Fecal Microbiota, Forage Nutrients, and Metabolic Responses of Horses Grazing Warm- and Cool-Season Grass Pastures
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Grazing Systems
2.2. Animal and Grazing Management
2.3. Fecal and Blood Sample Collection
2.4. Forage Sampling
2.5. Analysis of Plasma Glucose and Insulin
2.6. Fecal Sample Analyses
2.7. Sequence and Statistical Analysis
3. Results
3.1. Initial 16s rRNA Sequence Analysis
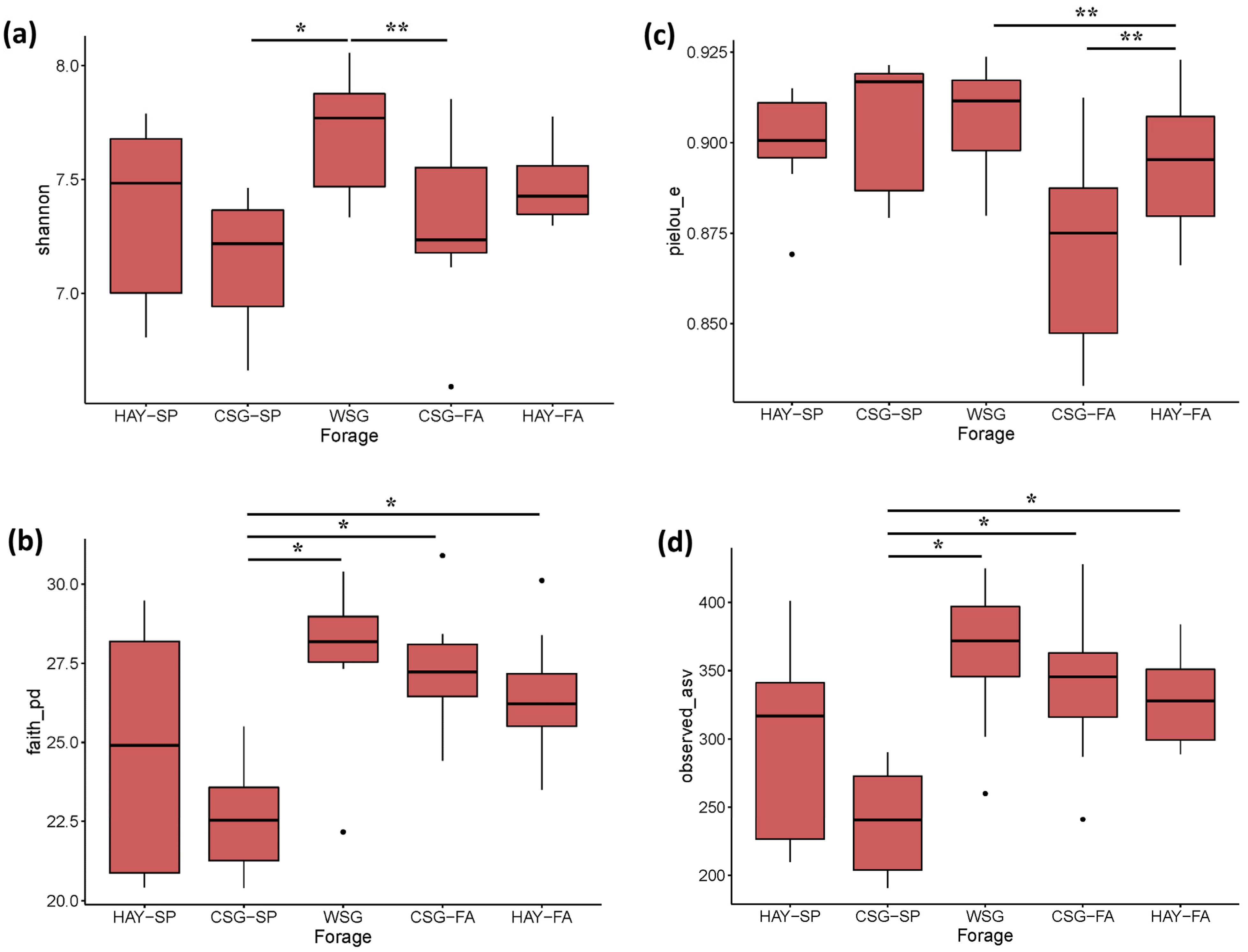
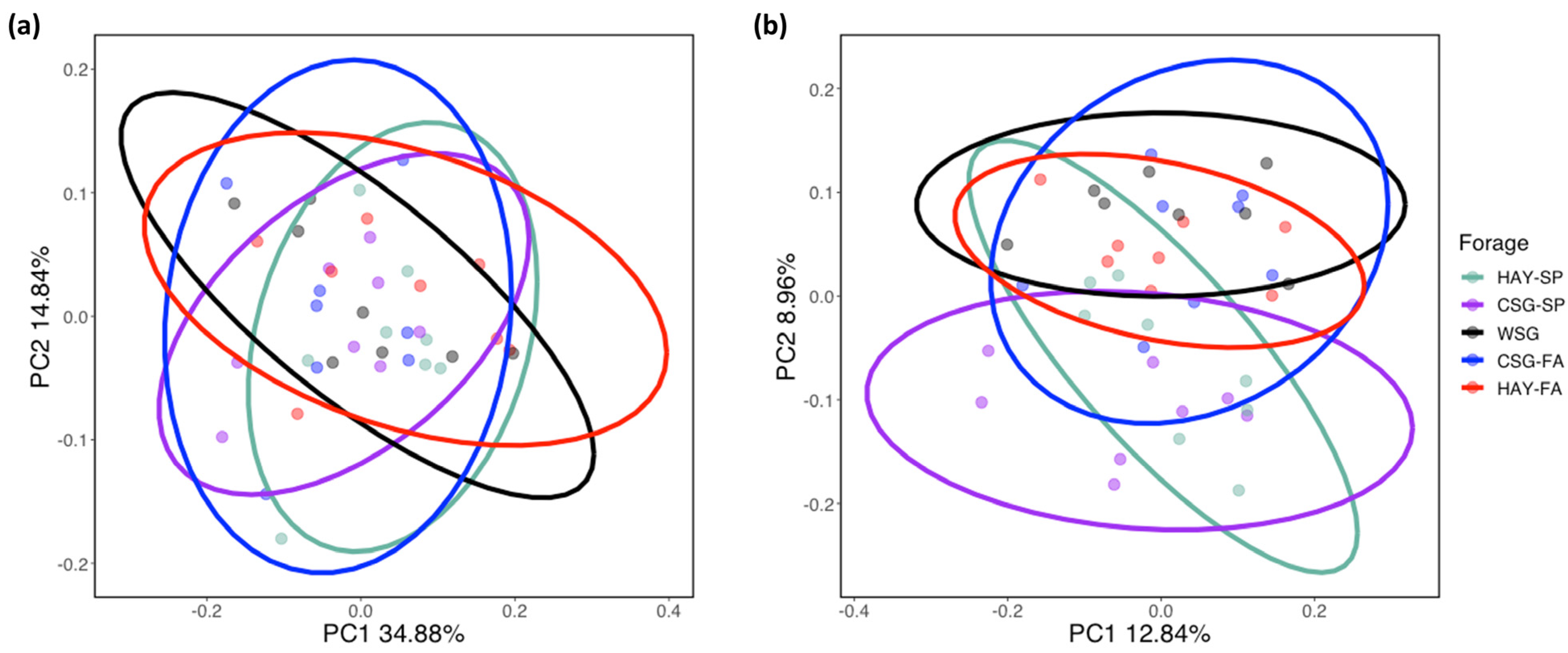
3.2. Diversity Analyses
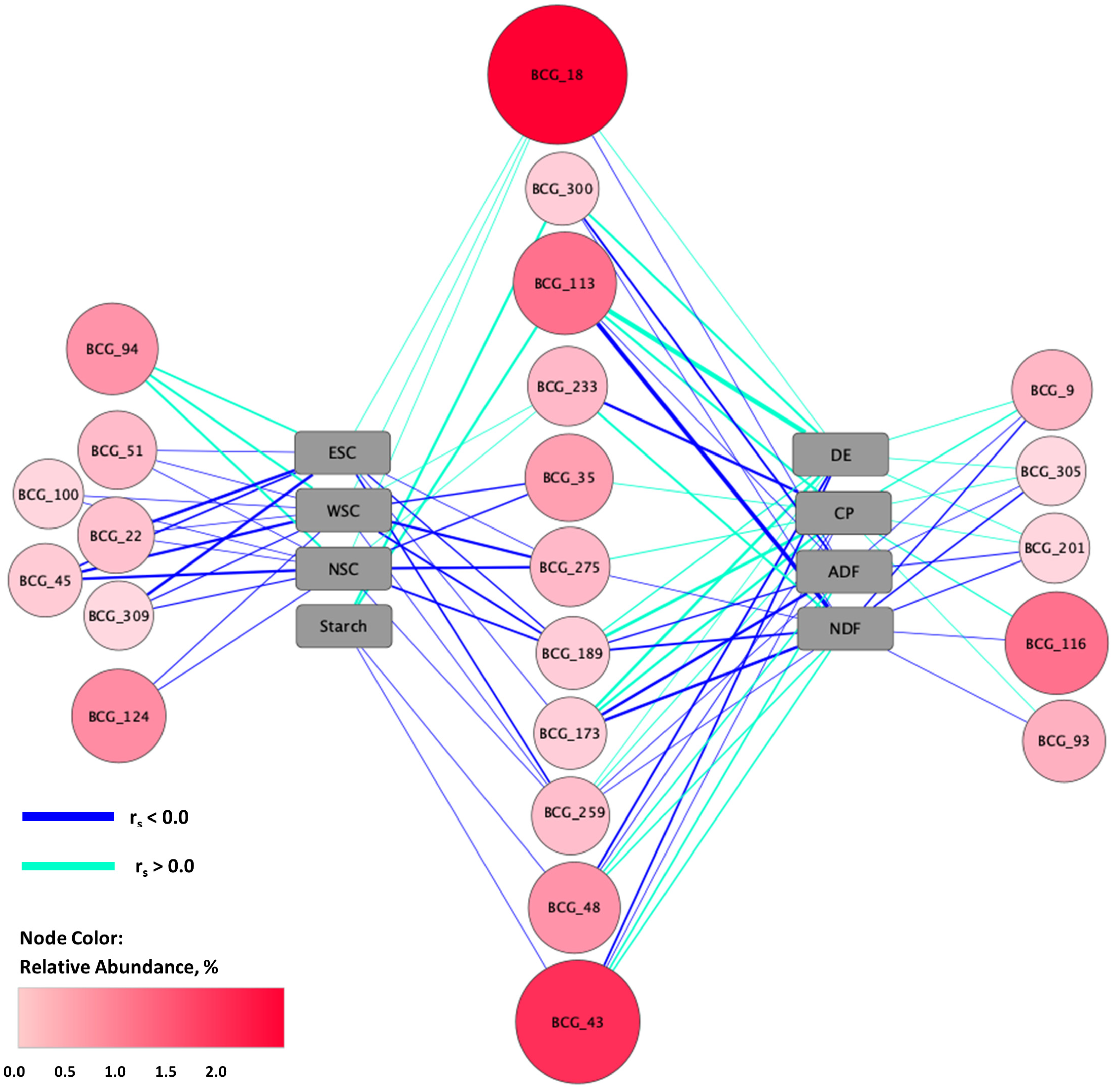
3.3. Differential Abundance
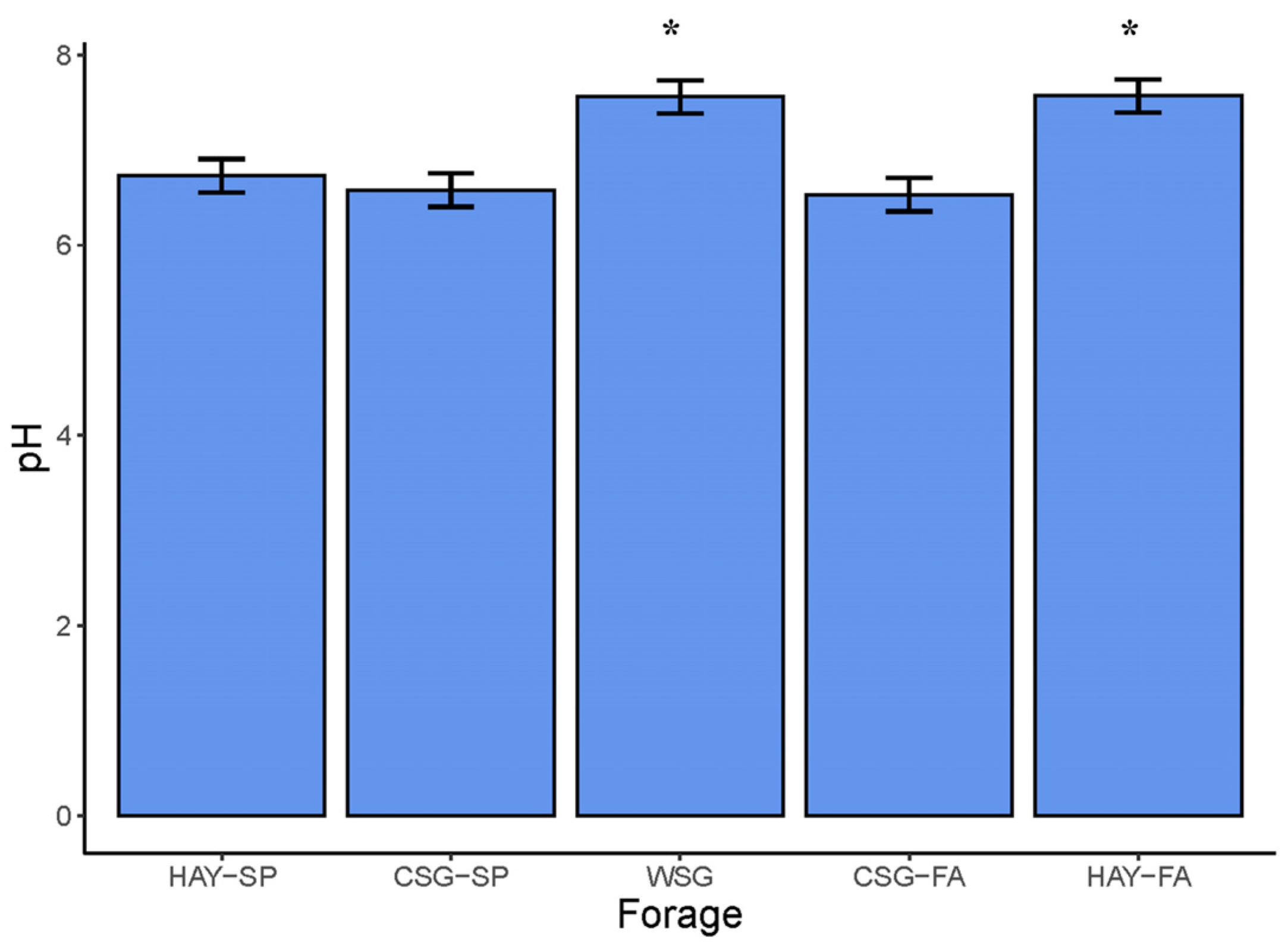
3.4. Fecal pH and Fermentation Metabolites
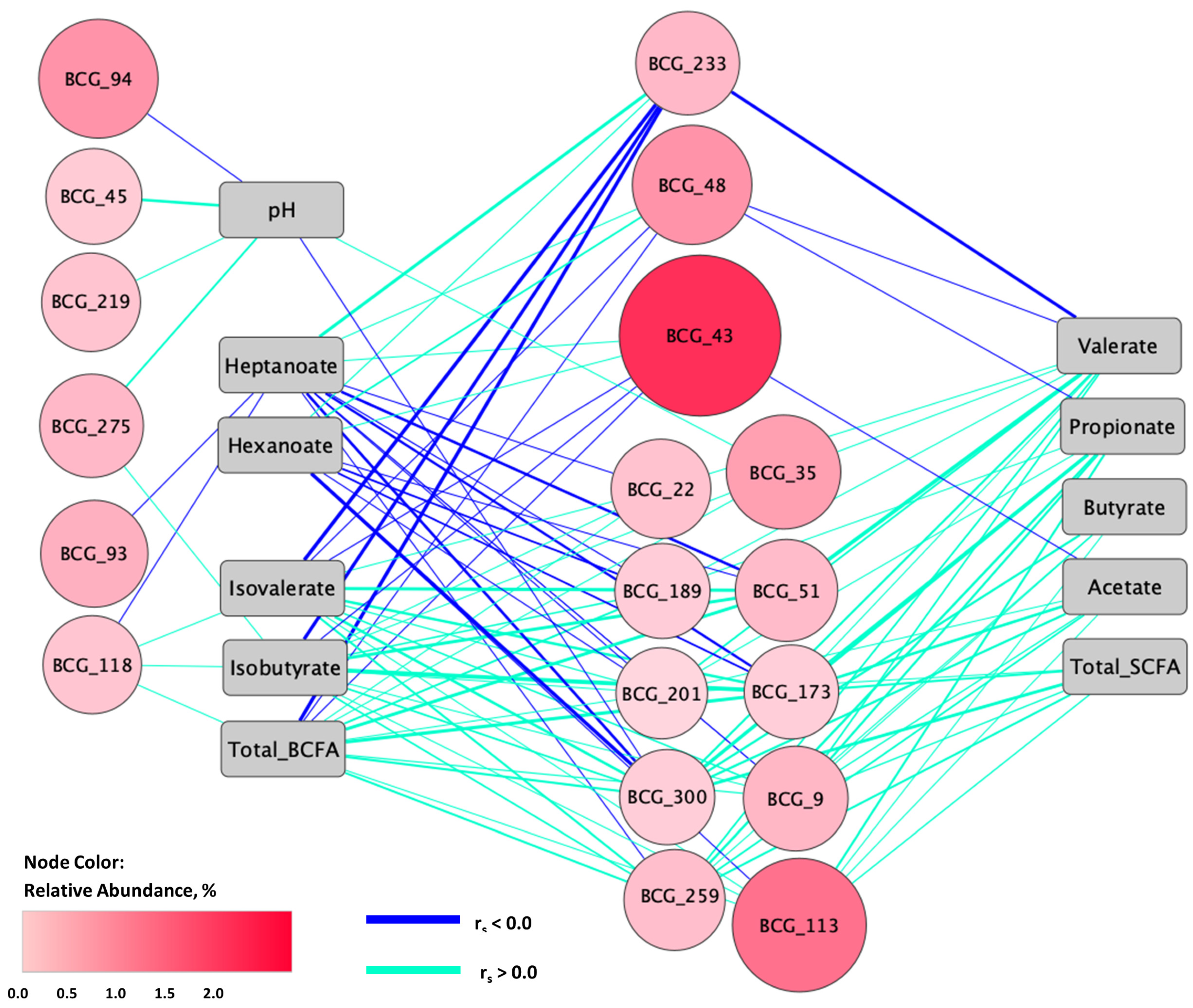
3.5. Relationships between Fecal Microbiota and Metabolites
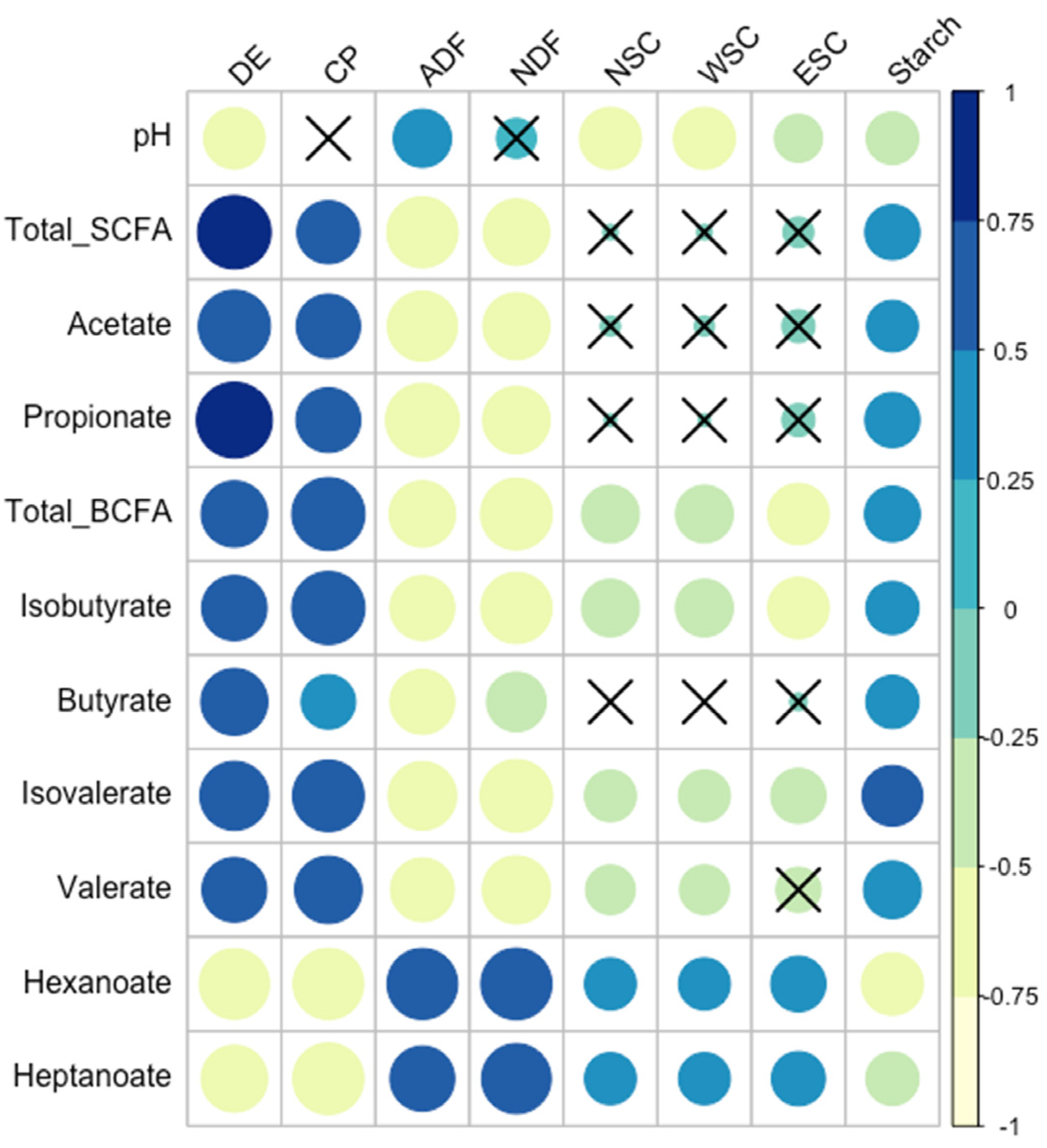
3.6. Relationships between Fecal Microbiota and Forage Nutrients
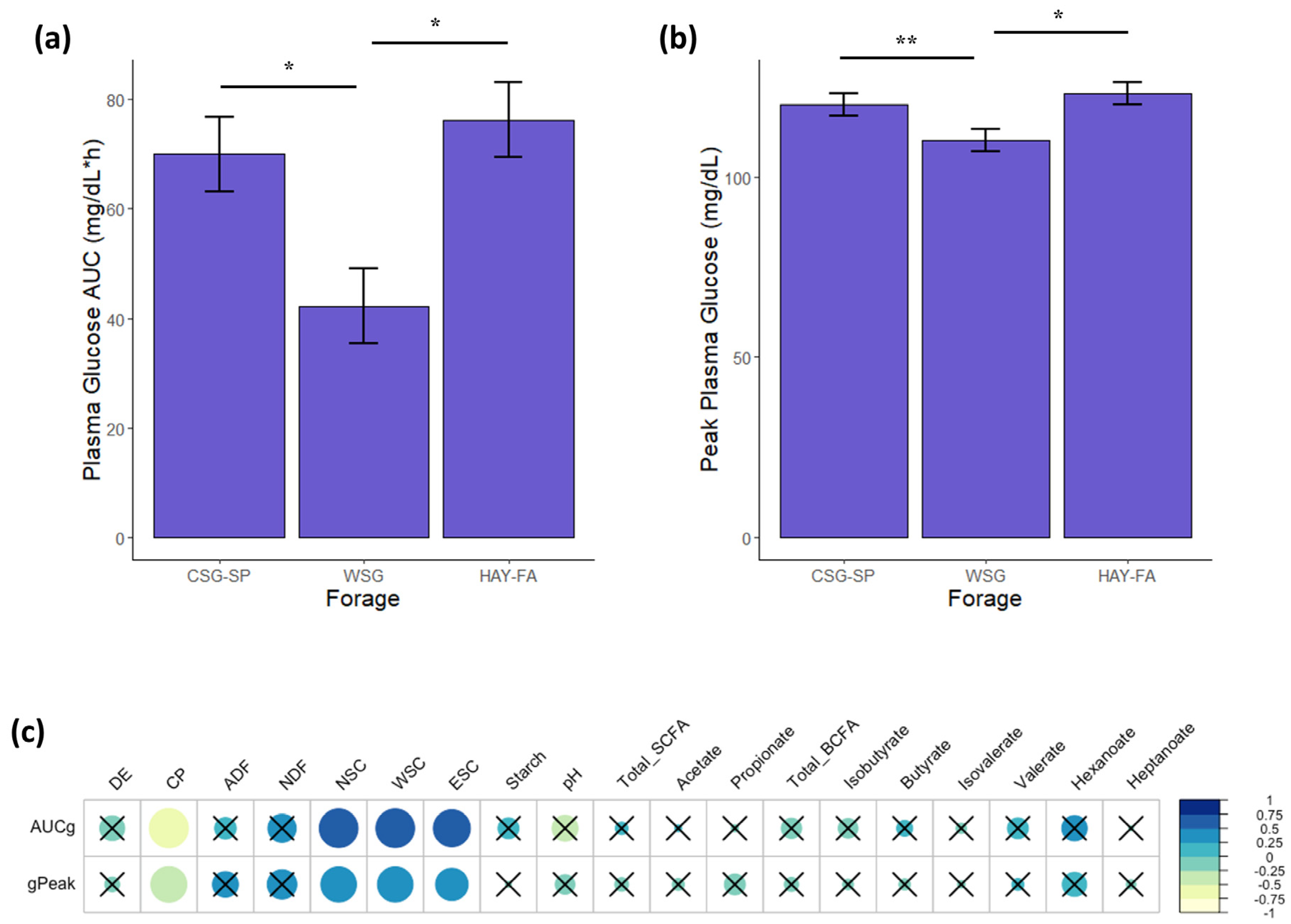
3.7. Blood Samples and Glycemic/Insulinemic Responses
3.8. Relationships between Glucose Metabolism and the Fecal Microbiota
4. Discussion
4.1. Forage Type and the Microbiome
4.2. Forage Nutrients and the Microbiome
4.3. Fecal pH and Fermentation Metabolites
4.4. Glucose and Insulin Metabolism
4.5. Additional Considerations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DeBoer, M.L.; Sheaffer, C.C.; Grev, A.M.; Catalano, D.N.; Wells, M.S.; Hathaway, M.R.; Martinson, K.L. Yield, nutritive value, and preference of annual warm-season grasses grazed by horses. Agron. J. 2017, 10, 2136–2148. [Google Scholar] [CrossRef]
- Weinert-Nelson, J.R.; Meyer, W.A.; Williams, C.A. Yield, Nutritive Value, and Horse Condition in Integrated Crabgrass and Cool-Season Grass Rotational Grazing Pasture Systems. Transl. Anim. Sci. 2021, 5, taxb208. [Google Scholar] [CrossRef]
- Ritz, K.E.; Heins, B.J.; Moon, R.; Sheaffer, C.; Weyers, S.L. Forage yield and nutritive value of cool-season and warm-season forages for grazing organic dairy cattle. Agronomy 2020, 10, 1963. [Google Scholar] [CrossRef]
- Chatterton, N.J.; Harrison, P.A.; Bennett, J.H.; Asay, K.H. Carbohydrate partitioning in 185 accessions of gramineae grown under warm and cool temperatures. J. Plant Physiol. 1989, 134, 169–179. [Google Scholar] [CrossRef]
- Jensen, K.B.; Harrison, P.; Chatterton, N.J.; Bushman, B.S.; Creech, J.E. Seasonal trends in nonstructural carbohydrates in cool-and warm-season grasses. Crop Sci. 2014, 54, 2328–2340. [Google Scholar] [CrossRef]
- Frank, N.; Geor, R.J.; Bailey, S.R.; Durham, A.E.; Johnson, P.J. Equine metabolic syndrome. J. Vet. Intern. Med. 2010, 24, 467–475. [Google Scholar] [CrossRef]
- Durham, A.E.; Frank, N.; McGowan, C.M.; Menzies-Gow, N.J.; Roelfsema, E.; Vervuert, I.; Feige, K.; Fey, K. ECEIM consensus statement on equine metabolic syndrome. J. Vet. Intern. Med. 2019, 33, 335–349. [Google Scholar] [CrossRef]
- Cavallini, D.; Penazzi, L.; Valle, E.; Raspa, F.; Bergero, D.; Formigoni, A.; Fusaro, I. When changing the hay makes a difference: A series of case reports. J. Equine Vet. Sci. 2022, 113, 103940. [Google Scholar] [CrossRef]
