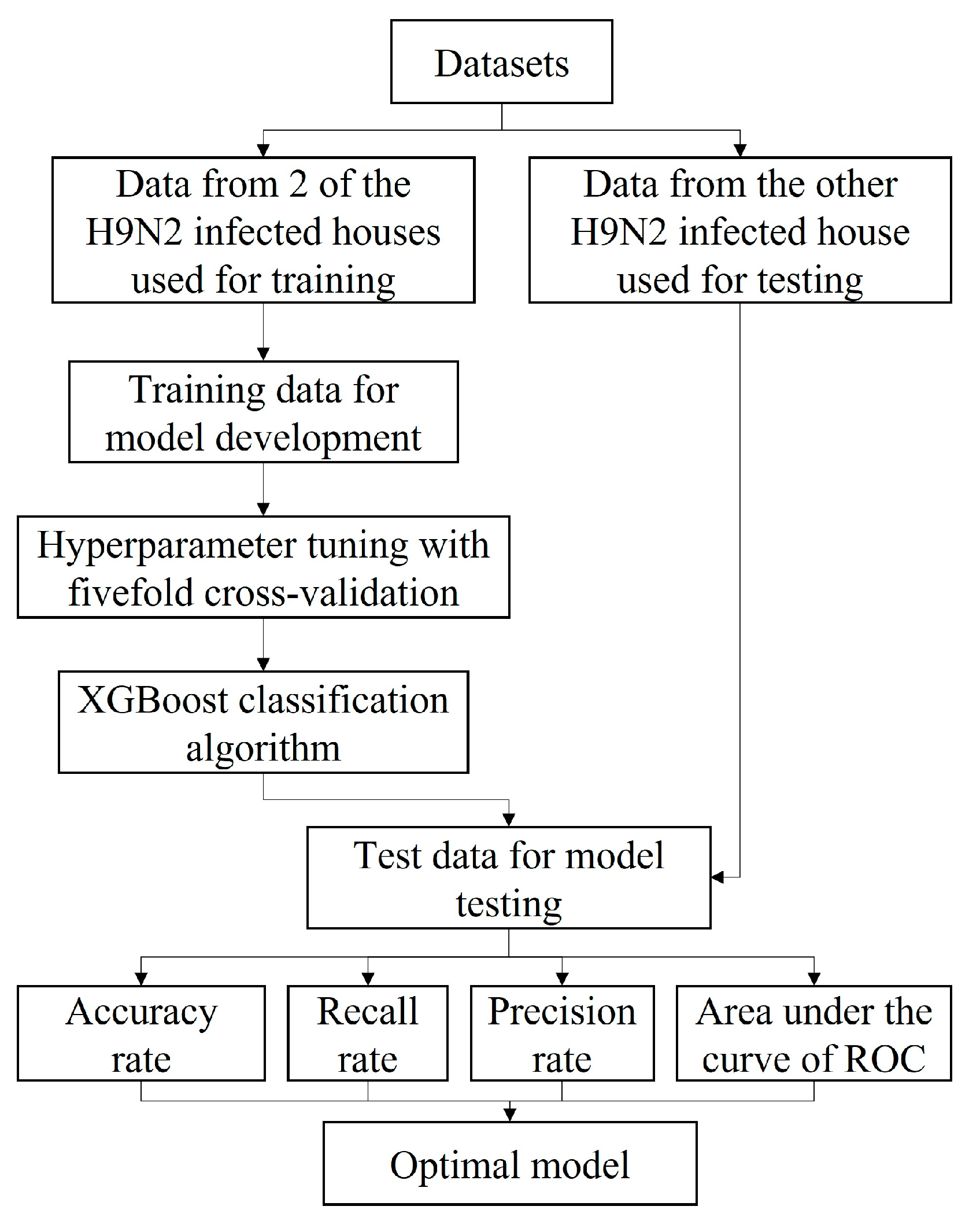
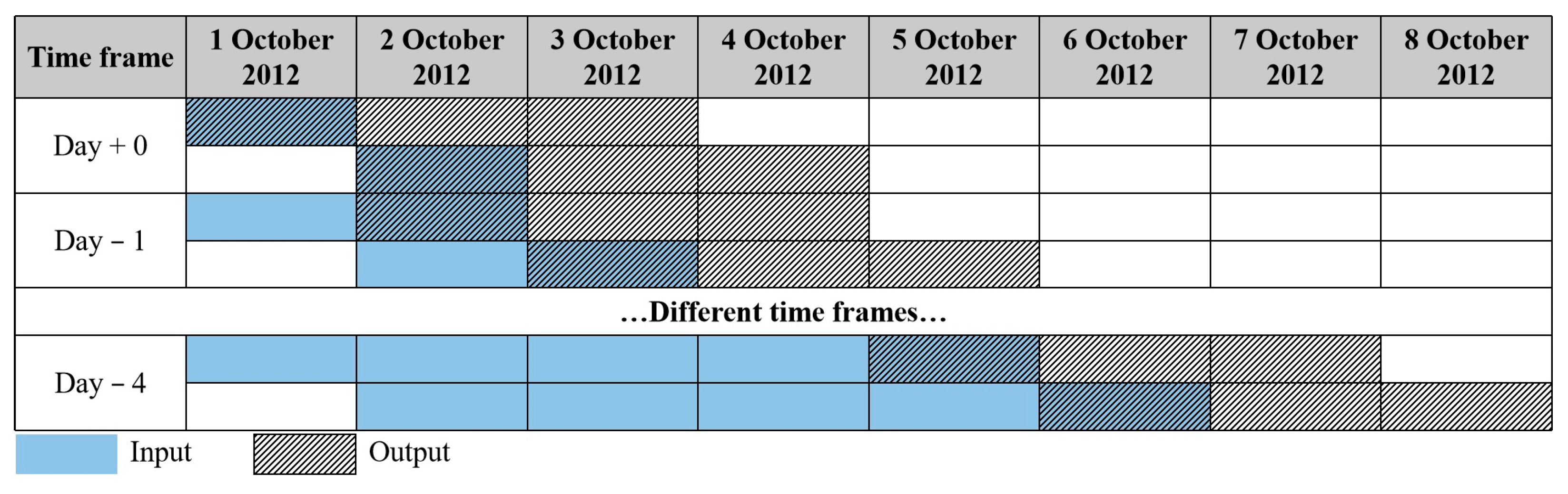
2.3. Framework Design
2.4. Evaluation Criteria
- (1)
- Accuracy rate (ACC)
- (2)
- Recall rate (RR)
- (3)
- Precision rate (PR)
3. Results
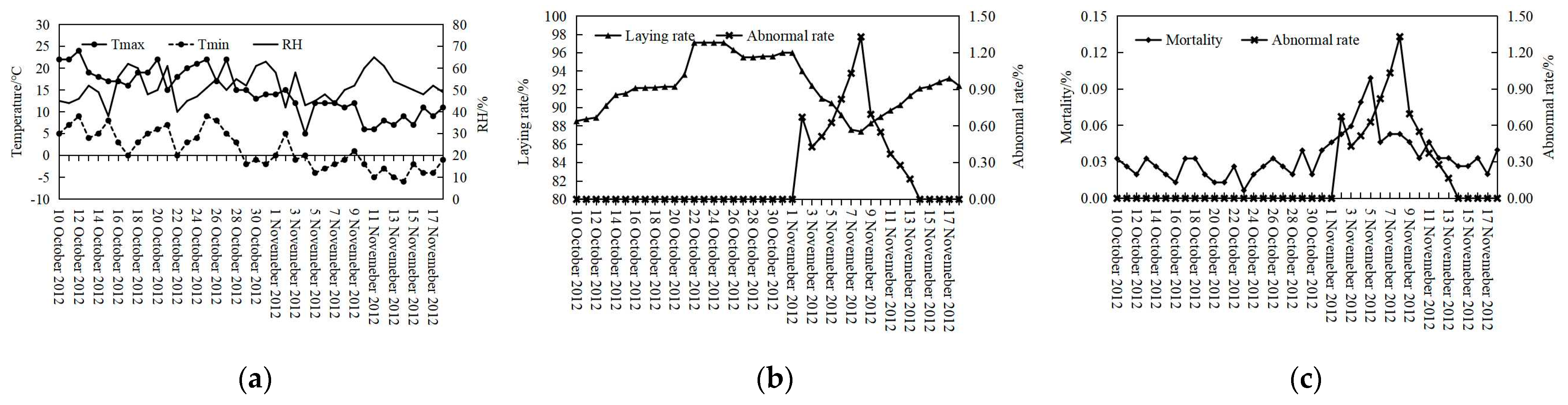
3.1. Descriptive Statistic
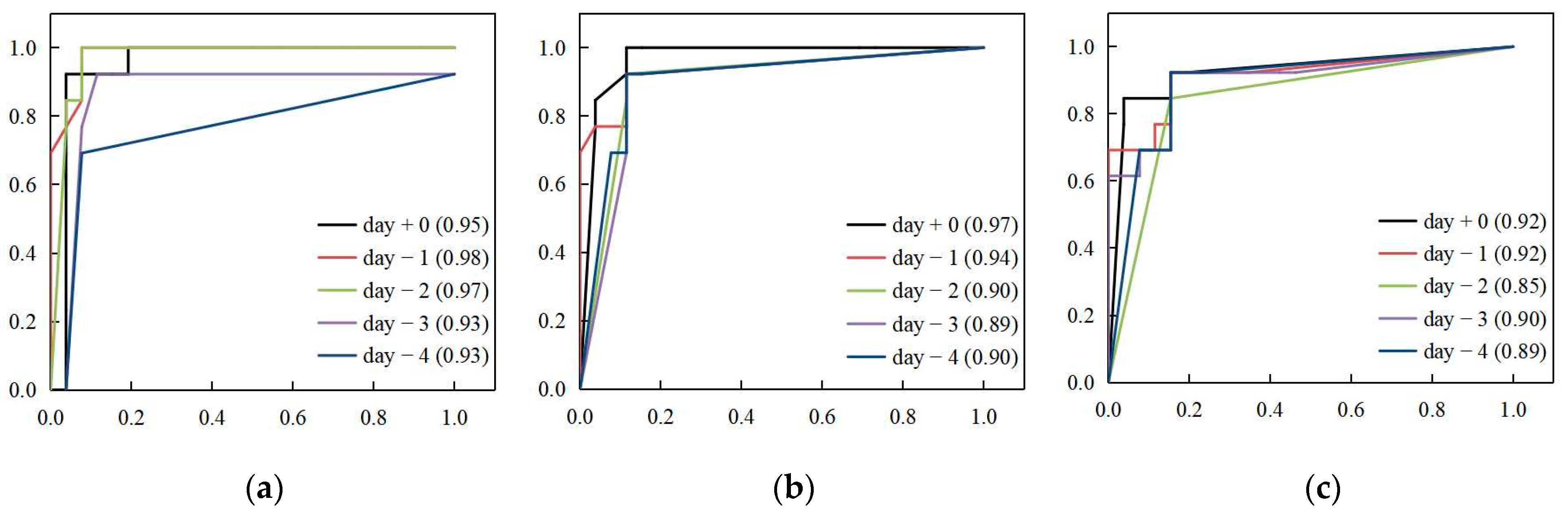
3.2. Predictive Performance Comparison
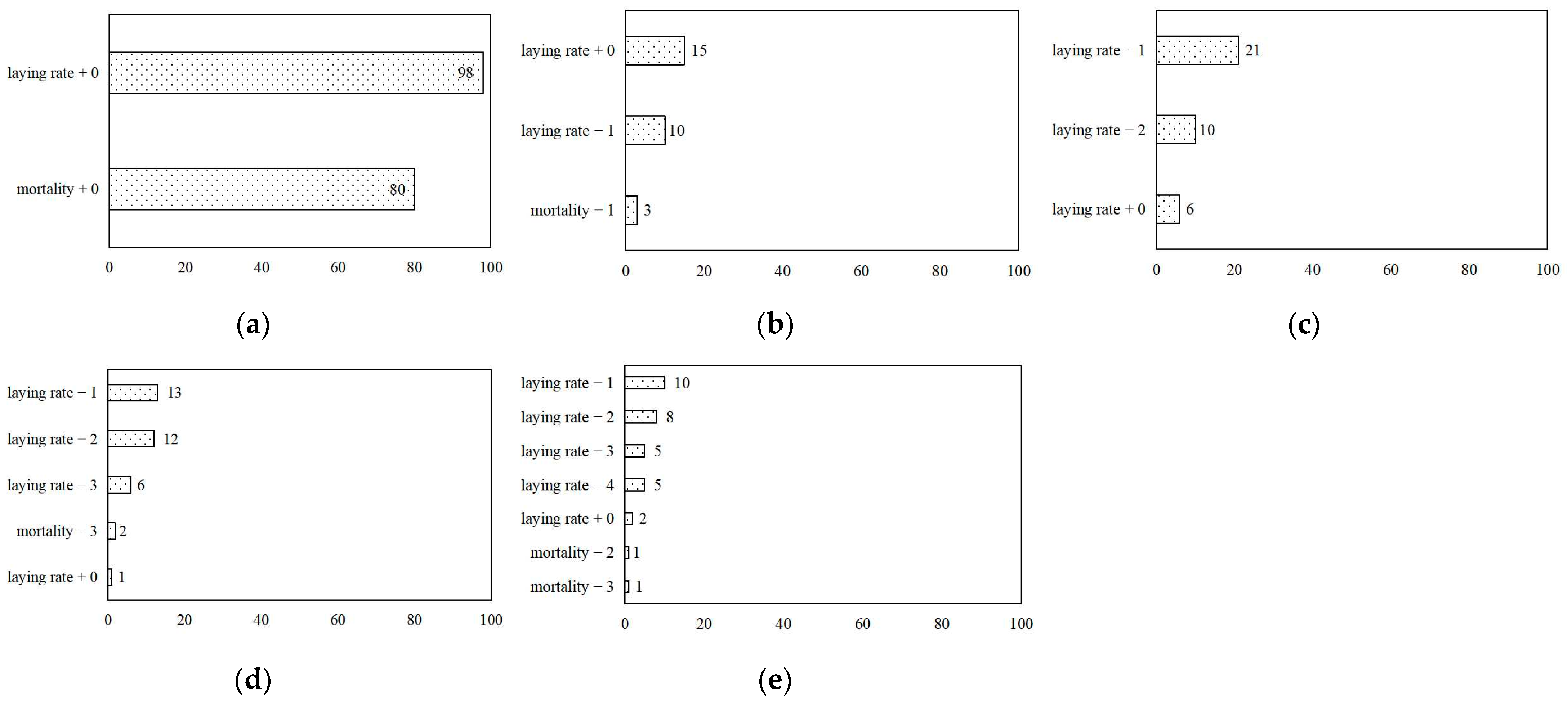
3.3. Feature Importance
4. Discussion
4.1. Relationship between Environment and Production Variables and H9N2 Status
4.2. Framework Performance and Model Interpretation
4.3. Potential Applications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- da Silva, M.C.; Schaefer, R.; Gava, D.; Souza, C.K.; da Silva Vaz, I., Jr.; Bastos, A.P.; Venancio, E.J. Production and application of anti-nucleoprotein IgY antibodies for influenza A virus detection in swine. J. Immunol. Methods 2018, 461, 100–105. [Google Scholar] [CrossRef]
- Kaur, H.; Garg, S.; Joshi, H.; Ayaz, S.; Sharma, S.; Bhandari, M. A review: Epidemics and pandemics in human history. Int. J. Pharma Res. Health Sci. 2020, 8, 3139–3142. [Google Scholar] [CrossRef]
- Transboundary Animal Diseases. Available online: https://www.fao.org/emergencies/emergency-types/transboundary-animaldiseases/en/ (accessed on 21 July 2022).
- Liang, W.S.; He, Y.C.; Wu, H.D.; Li, Y.T.; Shih, T.H.; Kao, G.S.; Guo, H.Y.; Chao, D.Y. Ecological factors associated with persistent circulation of multiple highly pathogenic avian influenza viruses among poultry farms in Taiwan during 2015–2017. PLoS ONE 2020, 15, e0236581. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.J.; Skehel, J.J.; McCauley, J. Characterization of influenza virus RNA complete transcripts. Virology 1982, 116, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Kargarfard, F.; Sami, A.; Ebrahimie, E. Knowledge discovery and sequence-based prediction of pandemic influenza using an integrated classification and association rule mining (CBA) algorithm. J. Biomed. Inform. 2015, 57, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Kargarfard, F.; Sami, A.; Mohammadi-Dehcheshmeh, M.; Ebrahimie, E. Novel approach for identification of influenza virus host range and zoonotic transmissible sequences by determination of host-related associative positions in viral genome segments. BMC Genomics. 2016, 17, 925. [Google Scholar] [CrossRef]
