Is Penguin Circovirus Circulating Only in the Antarctic Circle? Lack of Viral Detection in Namibia
Abstract
Simple Summary
Abstract
1. Introduction
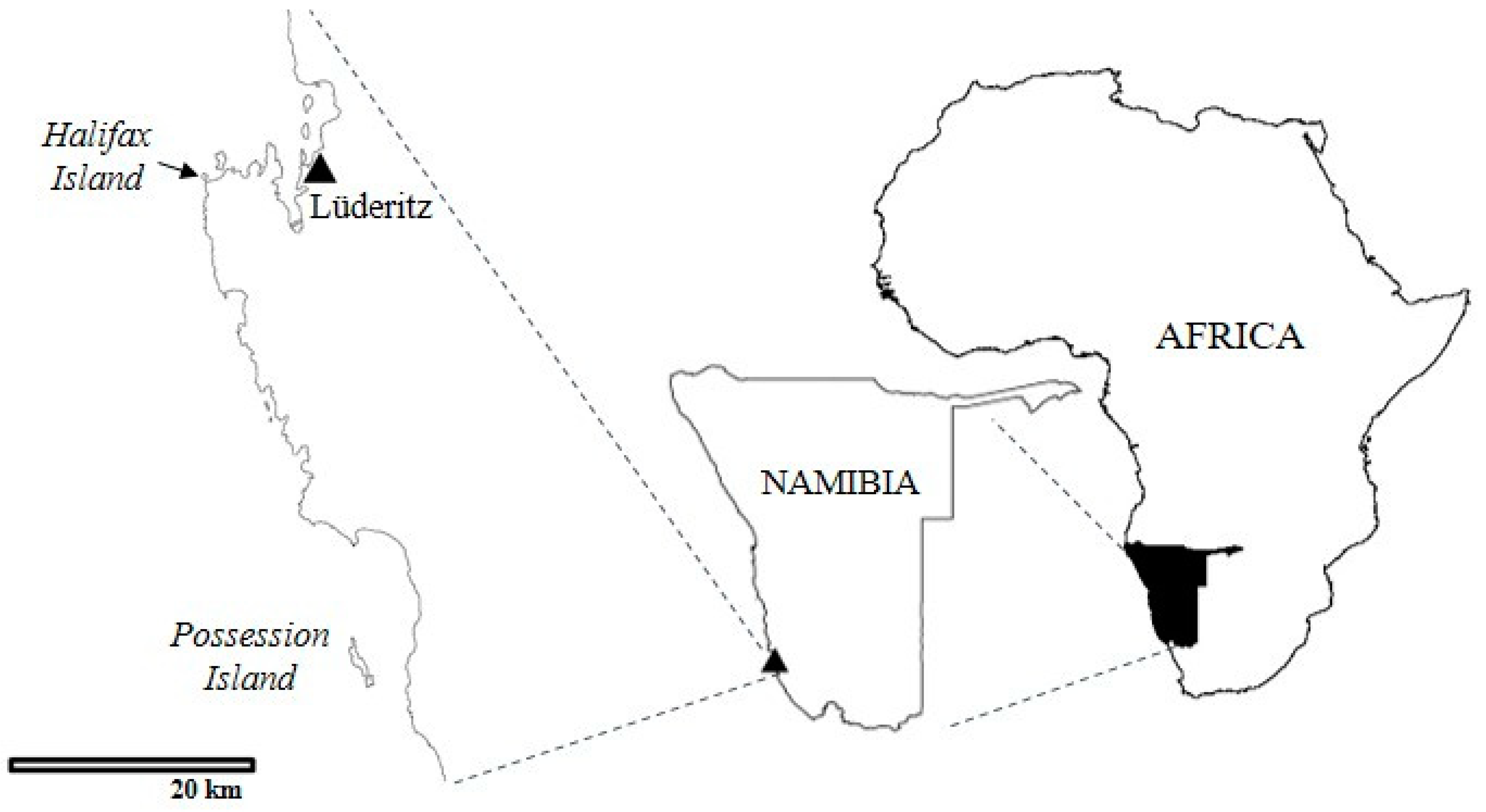
2. Materials and Methods
2.1. Sampling
2.2. PenCV Molecular Testing
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Franzo, G.; Segalés, J. Circoviruses (Circoviridae). In Encyclopedia of Virology; Bamford, D.H., Zuckerman, M., Eds.; Elsevier Science: Amsterdam, The Netherlands, 2020; pp. 182–192. ISBN 9780128145166. [Google Scholar]
- Opriessnig, T.; Karuppannan, A.K.; Castro, A.M.M.G.; Xiao, C.T. Porcine Circoviruses: Current Status, Knowledge Gaps and Challenges. Virus Res. 2020, 286, 198044. [Google Scholar] [CrossRef] [PubMed]
- Todd, D. Avian Circovirus Diseases: Lessons for the Study of PMWS. Vet. Microbiol. 2004, 98, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Segalés, J.; Kekarainen, T.; Cortey, M. The Natural History of Porcine Circovirus Type 2: From an Inoffensive Virus to a Devastating Swine Disease? Vet. Microbiol. 2013, 165, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Molini, U.; Coetzee, L.M.; Hemberger, M.Y.; Khaiseb, S.; Cattoli, G.; Dundon, W.G.; Franzo, G. The Oryx Antelope (Oryx Gazella): An Unexpected Host for Porcine Circovirus-2 (PCV-2). Pathogens 2021, 10, 1402. [Google Scholar] [CrossRef] [PubMed]
- Molini, U.; Franzo, G.; Gous, L.; Moller, S.; Hemberger, Y.M.; Chiwome, B.; Marruchella, G.; Khaiseb, S.; Cattoli, G.; Dundon, W.G. Three Different Genotypes of Porcine Circovirus 2 (PCV-2) Identified in Pigs and Warthogs in Namibia. Arch. Virol. 2021, 166, 1723–1728. [Google Scholar] [CrossRef] [PubMed]
