Population Genetic Study on the European Flounder (Platichthys flesus) from the Southern Baltic Sea Using SNPs and Microsatellite Markers
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
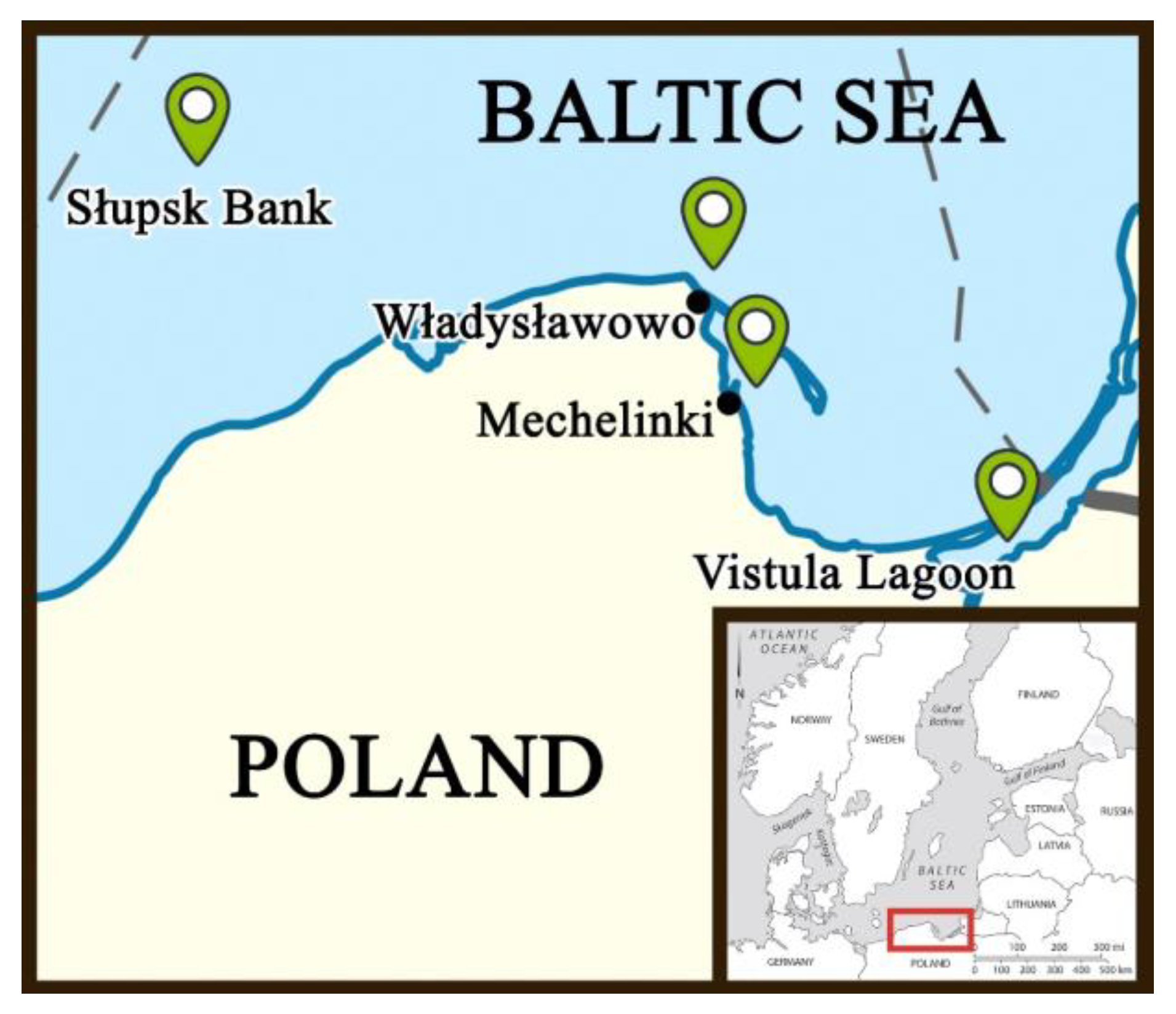
2.1. Fish Sampling and Morphological Data Collection
2.2. DNA Isolation, Microsatellite DNA Markers Amplification and Genotyping
2.3. Amplification and Genotyping of SNP Markers
2.4. Data Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zander, C.D.; Whitehead, P.J.P.; Bauchot, M.L.; Hureau, J.C.; Nielsen, J.; Tortonese, E. Fishes of the North-Eastern Atlantic and the Mediterranean; United Nations Educational Scientific and Cultural Organization: Paris, France, 1986; Volume 3. [Google Scholar]
- Haase, K.; Orio, A.; Pawlak, J.; Pachur, M.; Casini, M. Diet of dominant demersal fish species in the Baltic Sea: Is flounder stealing benthic food from cod? Mar. Ecol. Prog. Ser. 2020, 645, 159–170. [Google Scholar] [CrossRef]
- Florin, A.B. Flatfishes in the Baltic Sea. A review of biology and fishery with a focus on Swedish conditions. Finfo 2005, 14, 1–45. [Google Scholar]
- Nissling, A.; Nyberg, S.; Petereit, C. Egg buoyancy of flounder, Platichthys flesus, in the Baltic Sea—Adaptation to salinity and implications for egg survival. Fish. Res. 2017, 191, 179–189. [Google Scholar] [CrossRef]
- Jokinen, H.; Momigliano, P.; Merilä, J. From ecology to genetics and back: The tale of two flounder species in the Baltic Sea. ICES J. Mar. Sci. 2019, 76, 2267–2275. [Google Scholar] [CrossRef]
- Momigliano, P.; Denys, G.P.J.; Jokinen, H.; Merilä, J. Platichthys solemdali sp. nov. (Actinopterygii, Pleuronectiformes): A New Flounder Species From the Baltic Sea. Front. Mar. Sci. 2018, 5, 225. [Google Scholar] [CrossRef]
- FAO. Food and Agriculture Organization of the United Nations. 2022. Available online: http://www.fao.org/fishery/about/en (accessed on 1 August 2022).
- ICES Baltic Fisheries Assessment Working Group (WGBFAS). ICES Scientific Reports; ICES Baltic Fisheries Assessment Working Group (WGBFAS): Storr-Paulsen, Denmark, 2022; Volume 4, 659p. [Google Scholar] [CrossRef]
- Momigliano, P.; Jokinen, H.; Calboli, F.; Aro, E.; Merilä, J. Cryptic temporal changes in stock composition explain the decline of a flounder (Platichthys spp.) assemblage. Evol. Appl. 2018, 12, 549–559. [Google Scholar] [CrossRef]
- Hemmer-Hansen, J.; Nielsen, E.E.; Frydenberg, J.; Loeschcke, V. Adaptive divergence in a high gene flow environment: Hsc70 variation in the European flounder (Platichthys flesus L.). Heredity 2007, 99, 592–600. [Google Scholar] [CrossRef]
- Larsen, P.F.; Nielsen, E.E.; Williams, T.; Hemmer-Hansen, J.; Chipman, J.K.; Kruhøffer, M.; Grønkjær, P.; George, S.G.; Dyrskjøt, L.; Loeschcke, V. Adaptive differences in gene expression in European flounder (Platichthys flesus). Mol. Ecol. 2007, 16, 4674–4683. [Google Scholar] [CrossRef]
- Svedäng, H.; Hornborg, S. Selective fishing induces density-dependent growth. Nat. Commun. 2014, 5, 4152. [Google Scholar] [CrossRef]