- Lowie, T.; Callens, J.; Maris, J.; Ribbens, S.; Pardon, B. Decision tree analysis for pathogen identification based on circumstantial factors in outbreaks of bovine respiratory disease in calves. Prev. Vet. Med. 2021, 196, 105469. [Google Scholar] [CrossRef]
- Souley Kouato, B.; De Clercq, K.; Abatih, E.; Dal Pozzo, F.; King, D.P.; Thys, E.; Marichatou, H.; Saegerman, C. Review of epidemiological risk models for foot-and-mouth disease: Implications for prevention strategies with a focus on Africa. PLoS ONE 2018, 13, e0208296. [Google Scholar] [CrossRef]
- Nasiri, A.; Yoder, J.; Zhao, Y.; Hawkins, S.; Prado, M.; Gan, H. Pose estimation-based lameness recognition in broiler using CNN-LSTM network. Comput. Electron. Agric. 2022, 197, 106931. [Google Scholar] [CrossRef]
- Wiemken, T.L.; Kelley, R.R. Machine learning in epidemiology and health outcomes research. Annu. Rev. Public Health 2020, 41, 21–36. [Google Scholar] [CrossRef]
- Punyapornwithaya, V.; Klaharn, K.; Arjkumpa, O.; Sansamur, C. Exploring the predictive capability of machine learning models in identifying foot and mouth disease outbreak occurrences in cattle farms in an endemic setting of Thailand. Prev. Vet. Med. 2022, 207, 105706. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, M.; Aghagolzadeh, P.; Shamabadi, N.; Tahmasebi, A.; Alsharifi, M.; Adelson, D.L.; Hemmatzadeh, F.; Ebrahimie, E. Understanding the underlying mechanism of HA-subtyping in the level of physic-chemical characteristics of protein. PLoS ONE 2014, 9, e96984. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Kephart, G.; Juarez-Colunga, E. Predicting COVID-19 mortality risk in Toronto, Canada: A comparison of tree-based and regression-based machine learning methods. BMC Med. Res. Methodol. 2021, 21, 267. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.L.; An, S.Y.; Qiao, B.J.; Guan, P.; Huang, D.S.; Wu, W. A data-driven interpretable ensemble framework based on tree models for forecasting the occurrence of COVID-19 in the USA. Environ. Sci. Pollut. Res. 2023, 30, 12648–13659. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Xu, F.; Zhou, Y.; Zhang, X.S. Pig-vet: A web-based expert system for pig disease diagnosis. Expert Syst. Appl. 2005, 29, 93–103. [Google Scholar] [CrossRef]