- Bott, R.C.; Greene, E.A.; Koch, K.; Martinson, K.L.; Siciliano, P.D.; Williams, C.; Trottier, N.L.; Burk, A.; Swinker, A. Production and environmental implications of equine grazing. J. Equine Vet. Sci. 2013, 33, 1031–1043. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Boston, R.C.; Stefanovski, D.; Kronfeld, D.S.; Harris, P.A. Obesity and diet affect glucose dynamics and insulin sensitivity in Thoroughbred geldings. J. Anim. Sci. 2003, 81, 2333–2342. [Google Scholar] [CrossRef]
- Treiber, K.H.; Boston, R.C.; Kronfeld, D.S.; Staniar, W.B.; Harris, P.A. Insulin resistance and compensation in Thoroughbred weanlings adapted to high-glycemic meals. J. Anim. Sci. 2005, 83, 2357–2364. [Google Scholar] [CrossRef]
- Jacob, S.I.; Geor, R.J.; Weber, P.S.D.; Harris, P.A.; McCue, M.E. Effect of age and dietary carbohydrate profiles on glucose and insulin dynamics in horses. Equine Vet. J. 2018, 50, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, M.L.; Pleasant, R.S.; Crisman, M.V.; Were, S.R.; Milton, S.C.; Swecker, W.S. Effects of high and moderate non-structural carbohydrate hay on insulin, glucose, triglyceride, and leptin concentrations in overweight Arabian geldings. J. Anim. Physiol. Anim. Nutr. 2012, 96, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Suagee, J.K.; Corl, B.A.; Swyers, K.L.; Smith, T.L.; Flinn, C.D.; Geor, R.J. A 90-day adaptation to a high glycaemic diet alters postprandial lipid metabolism in non-obese horses without affecting peripheral insulin sensitivity. J. Anim. Phys. Anim. Nutr. 2013, 97, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Macon, E.L.; Harris, P.; Barker, V.D.; Adams, A.A. Seasonal insulin responses to the oral sugar test in healthy and insulin dysregulated horses. J. Equine Vet. Sci. 2022, 113, 103945. [Google Scholar] [CrossRef]
- Zhang, C.; Li, S.; Yang, L.; Huang, P.; Li, W.; Wang, S.; Zhao, G.; Zhang, M.; Pang, X.; Yan, Z. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat. Commun. 2013, 4, 2163. [Google Scholar] [CrossRef]
- Dougal, K.; de la Fuente, G.; Harris, P.A.; Girdwood, S.E.; Pinloche, E.; Geor, R.J.; Nielsen, B.D.; Schott, H.C.; Elzinga, S.; Newbold, C.J. Characterisation of the faecal bacterial community in adult and elderly horses fed a high fibre, high oil or high starch diet using 454 pyrosequencing. PLoS ONE 2014, 9, e87424. [Google Scholar] [CrossRef]
- Julliand, V.; Grimm, P. The impact of diet on the hindgut microbiome. J. Equine Vet. Sci. 2017, 52, 23–28. [Google Scholar] [CrossRef]
- Garber, A.; Hastie, P.; Murray, J.A. Factors influencing equine gut microbiota: Current knowledge. J. Equine Vet. Sci. 2020, 88, 102943. [Google Scholar] [CrossRef]
- Raspa, F.; Tarantola, M.; Muca, E.; Bergero, D.; Soglia, D.; Cavallini, D.; Vervuert, I.; Bordin, C.; De Palo, P.; Valle, E. Does Feeding Management Make a Difference to Behavioural Activities and Welfare of Horses Reared for Meat Production? Animals 2022, 12, 1740. [Google Scholar] [CrossRef]