- NMFRI. A Study Programme on the Marine Environment of the Puck Bay, with Particular Emphasis on Factors Relevant to Fisheries in 2019–2021; NMFRI: Gdynia, Poland, 2020. [Google Scholar]
- Hemmer-Hansen, J.; Nielsen, E.E.; Grønkjaer, P.; Loeschcke, V. Evolutionary mechanisms shaping the genetic population structure of marine fishes; lessons from the European flounder (Platichthys flesus L.). Mol. Ecol. 2007, 16, 3104–3118. [Google Scholar] [CrossRef]
- Florin, A.-B.; Höglund, J. Population structure of flounder (Platichthys flesus) in the Baltic Sea: Differences among demersal and pelagic spawners. Heredity 2008, 101, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Tysklind, N.; Neuparth, T.; Ashcroft, G.R.; Taylor, M.I.; Lyons, B.P.; McCarthy, I.D.; Carvalho, G.R. Isolation and characterization of 28 new microsatellite markers for European flounder (Platichthys flesus L.). Mol. Ecol. Resour. 2009, 9, 1065–1068. [Google Scholar] [CrossRef] [PubMed]
- Pédron, N.; Morvezen, R.; Le Moan, A.; Guinand, B.; Zambonino-Infante, J.-L.; Laroche, J.; Charrier, G. New set of candidate gene SNPs and microsatellites to disentangle selective and neutral processes shaping population responses of European flounder (Platichthys flesus) to anthropogenic stress and contrasted environments. Conserv. Genet. Resour. 2015, 7, 823–826. [Google Scholar] [CrossRef]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Nakano, S.-I.; Fujimoto, M.; Hara, H.; Sugimoto, N. Nucleic acid duplex stability: Influence of base composition on cation effects. Nucleic Acids Res. 1999, 27, 2957–2965. [Google Scholar] [CrossRef] [PubMed]
- Momigliano, P.; Jokinen, H.; Fraimout, A.; Florin, A.-B.; Norkko, A.; Merilä, J. Extraordinarily rapid speciation in a marine fish. Proc. Natl. Acad. Sci. USA 2017, 114, 6074–6079. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Peakall, R.O.D.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Yeh, F.C.; Boylet, J.B. Population genetic analysis of codominant and dominant markers and quantitative traits. Belg. J. Bot. 1997, 129, 157. [Google Scholar]
- Rousset, F. GenePop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Rice, W.R. Analyzing tables of statistical tests. Evolution 1989, 43, 223. [Google Scholar] [CrossRef] [PubMed]
- Piry, S.; Alapetite, A.; Cornuet, J.-M.; Paetkau, D.; Baudouin, L.; Estoup, A. GeneClass2: A Software for Genetic Assignment and First-Generation Migrant Detection. J. Hered. 2004, 95, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T.; Manlove, K.R.; Taper, M.L. ONCOR: A Computer Program for Genetic Stock Identification; Department of Ecology, Montana State University: Bozeman, MT, USA, 2007; Available online: https://www.montana.edu/kalinowski/software/oncor.html (accessed on 15 June 2022).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Duchesne, P.; Turgeon, J. FLOCK provides reliable solutions to the “number of populations” problem. J. Hered. 2012, 103, 734–743. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Langella, O. Populations 1.2.28. Logiciel de Génétique des Populations. Laboratoire Populations, Génétique et Évolution, CNRS UPR 9034, Gif-sur-Yvette. Available online: http://bioinformatics.org/~tryphon/populations/#ancre_bibliographie (accessed on 15 May 2022).