- Zeineldin, M.M.; El-Raof, Y.M.A.; El-Attar, H.A.; Ghanem, M.M. Lung ultrasonography and computer-aided scoring system as a diagnostic aid for bovine respiratory disease in feedlot cattle. Glob. Vet. 2016, 17, 588–594. [Google Scholar] [CrossRef]
- Katris, C. A time series-based statistical approach for outbreak spread forecasting: Application of COVID-19 in Greece. Expert Syst. Appl. 2021, 166, 114077. [Google Scholar] [CrossRef]
- Assad, D.B.N.; Cara, J.; Ortega-Mier, M. Comparing short-term univariate and multivariate time-series forecasting models in infectious disease outbreak. Bull. Math. Biol. 2023, 85, 9. [Google Scholar] [CrossRef]
- Wang, L.Y.; Wang, X.Y.; Chen, A.X.; Jin, X.; Che, H.L. Prediction of type 2 diabetes risk and its effect evaluation based on the XGBoost model. Healthcare 2020, 8, 247. [Google Scholar] [CrossRef]
- Wang, K.; Tian, Y.A.; Liu, S.S.; Zhang, Z.Y.; Shen, L.L.; Meng, D.Q.; Li, J. Risk factors and predictive model for dermatomyositis associated with rapidly progressive interstitial lung disease. Pharm. Pers. Med. 2022, 15, 775–783. [Google Scholar] [CrossRef]
- Huang, L.; Liu, Y.; Huang, W.; Dong, Y.; Ma, H.; Wu, K.; Guo, A. Combining random forest and XGBoost methods in detecting early and mid-term winter wheat stripe rust using canopy level hyperspectral measurements. Agriculture 2022, 12, 74. [Google Scholar] [CrossRef]
- Zhang, G.H.; Qu, Y.J.; Niu, Y.J.; Zhang, H.X.; Sun, Q.Q.; Liu, X.P.; Li, Y.; Zhang, H.; Liu, M.D. Difference in pathogenicity of 2 strains of avian leukosis virus subgroup J in broiler chicken. Poult. Sci. 2019, 98, 2772–2780. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Liu, A.J.; Zhang, F.M.; Ling, Y.; Ou, C.B.; Hou, N.; He, C. Co-infection of broilers with Ornithobacterium rhinotracheale and H9N2 avian influenza virus. BMC Vet. Res. 2012, 8, 104. [Google Scholar] [CrossRef] [PubMed]
- Linke, S.; Neubauer, K.; Dorner, M.B.; Dorner, B.G.; Pauli, G.; Schweiger, B. Generation and characterisation of monoclonal antibodies against influenza virus A, subtype H5N1. J. Virol. Methods 2011, 175, 85–94. [Google Scholar] [CrossRef]