- Molini, U.; Coetzee, L.M.; van Zyl, L.; Khaiseb, S.; Cattoli, G.; Dundon, W.G.; Franzo, G. Molecular Detection and Genetic Characterization of Porcine Circovirus 2 (PCV-2) in Black-Backed Jackal (Lupulella Mesomelas) in Namibia. Animals 2022, 12, 620. [Google Scholar] [CrossRef] [PubMed]
- Sherley, R.B.; Crawford, R.J.M.; de Blocq, A.D.; Dyer, B.M.; Geldenhuys, D.; Hagen, C.; Kemper, J.; Makhado, A.B.; Pichegru, L.; Tom, D.; et al. The Conservation Status and Population Decline of the African Penguin Deconstructed in Space and Time. Ecol. Evol. 2020, 10, 8506–8516. [Google Scholar] [CrossRef] [PubMed]
- Morandini, V.; Dugger, K.M.; Ballard, G.; Elrod, M.; Schmidt, A.; Ruoppolo, V.; Lescroël, A.; Jongsomjit, D.; Massaro, M.; Pennycook, J.; et al. Identification of a Novel Adélie Penguin Circovirus at Cape Crozier (Ross Island, Antarctica). Viruses 2019, 11, 1088. [Google Scholar] [CrossRef] [PubMed]
- Levy, H.; Fiddaman, S.R.; Djurhuus, A.; Black, C.E.; Kraberger, S.; Smith, A.L.; Hart, T.; Varsani, A. Identification of Circovirus Genome in a Chinstrap Penguin (Pygoscelis Antarcticus) and Adélie Penguin (Pygoscelis Adeliae) on the Antarctic Peninsula. Viruses 2020, 12, 858. [Google Scholar] [CrossRef] [PubMed]
- Kane, O.J.; Smith, J.R.; Dee Boersma, P.; Parsons, N.J.; Strauss, V.; Garcia-Borboroglu, P.; Villanueva, C. Feather-Loss Disorder in African and Magellanic Penguins. Available online: https://www.jstor.org/stable/40891056 (accessed on 1 March 2023).