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimatorv2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 2013, 14, 209–214. [Google Scholar] [CrossRef]
- Waples, R.S.; Do, C. Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: A largely untapped resource for applied conservation and evolution. Evol. Appl. 2009, 3, 244–262. [Google Scholar] [CrossRef] [PubMed]
- Piry, S.; Luikart, G.; Cornuet, J.-M. Bottleneck: A computer program for detecting recent reductions in effective population size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Costopoulos, C.; Fonds, M. Proximate body composition and energy content of plaice (Pleuronectes platessa) in relation to the condition factor. Neth. J. Sea Res. 1989, 24, 45–55. [Google Scholar] [CrossRef]
- Giguère, A.; Campbell, P.G.; Hare, L.; McDonald, D.G.; Rasmussen, J.B. Influence of lake chemistry and fish age on cadmium, copper, and zinc concentrations in various organs of indigenous yellow perch (Perca flavescens). Can. J. Fish. Aquat. Sci. 2004, 61, 1702–1716. [Google Scholar] [CrossRef]
- Chapuis, M.-P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2006, 24, 621–631. [Google Scholar] [CrossRef]
- Huang, K.; Ritland, K.; Dunn, D.W.; Qi, X.; Guo, S.; Li, B. Estimating relatedness in the presence of null alleles. Genetics 2015, 202, 247–260. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; David, A.; Briscoe, D.A. Introduction to Conservation Genetics, 2nd ed.; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Bonanomi, S.; Pellissier, L.; Therkildsen, N.O.; Hedeholm, R.B.; Retzel, A.; Meldrup, D.; Olsen, S.M.; Nielsen, A.; Pampoulie, C.; Hemmer-Hansen, J.; et al. Archived DNA reveals fisheries and climate induced collapse of a major fishery. Sci. Rep. 2015, 5, 15395. [Google Scholar] [CrossRef]
- Hauser, L.; Adcock, G.J.; Smith, P.J.; Ramírez, J.H.B.; Carvalho, G.R. Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus). Proc. Natl. Acad. Sci. USA 2002, 99, 11742–11747. [Google Scholar] [CrossRef]
- Hutchinson, W.F.; Van Oosterhout, C.; Rogers, S.I.; Carvalho, G.R. Temporal analysis of archived samples indicates marked genetic changes in declining North Sea cod (Gadus morhua). Proc. R. Soc. B Boil. Sci. 2003, 270, 2125–2132. [Google Scholar] [CrossRef]
- Hoarau, G.; Boon, E.; Jongma, D.N.; Ferber, S.; Palsson, J.; Van der Veer, H.W.; Rijnsdorp, A.D.; Stam, W.T.; Olsen, J.L. Low effective population size and evidence for inbreeding in an overexploited flatfish, plaice (Pleuronectes platessa L.). Proc. R. Soc. B Boil. Sci. 2005, 272, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Sterner, T. Unobserved diversity, depletion and irreversibility The importance of subpopulations for management of cod stocks. Ecol. Econ. 2007, 61, 566–574. [Google Scholar] [CrossRef]