- Gail, M.H.; Altman, D.G.; Cadarette, S.M.; Collins, G.; Evans, S.J.; Sekula, P.; Williamson, E.; Woodward, M. Design choices for observational studies of the effect of exposure on disease incidence. BMJ Open 2019, 9, e031031. [Google Scholar] [CrossRef]
- Chen, M.H.; Liu, Q.Y.; Chen, S.H.; Liu, Y.C.; Zhang, C.H.; Liu, R.H. XGBoost-based algorithm interpretation and application on post-fault transient stability status prediction of power system. IEEE Access 2019, 7, 13149–13158. [Google Scholar] [CrossRef]
- Fan, J.L.; Wang, X.K.; Wu, L.F.; Zhou, H.M.; Zhang, F.C.; Yu, X.; Lu, X.H.; Xiang, Y.Z. Comparison of support vector machine and extreme gradient boosting for predicting daily global solar radiation using temperature and precipitation in humid subtropical climates: A case study in China. Energy Convers. Manag. 2018, 164, 102–111. [Google Scholar] [CrossRef]
- Omotehinwa, T.O.; Oyewola, D.O. Hyperparameter optimization of ensemble models for spam email detection. Appl. Sci. 2023, 13, 1971. [Google Scholar] [CrossRef]
- Liu, Y.; Zhuang, Y.R.; Ji, B.Y.; Zhang, G.Q.; Rong, L.; Teng, G.H.; Wang, C.Y. Prediction of laying hen house odor concentrations using machine learning models based on small sample data. Comput. Electron. Agric. 2022, 195, 106849. [Google Scholar] [CrossRef]
- Hosmer, D.W., Jr.; Lemeshow, S.; Sturdivant, R.X. Applied Logistic Regression, 3rd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; pp. 153–225. [Google Scholar]
- Xu, H.Y.; Lu, Y.X.; Li, D.; Yan, C.Y.; Jiang, Y.R.; Hu, Z.; Zhang, Z.P.; Du, R.R.; Zhao, X.L.; Zhang, Y.; et al. Probiotic mediated intestinal microbiota and improved performance, egg quality and ovarian immune function of laying hens at different laying stage. Front. Microbiol. 2023, 14, 1041072. [Google Scholar] [CrossRef]
- Adlhoch, C.; Brouwer, A.; Kuiken, T.; Miteva, A.; Mulatti, P.; Smietanka, K.; Staubach, C.; Gogin, A.; Guajardo, I.M.; Baldinelli, F. Avian influenza overview August–November 2018. EFSA J. 2018, 16, 5573. [Google Scholar] [CrossRef]