- Barbosa, A.; Colominas-Ciuró, R.; Coria, N.; Centurión, M.; Sandler, R.; Negri, A.; Santos, M. First Record of Feather-Loss Disorder in Antarctic Penguins. Antarct. Sci. 2014, 27, 69–70. [Google Scholar] [CrossRef]
- Smeele, Z.E.; Ainley, D.G.; Varsani, A. Viruses Associated with Antarctic Wildlife: From Serology Based Detection to Identification of Genomes Using High Throughput Sequencing. Virus Res. 2018, 243, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Cameron, A.R.; Baldock, F.C. A New Probability Formula for Surveys to Substantiate Freedom from Disease. Prev. Vet. Med. 1998, 34, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Legnardi, M.; Centelleghe, C.; Tucciarone, C.M.; Cecchinato, M.; Cortey, M.; Segalés, J.; Drigo, M. Development and Validation of Direct PCR and Quantitative PCR Assays for the Rapid, Sensitive, and Economical Detection of Porcine Circovirus 3. J. Vet. Diagn. Investig. 2018, 30, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, B.; Depner, K.; Schirrmeier, H.; Beer, M. A Universal Heterologous Internal Control System for Duplex Real-Time RT-PCR Assays Used in a Detection System for Pestiviruses. J. Virol. Methods 2006, 136, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.-L.; Lu, S.-S.; Wei, W.-K.; Lv, D.-H.; Wen, X.-H.; Zhai, Q.; Chen, Q.-L.; Sun, Y.-W.; Xi, Y. Reservoirs of Porcine Circoviruses: A Mini Review. Front. Vet. Sci. 2019, 6, 319. [Google Scholar] [CrossRef] [PubMed]
- Abadie, J.; Nguyen, F.; Groizeleau, C.; Amenna, N.; Fernandez, B.; Guereaud, C.; Guigand, L.; Robart, P.; Lefebvre, B.; Wyers, M. Pigeon Circovirus Infection: Pathological Observations and Suggested Pathogenesis. Avian Pathol. 2001, 30, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Duchatel, J.P.; Todd, D.; Willeman, C.; Losson, B. Quantification of Pigeon Circovirus in Serum, Blood, Semen and Different Tissues of Naturally Infected Pigeons Using a Real-Time Polymerase Chain Reaction. Avian Pathol. 2009, 38, 143–148. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roberts, L.C.; Molini, U.; Coetzee, L.M.; Khaiseb, S.; Roux, J.-P.; Kemper, J.; Roberts, D.G.; Ludynia, K.; Doherr, M.; Abernethy, D.; et al. Is Penguin Circovirus Circulating Only in the Antarctic Circle? Lack of Viral Detection in Namibia. Animals 2023, 13, 1449. https://doi.org/10.3390/ani13091449
Roberts LC, Molini U, Coetzee LM, Khaiseb S, Roux J-P, Kemper J, Roberts DG, Ludynia K, Doherr M, Abernethy D, et al. Is Penguin Circovirus Circulating Only in the Antarctic Circle? Lack of Viral Detection in Namibia. Animals. 2023; 13(9):1449. https://doi.org/10.3390/ani13091449
Chicago/Turabian StyleRoberts, Laura C., Umberto Molini, Lauren M. Coetzee, Siegfried Khaiseb, Jean-Paul Roux, Jessica Kemper, David G. Roberts, Katrin Ludynia, Marcus Doherr, Darrell Abernethy, and et al. 2023. "Is Penguin Circovirus Circulating Only in the Antarctic Circle? Lack of Viral Detection in Namibia" Animals 13, no. 9: 1449. https://doi.org/10.3390/ani13091449
APA StyleRoberts, L. C., Molini, U., Coetzee, L. M., Khaiseb, S., Roux, J.-P., Kemper, J., Roberts, D. G., Ludynia, K., Doherr, M., Abernethy, D., & Franzo, G. (2023). Is Penguin Circovirus Circulating Only in the Antarctic Circle? Lack of Viral Detection in Namibia. Animals, 13(9), 1449. https://doi.org/10.3390/ani13091449