- Hutchinson, W.F. The dangers of ignoring stock complexity in fishery management: The case of the North Sea cod. Biol. Lett. 2008, 4, 693–695. [Google Scholar] [CrossRef] [PubMed]
- Lindegren, M.; Waldo, S.; Nilsson, P.A.; Svedäng, H.; Persson, A. Towards sustainable fisheries of the Öresund cod (Gadus morhua) through sub-stock-specific assessment and management recommendations. ICES J. Mar. Sci. 2013, 70, 1140–1150. [Google Scholar] [CrossRef]
- Calvès, I.; Lavergne, E.; Meistertzheim, A.; Charrier, G.; Cabral, H.; Guinand, B.; Quiniou, L.; Laroche, J. Genetic structure of European flounder Platichthys flesus: Effects of both the southern limit of the species’ range and chemical stress. Mar. Ecol. Prog. Ser. 2013, 472, 257–273. [Google Scholar] [CrossRef]
- Larsson, L.C.; Laikre, L.; André, C.; Dahlgren, T.G.; Ryman, N. Temporally stable genetic structure of heavily exploited Atlantic herring (Clupea harengus) in Swedish waters. Heredity 2009, 104, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Cuveliers, E.L.; Volckaert, F.A.M.; Rijnsdorp, A.D.; Larmuseau, M.H.D.; Maes, G.E. Temporal genetic stability and high effective population size despite fisheries-induced life-history trait evolution in the North Sea sole. Mol. Ecol. 2011, 20, 3555–3568. [Google Scholar] [CrossRef] [PubMed]
- Sekino, M.; Hara, M.; Taniguchi, N. Loss of microsatellite and mitochondrial DNA variation in hatchery strains of Japanese flounder Paralichthys olivaceus. Aquaculture 2002, 213, 101–122. [Google Scholar] [CrossRef]
- Xu, D.; Li, S.; Lou, B.; Zhang, Y.; Zhan, W.; Shi, H. Genetic diversity in two Japanese flounder populations from China seas inferred using microsatellite markers and COI sequences. Chin. J. Oceanol. Limnol. 2012, 30, 604–610. [Google Scholar] [CrossRef]
- An, H.S.; Nam, M.M.; Myeong, J.I.; An, C.M. Genetic diversity and differentiation of the Korean starry flounder (Platichthys stellatus) between and within cultured stocks and wild populations inferred from microsatellite DNA analysis. Mol. Biol. Rep. 2014, 41, 7281–7292. [Google Scholar] [CrossRef]
- Pan, T.; Zhang, Y.; Gao, T.; Li, F. Genetic diversity of Pleuronectes yokohamae population revealed by fluorescence microsatellite labeled. Biochem. Syst. Ecol. 2014, 55, 118–124. [Google Scholar] [CrossRef]
- Poćwierz-Kotus, A.; Kijewska, A.; Petereit, C.; Bernaś, R.; Więcaszek, B.; Arnyasi, M.; Lien, S.; Kent, M.; Wenne, R. Genetic differentiation of brackish water populations of cod Gadus morhua in the southern Baltic, inferred from genotyping using SNP-arrays. Mar. Genom. 2015, 19, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Olsen, E.M.; Knutsen, H.; Gjøsaeter, J.; Jorde, P.E.; Knutsen, J.A.; Stenseth, N.C. Small-scale biocomplexity in coastal Atlantic cod supporting a Darwinian perspective on fisheries management. Evol. Appl. 2008, 1, 524–533. [Google Scholar] [CrossRef]