- Raspa, F.; Vervuert, I.; Capucchio, M.T.; Colombino, E.; Bergero, D.; Forte, C.; Greppi, M.; Cavallarin, L.; Giribaldi, M.; Antoniazzi, S.; et al. A high-starch vs. high-fibre diet: Effects on the gut environment of the different intestinal compartments of the horse digestive tract. BMC Vet. Res. 2022, 18, 187. [Google Scholar] [CrossRef]
- Garber, A.; Hastie, P.; McGuinness, D.; Malarange, P.; Murray, J.A. Abrupt dietary changes between grass and hay alter faecal microbiota of ponies. PLoS ONE 2020, 15, e0237869. [Google Scholar] [CrossRef]
- Sorensen, R.J.; Drouillard, J.S.; Douthit, T.L.; Ran, Q.; Marthaler, D.G.; Kang, Q.; Vahl, C.I.; Lattimer, J.M. Effect of hay type on cecal and fecal microbiome and fermentation parameters in horses. J. Anim. Sci. 2021, 99, skaa407. [Google Scholar] [CrossRef]
- Weinert-Nelson, J.R.; Biddle, A.S.; Williams, C.A. Fecal microbiome of horses transitioning between warm-season and cool-season grass pasture within integrated rotational grazing systems. Anim. Microbiome 2022, 4, 41. [Google Scholar] [CrossRef]
- Elzinga, S.E.; Weese, J.S.; Adams, A.A. Comparison of the fecal microbiota in horses with equine metabolic syndrome and metabolically normal controls fed a similar all-forage diet. J. Equine Vet. Sci. 2016, 44, 9–16. [Google Scholar] [CrossRef]
- Fitzgerald, D.M.; Spence, R.J.; Stewart, Z.K.; Prentis, P.J.; Sillence, M.N.; De Laat, M.A. The effect of diet change and insulin dysregulation on the faecal microbiome of ponies. J. Exper. Biol. 2020, 223, jeb219154. [Google Scholar] [CrossRef]
- Boshuizen, B.; Moreno de Vega, C.V.; De Maré, L.; De Meeûs, C.; De Oliveira, J.E.; Hosotani, G.; Gansemans, Y.; Deforce, D.; Van Nieuwerburgh, F.; Delesalle, C. Effects of aleurone supplementation on glucose-insulin metabolism and gut microbiome in untrained healthy horses. Front. Vet. Sci. 2021, 12, 642809. [Google Scholar] [CrossRef]
- Biddle, A.S.; Tomb, J.F.; Fan, Z. Microbiome and blood analyte differences point to community and metabolic signatures in lean and obese horses. Front. Vet. Sci. 2018, 5, 225. [Google Scholar] [CrossRef]
- Pan, F.; Zhang, L.; Li, M.; Hu, Y.; Zeng, B.; Yuan, H.; Zhao, L.; Zhang, C. Predominant gut Lactobacillus murinus strain mediates anti-inflammaging effects in calorie-restricted mice. Microbiome 2018, 6, 54. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, F.; Ding, X.; Wu, G.; Lam, Y.Y.; Wang, X.; Fu, H.; Xue, X.; Lu, C.; Ma, J.; et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018, 359, 1151–1156. [Google Scholar] [CrossRef]
- Office of the New Jersey State Climatologist at Rutgers University: Rutgers New Jersey Weather Network. Available online: https://www.njweather.org/data (accessed on 12 July 2021).
- Williams, C.A.; Kenny, L.B.; Weinert, J.R.; Sullivan, K.; Meyer, W.; Robson, M.G. Effects of 27 mo of rotational vs. continuous grazing on horse and pasture condition. Transl. Anim. Sci. 2020, 4, txaa084. [Google Scholar] [CrossRef] [PubMed]