- Pawar, S.S.; Sajjanar, B.; Lonkar, V.D.; Kurade, N.P.; Kadam, A.S.; Nirmal, A.V.; Brahmane, M.P.; Bal, S.K. Assessing and mitigating the impact of heat stress on poultry. Adv. Anim. Vet. Sci. 2016, 4, 332–341. [Google Scholar] [CrossRef]
- Dong, G.Y.; Cong, X.; Wang, C.M.; Wu, B.; Luo, J.; Zhang, H.; Nolte, D.L.; Deliberto, T.J.; Duan, M.X.; Ji, G.J.; et al. Reassortant H9N2 influenza viruses containing H5N1-like PB1 genes isolated from black-billed magpies in Southern China. PLoS ONE 2011, 6, e25808. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, J. H9N2 influenza virus in China: A cause of concern. Protein Cell 2015, 6, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.D.; Qi, L.; Gao, H.J.; Sun, H.L.; Pu, J.; Sun, Y.P.; Liu, J.H. Generation and protective efficacy of a cold-adapted attenuated avian H9N2 influenza vaccine. Sci. Rep. 2016, 6, 30382. [Google Scholar] [CrossRef]
- Gu, M.; Chen, H.Z.; Li, Q.H.; Huang, J.Q.; Zhao, M.J.; Gu, X.B.; Jiang, K.J.; Wang, X.Q.; Peng, D.X.; Liu, X.F. Enzootic genotype S of H9N2 avian influenza viruses donates internal genes to emerging zoonotic influenza viruses in China. Vet. Microbiol. 2014, 174, 309–315. [Google Scholar] [CrossRef]
- Cheung, C.L.; Vijaykrishna, D.; Smith, G.J.D.; Fan, X.H.; Zhang, J.X.; Bahl, J.; Duan, L.; Huang, K.; Tai, H.; Wang, J.; et al. Establishment of influenza A virus (H6N1) in minor poultry species in southern China. J. Virol. 2007, 81, 10402–10412. [Google Scholar] [CrossRef]
- Bi, Y.H.; Xiao, H.X.; Chen, Q.J.; Wu, Y.; Fu, L.F.; Quan, C.S.; Wong, G.; Liu, J.; Haywood, J.; Liu, Y.X.; et al. Changes in the length of the neuraminidase stalk region impact H7N9 virulence in mice. J. Virol. 2015, 90, 2142–2149. [Google Scholar] [CrossRef]
- Zhang, T.; Bi, Y.H.; Tian, H.A.; Li, X.W.; Liu, D.; Wu, Y.; Jin, T.; Wang, Y.; Chen, Q.J.; Chen, Z.; et al. Human infection with influenza virus A(H10N8) from live poultry markets, China, 2014. Emerg. Infect. Dis. 2014, 20, 2076–2079. [Google Scholar] [CrossRef]
- Kim, J.A.; Cho, S.H.; Kim, H.S.; Seo, S.H. H9N2 influenza viruses isolated from poultry in Korean live bird markets continuously evolve and cause the severe clinical signs in layers. Vet. Microbiol. 2006, 118, 169–176. [Google Scholar] [CrossRef]