- Hermant, M.; Lobry, J.; Bonhommeau, S.; Poulard, J.-C.; Le Pape, O. Impact of warming on abundance and occurrence of flatfish populations in the Bay of Biscay (France). J. Sea Res. 2010, 64, 45–53. [Google Scholar] [CrossRef]
- Marchand, J.; Evrard, E.; Guinand, B.; Cachot, J.; Quiniou, L.; Laroche, J. Genetic polymorphism and its potential relation to environmental stress in five populations of the European flounder Platichthys flesus, along the French Atlantic coast. Mar. Environ. Res. 2010, 70, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Borsa, P.; Blanquer, A.; Berrebi, P. Genetic structure of the flounders Platichthys flesus and P. stellatus at different geographic scales. Mar. Biol. 1997, 129, 233–246. [Google Scholar] [CrossRef]
- Pédron, N.; Laroche, J.; Aboim, M.A.; Tanner, S.E.; Reis-Santos, P.; Vasconcelos, R.P. Genetic variation across the geographical range of the European flounder, with a focus on southern peripheral populations. Remerciements 2016, 51. Available online: https://theses.hal.science/tel-02928183 (accessed on 28 February 2023).
- Psuty, I.; Wilkońska, H. The stability of fish assemblages under unstable conditions: A ten-year series from the Polish part of the Vistula Lagoon. Fish Aquat. Sci. 2009, 17, 65–76. [Google Scholar] [CrossRef]
- Turner, T.F.; Wares, J.P.; Gold, J.R. Genetic effective size is three orders of magnitude smaller than adult census size in an abundant, estuarine-dependent marine fish (Sciaenops ocellatus). Genetics 2002, 162, 1329–1339. [Google Scholar] [CrossRef]
- Hauser, L.; Carvalho, G.R. Paradigm shifts in marine fisheries genetics: Ugly hypotheses slain by beautiful facts. Fish 2008, 9, 333–362. [Google Scholar] [CrossRef]
- Ruggeri, P.; Splendiani, A.; Bonanomi, S.; Arneri, E.; Cingolani, N.; Santojanni, A.; Belardinelli, A.; Giovannotti, M.; Caputo, V. Temporal genetic variation as revealed by a microsatellite analysis of European sardine (Sardina pilchardus) archived samples. Can. J. Fish. Aquat. Sci. 2012, 69, 1698–1709. [Google Scholar] [CrossRef]
- Ruggeri, P.; Splendiani, A.; Di Muri, C.; Fioravanti, T.; Santojanni, A.; Leonori, I.; De Felice, A.; Biagiotti, I.; Carpi, P.; Arneri, E.; et al. Coupling demographic and genetic variability from archived collections of European anchovy (Engraulis encrasicolus). PLoS ONE 2016, 11, e0151507. [Google Scholar] [CrossRef]
- Zhivotovsky, L.A.; Teterina, A.A.; Mukhina, N.V.; Stroganov, A.N.; Rubtsova, G.A.; Afanasiev, K.I. Effects of genetic drift in a small population of Atlantic cod (Gadus morhua kildinensis Derjugin) landlocked in a meromictic lake: Genetic variation and conservation measures. Conserv. Genet. 2015, 17, 229–238. [Google Scholar] [CrossRef]
- Leberg, P.L. Effects of population bottlenecks on genetic diversity as measured by allozyme electrophoresis. Evolution 1992, 46, 477–494. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.; Wright, J.M. Microsatellite DNA in fishes. Rev. Fish Biol. Fish. 1997, 7, 331–363. [Google Scholar] [CrossRef]