- Henneke, D.R.; Potter, G.D.; Kreider, J.L.; Yeates, B.F. Relationship between condition score, physical measurements and body fat percentage in mares. Equine Vet. J. 1983, 15, 371–372. [Google Scholar] [CrossRef]
- Schuver, A.; Frank, N.; Chameroy, K.A.; Elliott, S.B. Assessment of insulin and glucose dynamics by using an oral sugar test in horses. J. Equine Vet. Sci. 2014, 34, 465–470. [Google Scholar] [CrossRef]
- Tinworth, K.D.; Wynn, P.C.; Boston, R.C.; Harris, P.A.; Sillence, M.N.; Thevis, M.; Thomas, A.; Noble, G.K. Evaluation of commercially available assays for the measurement of equine insulin. Dom. Anim. Endocrin. 2011, 41, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.A.; Kenny, L.B.; Burk, A.O. Effects of grazing system, season, and forage carbohydrates on glucose and insulin dynamics of the grazing horse. J. Anim. Sci. 2019, 97, 2541–2554. [Google Scholar] [CrossRef] [PubMed]
- Honarbakhsh, M.; Ericsson, A.; Zhong, G.; Isoherranen, N.; Zhu, C.; Bromberg, Y.; Van Buiten, C.; Malta, K.; Joseph, L.; Sampath, H.; et al. Impact of vitamin A transport and storage on intestinal retinoid homeostasis and functions. J. Lipid Res. 2021, 62, 100046. [Google Scholar] [CrossRef]
- García-Villalba, R.; Giménez-Bastida, J.A.; García-Conesa, M.T.; Tomás-Barberán, F.A.; Carlos Espín, J.; Larrosa, M. Alternative method for gas chromatography-mass spectrometry analysis of short-chain fatty acids in faecal samples. J. Sep. Sci. 2012, 35, 1906–1913. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible; interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- McDonald, D.; Clemente, J.C.; Kuczynski, J.; Rideout, J.R.; Stombaugh, J.; Wendel, D.; Wilke, A.; Huse, S.; Hufnagle, J.; Meyer, F.; et al. The Biological Observation Matrix (BIOM) format or: How I learned to stop worrying and love the ome-ome. Gigascience 2012, 1, 7. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stakebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York City, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423, 623–656. [Google Scholar] [CrossRef]
- Kruskal, W.H.; Wallis, W.A. Use of ranks in one-criterion variance analysis. J. Amer. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Pielou, E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Faith, D. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- McKinney, W. Data structures for statistical computing in python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 21–23 July 2010; van der Walt, S., Millman, J., Eds.; pp. 56–61. [Google Scholar]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 27. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and uqalitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Hamady, M.; Lozupone, C.; Knight, R. Fast unifrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequening and PhyloChip data. ISME J. 2010, 4, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Chang, Q.; Luan, Y.; Sun, F. Variance adjusted weighted UniFrac: A powerful beta diversity measure for comparing communities based on phylogeny. BMC Bioinform. 2011, 12, 118. [Google Scholar] [CrossRef]