- La, A.; Zhang, Q.; Cicek, N.; Coombs, K.M. Current understanding of the airborne transmission of important viral animal pathogens in spreading disease. Biosyst. Eng. 2022, 224, 92–117. [Google Scholar] [CrossRef]
- Fuentes, S.; Gonzalez Viejo, C.; Cullen, B.; Tongson, E.; Chauhan, S.S.; Dunshea, F.R. Artificial intelligence applied to a robotic dairy farm to model milk productivity and quality based on cow data and daily environmental parameters. Sensors 2020, 20, 2975. [Google Scholar] [CrossRef] [PubMed]
- Thabtah, F.; Hammoud, S.; Kamalov, F.; Gonsalves, A. Data imbalance in classification: Experimental evaluation. Inf. Sci. 2020, 513, 429–441. [Google Scholar] [CrossRef]
- Zhu, X.Q.; Wu, X.D. Class noise vs. attribute noise: A quantitative study of their impacts. Artif. Intell. Rev. 2004, 22, 177–210. [Google Scholar] [CrossRef]
- Ji, B.Y.; Banhazi, T.; Phillips, C.J.C.; Wang, C.Y.; Li, B.M. A machine learning framework to predict the next month’s daily milk yield, milk composition and milking frequency for cows in a robotic dairy farm. Biosyst. Eng. 2022, 216, 186–197. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Mohammadi-Dehcheshmeh, M.; Ebrahimie, E.; Petrovski, K.R. Comprehensive analysis of machine learning models for prediction of sub-clinical mastitis: Deep Learning and Gradient-Boosted Trees outperform other models. Comput. Biol. Med. 2019, 114, 103456. [Google Scholar] [CrossRef]
- Zhang, B.; Dong, X.; Hu, Y.; Jiang, X.; Li, G. Classification and prediction of spinal disease based on the SMOTE-RFE-XGBoost model. PeerJ. Comput. Sci. 2023, 9, e1280. [Google Scholar] [CrossRef]
- Taniya. Machine Learning Algorithms: A Comparison of Different Algorithms and When to Use Them. Medium. 2018. Available online: https://medium.com/@taniyaghosh29/machine-learning-algorithms-what-are-the-differences-9b71df4f248f (accessed on 27 May 2018).
- Adediran, S.A.; Ratkowsky, D.A.; Donaghy, D.J.; Malau-Aduli, A.E.O. Comparative evaluation of a new lactation curve model for pasture-based Holstein-Friesian dairy cows. J. Dairy Sci. 2012, 95, 5344–5356. [Google Scholar] [CrossRef]
- Kokate, L.; Dongre, V.; Khandait, V.; Kale, S. Modeling of lactation curves for prediction of standard lactation milk yield in Marathwadi buffaloes. Indian J. Anim. Sci. 2019, 89, 909–911. [Google Scholar] [CrossRef]