- Karlsson, S.; Mork, J. Deviation from Hardy–Weinberg equilibrium, and temporal instability in allele frequencies at microsatellite loci in a local population of Atlantic cod. ICES J. Mar. Sci. 2005, 62, 1588–1596. [Google Scholar] [CrossRef]
- Atarhouch, T.; Rüber, L.; Gonzalez, E.G.; Albert, E.M.; Rami, M.; Dakkak, A.; Zardoya, R. Signature of an early genetic bottleneck in a population of Moroccan sardines (Sardina pilchardus). Mol. Phylogenet. Evol. 2006, 39, 373–383. [Google Scholar] [CrossRef]
- Tzika, A.C.; Rosa, S.F.P.; Fabiani, A.; Snell, H.L.; Snell, H.M.; Marquez, C.; Tapia, W.; Rassmann, K.; Gentile, G.; Milinkovitch, M.C. Population genetics of Galápagos land iguana (genus Conolophus) remnant populations. Mol. Ecol. 2008, 17, 4943–4952. [Google Scholar] [CrossRef]

| Sampling Site | Ao | Ae | Ho | He | P | I | PIC | Fis |
|---|---|---|---|---|---|---|---|---|
| Władysławowo | 18.8 | 9.6 | 0.815 | 0.847 | 0.121 | 2.697 | 0.887 | 0.031 |
| Mechelinki | 17.9 | 9.6 | 0.767 | 0.841 | 0.045 a | 2.656 | 0.835 | 0.074 a |
| Słupsk Bank | 19.4 | 10.8 | 0.751 | 0.844 | 0.005 b | 2.768 | 0.903 | 0.093 b |
| Vistula Lagoon | 18.8 | 10.1 | 0.801 | 0.844 | 0.214 | 2.579 | 0.832 | 0.036 |
| Fish Group | He | IAM | TPM | SMM | M | |||
|---|---|---|---|---|---|---|---|---|
| Heq | P | Heq | P | Heq | P | |||
| Władysławowo | 0.847 | 0.820 | 0.049 | 0.846 | 0.578 | 0.858 | 0.986 | 0.600 |
| Mechelinki | 0.767 | 0.813 | 0.039 | 0.840 | 0.638 | 0.855 | 0.996 | 0.574 |
| Słupsk Bank | 0.844 | 0.812 | 0.001 | 0.836 | 0.406 | 0.850 | 0.968 | 0.647 |
| Vistula Lagoon | 0.653 | 0.817 | 0.044 | 0.843 | 0.658 | 0.856 | 0.991 | 0.587 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuciński, M.; Jakubowska-Lehrmann, M.; Góra, A.; Mirny, Z.; Nadolna-Ałtyn, K.; Szlinder-Richert, J.; Ocalewicz, K. Population Genetic Study on the European Flounder (Platichthys flesus) from the Southern Baltic Sea Using SNPs and Microsatellite Markers. Animals 2023, 13, 1448. https://doi.org/10.3390/ani13091448
Kuciński M, Jakubowska-Lehrmann M, Góra A, Mirny Z, Nadolna-Ałtyn K, Szlinder-Richert J, Ocalewicz K. Population Genetic Study on the European Flounder (Platichthys flesus) from the Southern Baltic Sea Using SNPs and Microsatellite Markers. Animals. 2023; 13(9):1448. https://doi.org/10.3390/ani13091448
Chicago/Turabian StyleKuciński, Marcin, Magdalena Jakubowska-Lehrmann, Agnieszka Góra, Zuzanna Mirny, Katarzyna Nadolna-Ałtyn, Joanna Szlinder-Richert, and Konrad Ocalewicz. 2023. "Population Genetic Study on the European Flounder (Platichthys flesus) from the Southern Baltic Sea Using SNPs and Microsatellite Markers" Animals 13, no. 9: 1448. https://doi.org/10.3390/ani13091448
APA StyleKuciński, M., Jakubowska-Lehrmann, M., Góra, A., Mirny, Z., Nadolna-Ałtyn, K., Szlinder-Richert, J., & Ocalewicz, K. (2023). Population Genetic Study on the European Flounder (Platichthys flesus) from the Southern Baltic Sea Using SNPs and Microsatellite Markers. Animals, 13(9), 1448. https://doi.org/10.3390/ani13091448