- Chen, J.; Bittinger, K.; Charlson, E.S.; Hofmann, C.; Lewis, J.; Wu, G.D.; Collman, G.; Bushman, F.D.; Li, H. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 2012, 28, 2106–2113. [Google Scholar] [CrossRef]
- McDonald, D.; Vázquez -Baeza, Y.; Koslicki, D.; McClelland, J.; Reeve, N.; Xu, Z.; Gonzalez, A.; Knight, R. Striped UniFrac: Enebling microbiome analysis at unprecedented scale. Nat. Methods 2018, 15, 847–848. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Hagberg, A.A.; Shult, D.A.; Swart, P.J. Exploring network structure, dynamics, and function using NetworkX. In Proceedings of the 7th Python in Science Conference, Pasedena, CA, USA, 19–24 August 2008; Varoquaux, G., Vaught, T., Millman, J., Eds.; pp. 11–15. [Google Scholar]
- Shaffer, M.; Thurimella, K.; Sterrett, J.D.; Lozupone, C.A. SCNIC: Sparse correlation network investigation for compositional data. Mol. Ecol. Resour. 2022, 23, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, B.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar] [CrossRef]
- Bukolich, N.A.; Dillon, M.; Bolyen, E.; Kaehler, B.D.; Huttley, G.A.; Caparaso, J.G. q2-sample-classifier: Machine learning tools for microbiome classification and regression. J. Open Res. Soft. 2018, 3, 934. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, M.; Wang, S.; Han, R.; Cao, Y.; Hua, W.; Mao, Y.; Zhang, X.; Pang, X.; Wei, C.; et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2013, 4, 232. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, K.; Peplies, J.; Glockner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl. Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Pablo, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucl. Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caparaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-eature-classifier plugin. Microbiome 2018, 1, 90. [Google Scholar] [CrossRef]
- Shiang, K.D. The SAS calculations of areas under the curve (AUC) for multiple metabolic readings. In Proceedings of the SAS Conference Proceedings: Western Users of SAS Software, Pasedena, CA, USA, 13–15 October 2004; pp. 1–14. [Google Scholar]
- Treiber, K.H.; Kronfeld, D.S.; Hess, T.M.; Boston, R.C.; Staniar, W.B.; Harris, P.A. Use of proxies and reference quintiles obtained from minimal model analysis for determination of insulin sensitivity and pancreatic beta-cell responsiveness in horses. Am. J. Vet. Res. 2005, 66, 2114–2121. [Google Scholar] [CrossRef]
- Staniar, W.B.; Cubitt, T.A.; George, L.A.; Harris, P.A.; Geor, R.J. Glucose and insulin responses to different dietary energy sources in Thoroughbred broodmares grazing cool season pasture. Livest. Sci. 2007, 111, 164–171. [Google Scholar] [CrossRef]
- Biddle, A.S.; Stewart, L.; Blanchard, J.; Leschine, S. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in Diverse Gut Communities. Diversity 2013, 5, 627–640. [Google Scholar] [CrossRef]