- García, M. Current and future vaccines and vaccination strategies against infectious laryngotracheitis (ILT) respiratory disease of poultry. Vet. Microbiol. 2017, 206, 157–162. [Google Scholar] [CrossRef]
- Bell, J.G. Factors limiting production efficiency and profitability from smallholder poultry production. World’s Poult. Sci. J. 2009, 65, 207–210. [Google Scholar] [CrossRef]
| Variables | Variables |
|---|---|
| Date | Laying rate/% |
| Age/days | Mortality/% |
| Maximum temperature throughout the day (Tmax)/°C | Number of abnormal laying hens (Nab-hens) |
| Minimum temperature throughout the day (Tmin)/°C | Qualified rate of immunization (QRimmu)/% |
| Average relative humidity (RH)/% | Treatment for H9N2 infection |
| Variable | Unit | Max | Mean | Min | SD | |
|---|---|---|---|---|---|---|
| House 1 | Age | days | 199 | - | 161 | - |
| Tmax | °C | 24 | 15 | 5 | 5 | |
| Tmin | °C | 9 | 1 | −6 | 4 | |
| RH | % | 65 | 52 | 38 | 7 | |
| Laying rate | % | 97.1 | 92.5 | 87.4 | 2.9 | |
| Mortality | % | 0.10 | 0.03 | 0.01 | 0.02 | |
| Nab-hens | hens | 201 | - | 0 | - | |
| QRimmu | % | 90 | 90 | 90 | 0 | |
| House 2 | Age | days | 258 | - | 218 | - |
| Tmax | °C | 22 | 11 | 3 | 5 | |
| Tmin | °C | 9 | −2 | −10 | 4 | |
| RH | % | 65 | 49 | 35 | 8 | |
| Laying rate | % | 94.3 | 92.4 | 86.1 | 2.3 | |
| Mortality | % | 0.20 | 0.04 | 0.01 | 0.03 | |
| Nab-hens | hens | 265 | - | 0 | - | |
| QRimmu | % | 80 | 80 | 80 | 0 | |
| House 3 | Age | days | 266 | - | 222 | - |
| Tmax | °C | 12 | 4 | −4 | 5 | |
| Tmin | °C | 1 | −6 | −14 | 4 | |
| RH | % | 65 | 46 | 35 | 7 | |
| Laying rate | % | 93.4 | 91.4 | 84.1 | 2.7 | |
| Mortality | % | 0.13 | 0.05 | 0.01 | 0.03 | |
| Nab-hens | hens | 214 | - | 0 | - | |
| QRimmu | % | 80 | 80 | 80 | 0 | |
| Criteria | Predicted Future Days | Day + 0 | Day − 1 | Day − 2 | Day − 3 | Day − 4 |
|---|---|---|---|---|---|---|
| ACC/% | H9N2 status + 0 | 92.31 | 92.31 | 92.31 | 92.31 | 89.74 |
| H9N2 status + 1 | 89.74 | 89.74 | 87.18 | 89.74 | 89.74 | |
| H9N2 status + 2 | 84.62 | 84.62 | 84.62 | 84.62 | 84.62 | |
| RR/% | H9N2 status + 0 | 92.31 | 92.31 | 92.31 | 92.31 | 84.62 |