- Elliott, C.L.; Edwards, J.E.; Wilkinson, T.J.; Allison, G.G.; McCaffrey, K.; Scott, M.B.; Rees-Stevens, P.; Kingston-Smith, A.H.; Huws, S.A. Using ‘Omic approaches to compare temporal bacterial colonization of Lolium perenne, Lotus corniculatus, and Trifolium pratense in the rumen. Front. Microbiol. 2018, 9, 2184. [Google Scholar] [CrossRef]
- Palevich, N.; Maclean, P.H.; Kelly, W.J.; Leahy, S.C.; Rakonjac, J.; Atwood, G.T. Complete genome sequence of the polysaccharide-degrading rumen bacterium Pseudobutyvibrio xylanivorans MA3014 reveals an incomplete glycolytic pathway. Genome Biol. Evol. 2020, 12, 1566–1572. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, X.; Deng, L.; Chen, S.; Zhu, C.; Li, J. Effects of pasture grass, silage, and hay diet on equine fecal microbiota. Animals 2021, 11, 1330. [Google Scholar] [CrossRef]
- Warzecha, C.M.; Coverdale, J.A.; Janecka, J.E.; Leatherwood, J.L.; Pinchak, W.E.; Wickersham, T.A.; McCann, J.C. Influence of short-term dietary starch inclusion on the equine cecal microbiome. J. Anim. Sci. 2017, 95, 5077–5090. [Google Scholar] [CrossRef]
- Millinovich, G.J.; Burrell, P.C.; Pollitt, C.C.; Klieve, A.V.; Blackall, L.L.; Ouwerkerk, D.; Woodland, E.; Trott, D.J. Microbial ecology of the equine hindgut during oliofructose-induced laminitis. ISME J. 2008, 2, 1089–1100. [Google Scholar] [CrossRef]
- Tuniyazi, M.; He, J.; Guo, J.; Li, S.; Zhang, N.; Hu, X.; Fu, Y. Changes of microbial and metabolome of the equine hindgut during oligofructose-induced laminitis. BMC Vet. Res. 2021, 17, 11. [Google Scholar] [CrossRef] [PubMed]
- Solden, L.M.; Hoyt, D.W.; Collins, W.B.; Plank, J.E.; Daly, R.A.; Hildebrand, E.; Beavers, T.J.; Wolfe, R.; Nicora, C.D.; Purvine, S.O.; et al. New roles in hemicellulosic sugar fermentation for the uncultivated Bacteroidetes family BS11. ISME J. 2017, 11, 691–703. [Google Scholar] [CrossRef]
- Grimm, P.; Combes, S.; Pascal, G.; Cauquil, L.; Julliand, V. Dietary composition and yeast/microalgae combination supplementation modulate the microbial ecosystem in the caecum, colon and faeces of horses. Br. J. Nutr. 2020, 123, 372–382. [Google Scholar] [CrossRef]
- Stewart, H.L.; Engiles, J.B.; Stefanovski, D.; Southwood, L. Clinical and intestinal histologic features of horses treated for recurrent colic: 66 cases (2006–2015). J. Am. Vet. Med. Assoc. 2018, 252, 1279–1288. [Google Scholar] [CrossRef]
- Stewart, H.L.; Pitta, D.; Indugu, N.; Vecchiarelli, B.; Hennessy, M.L.; Engiles, J.B.; Southwood, L.L. Changes in the faecal bacterial microbiota during hospitalisation of horses with colic and the effect of different causes of colic. Equine Vet. J. 2020, 53, 1119–1131. [Google Scholar] [CrossRef]
- Cheng, W.; Lu, J.; Li, B.; Lin, W.; Zhang, Z.; Wei, X.; Sun, C.; Chi, M.; Bi, W.; Yang, B.; et al. Effect of functional oligosaccharides and ordinary dietary fiber on intestinal microbiota diversity. Front. Microbiol. 2017, 20, 1750. [Google Scholar] [CrossRef]