| H9N2 status + 1 | 92.31 | 92.31 | 84.62 | 92.31 | 92.31 | |
| H9N2 status + 2 | 84.62 | 84.62 | 84.62 | 84.62 | 84.62 | |
| PR/% | H9N2 status + 0 | 85.71 | 85.71 | 85.71 | 85.71 | 84.62 |
| H9N2 status + 1 | 80.00 | 80.00 | 78.57 | 80.00 | 80.00 | |
| H9N2 status + 2 | 73.33 | 73.33 | 73.33 | 73.33 | 73.33 |
| Laying Hen House | Tmax | Tmin | RH | Laying Rate | Mortality |
|---|---|---|---|---|---|
| House 1 | −0.61 ** | −0.45 ** | −0.01 | −0.58 ** | 0.67 ** |
| House 2 | −0.30 | −0.36 * | −0.31 * | −0.86 ** | 0.66 ** |
| House 3 | −0.67 ** | −0.52 ** | −0.21 | −0.87 ** | 0.83 ** |
| Predicted H9N2 Status | Warning Level | Meaning | H9N2 Development | |
|---|---|---|---|---|
| H9N2 Status + 0 | H9N2 Status + 1 | |||
| 0 | 0 | Green | Safe | None |
| 0 | 1 | Yellow | Low warning | Start |
| 1 | 1 | Red | High warning | Development |
| 1 | 0 | Yellow | Low warning | Nearly finish |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Zhuang, Y.; Yu, L.; Li, Q.; Zhao, C.; Meng, R.; Zhu, J.; Guo, X. A Machine Learning Framework Based on Extreme Gradient Boosting to Predict the Occurrence and Development of Infectious Diseases in Laying Hen Farms, Taking H9N2 as an Example. Animals 2023, 13, 1494. https://doi.org/10.3390/ani13091494
Liu Y, Zhuang Y, Yu L, Li Q, Zhao C, Meng R, Zhu J, Guo X. A Machine Learning Framework Based on Extreme Gradient Boosting to Predict the Occurrence and Development of Infectious Diseases in Laying Hen Farms, Taking H9N2 as an Example. Animals. 2023; 13(9):1494. https://doi.org/10.3390/ani13091494
Chicago/Turabian StyleLiu, Yu, Yanrong Zhuang, Ligen Yu, Qifeng Li, Chunjiang Zhao, Rui Meng, Jun Zhu, and Xiaoli Guo. 2023. "A Machine Learning Framework Based on Extreme Gradient Boosting to Predict the Occurrence and Development of Infectious Diseases in Laying Hen Farms, Taking H9N2 as an Example" Animals 13, no. 9: 1494. https://doi.org/10.3390/ani13091494
APA StyleLiu, Y., Zhuang, Y., Yu, L., Li, Q., Zhao, C., Meng, R., Zhu, J., & Guo, X. (2023). A Machine Learning Framework Based on Extreme Gradient Boosting to Predict the Occurrence and Development of Infectious Diseases in Laying Hen Farms, Taking H9N2 as an Example. Animals, 13(9), 1494. https://doi.org/10.3390/ani13091494