- Pi, Y.; Hu, J.; Bai, Y.; Wang, Z.; Wu, Y.; Ye, H.; Zhang, S.; Tao, S.; Xiao, Y.; Han, D.; et al. Effects of dietary fibers with different physicochemical properties on fermentation kinetics and microbial composition by fecal inoculum from lactating sows in vitro. J. Sci. Food Agric. 2020, 101, 907–917. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Ye, C.; Yuan, J.; Qin, S. Alginate oligosaccharide improves lipid metabolism and inflammation by modulating gut microbiota in high-fat diet fed mice. Appl. Microbiol. Biotech. 2020, 104, 3541–3554. [Google Scholar] [CrossRef]
- Ericsson, A.C.; Johnson, P.J.; Gieche, L.M.; Zobrist, C.; Bucy, K.; Townsend, K.S.; Martin, L.M.; LaCarrubba, A.M. The influence of diet change and oral metformin on blood glucose regulation and the fecal microbiota of healthy horses. Animals 2021, 11, 976. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef]
- Shin, N.R.; Lee, J.C.; Lee, H.Y.; Kim, M.S.; Whon, T.W.; Lee, M.S.; Bae, J.W. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 2014, 63, 727–735. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, W.; Wang, J.; Shi, J.; Sun, Y.; Wang, W.; Ning, G.; Liu, R.; Hong, J. Akkermansia muciniphila improves metabolic profiles by reducing inflammation in chow diet-fed mice. J. Mol. Endocrinol. 2017, 58, 1–14. [Google Scholar] [CrossRef]
- Zhai, R.; Xue, X.; Zhang, L.; Yang, X.; Zhao, L.; Zhang, C. Strain-specific anti-inflammatory properties of two Akkermansia muciniphila strains on chronic colitis in mice. Front. Cell. Infect. Microbiol. 2019, 9, 239. [Google Scholar] [CrossRef]
- Lindenberg, F.; Krych, L.; Fielden, J.; Kot, W.; Frøkiær, H.; Van Galen, G.; Nielsen, D.S.; Hansen, A.K. Expression of immune regulatory genes correlate with the abundance of specific Clostridiales and Verrucomicrobia species in the equine ileum and cecum. Sci. Rep. 2019, 9, 12674. [Google Scholar] [CrossRef]
- Lindenberg, F.C.; Lützhøft, D.O.; Krych, L.; Fielden, J.; Kot, W.; Frøkiær, H.; van Galen, G.; Nielsen, D.S.; Hansen, A.K. An oligosaccharide rich diet increases Akkermansia spp. bacteria in the equine microbiota. Front. Microbiol. 2021, 12, 666039. [Google Scholar] [CrossRef]
- Respondek, F.; Goachet, A.; Julliand, R.F.V. Effects of short-chain fructo-oligosaccharides on the microbial and biochemical profile of different segments of the gastro-intestinal tract in horses. Pferdeheilkunde 2008, 23, 146. [Google Scholar] [CrossRef]
- Respondek, F.; Meyers, K.; Smith, T.L.; Wagner, A.; Goer, R.J. Dietary supplementation with short-chain fructo-oligosaccharides improves insulin sensitivity in obese horses. J. Anim. Sci. 2011, 89, 77–83. [Google Scholar] [CrossRef]
- Uyeno, Y.; Kawashima, K.; Hasunuma, T.; Wakimoto, W.; Noda, M.; Nagashima, S.; Akiyama, K.; Tabata, M.; Kushibiki, S. Effects of cellooligosaccharide or a combination of cellooligosaccharide and live Clostridium butyricum culture on performance and intestinal ecology in Holstein calves fed milk or milk replacer. Livest. Sci. 2013, 153, 88–93. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Zeng, X.; Zhou, L.; Cao, G.; Yang, C. Effects of dietary supplementation of probiotic, Clostridium butyricum, on growth performance, immune response, intestinal barrier function, and digestive enzyme activity in broiler chickens challenged with Escherichia coli K88. J. Anim. Sci. Biotech. 2016, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, S.; Zheng, J.; Li, W.; Jiang, X.; Zhao, X.; Li, J.; Che, L.; Lin, Y.; Xu, S.; et al. Effects of dietary Clostridium butyricum supplementation on growth performance, intestinal development, and immune response of weaned piglets challenged with lipopolysaccharide. J. Anim. Sci. Biotech. 2016, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Urshel, K.L.; Lawrence, L.M. Amino acids and protein. In Equine Applied and Clinical Nutrition: Health, Welfare and Performance; Goer, R., Harris, P.A., Coenen, M., Eds.; Elsevier Health Sciences: London, UK, 2013; pp. 113–135. [Google Scholar]
- Diether, N.E.; Willing, B.P. Microbial fermentation of dietary protein: An important factor in diet–microbe–host interaction. Microorganisms 2019, 7, 19. [Google Scholar] [CrossRef]
- Hansen, N.C.; Avershina, E.; Mydland, L.T.; Næsset, J.A.; Austbø, D.; Moen, B.; Måge, I.; Rudi, K. High nutrient availability reduces the diversity and stability of the equine caecal microbiota. Microb. Ecol. Health Dis. 2015, 26, 27216. [Google Scholar] [CrossRef]
- Daly, K.; Proudman, C.J.; Duncan, S.H.; Flint, H.J.; Dyer, J.; Shirazi-Beechey, S.P. Alterations in microbiota and fermentation products in equine large intestine in response to dietary variation and intestinal disease. Br. J. Nutr. 2012, 107, 989–995. [Google Scholar] [CrossRef]
- Medina, M.; Girard, I.D.; Jacotot, E.; Julliand, V. Effect of a preparation of Saccharomyces cerevisiae on microbial profiles and fermentation patterns in the large intestine of horses fed a high fiber or a high starch diet. J Anim. Sci. 2002, 80, 2600–2609. [Google Scholar] [CrossRef]
- Jensen, R.; Austbø, D.; Blache, D.; Knudsen, K.B.; Tauson, A.-H. The effect of feeding barley or hay alone or in combination with molassed sugar beet pulp on the metabolic responses in plasma and caecum of horses. Anim. Feed Sci. Technol. 2016, 214, 53–65. [Google Scholar] [CrossRef]
- Garner, H.E.; Moore, J.N.; Johnson, J.H.; Clark, L.; Amend, J.F.; Tritschler, L.G.; Coffmann, J.R.; Sprouse, R.F.; Hutcheson, D.P.; Salem, C.A. Changes in the caecal flora associated with the onset of laminitis. Equine Vet. J. 1978, 10, 249–252. [Google Scholar] [CrossRef]
- Goodson, J.; Tyznik, W.J.; Cline, J.H.; Dehority, B.A. Effects of an abrupt diet change from hay to concentrate on microbial numbers and physical environment in the cecum of the pony. Appl. Environ. Microbiol. 1988, 54, 1946–1950. [Google Scholar] [CrossRef]
- Geor, R.J.; Harris, P.A. How to minimize gastrointestinal disease associated with carbohydrate nutrition in horses. In Proceedings of the Annual Convention of the AAEP, Orlando, FL, USA, 1–5 December 2007; pp. 178–185. [Google Scholar]
- Allison, S.D.; Martiny, J.B.H. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Moya, A.; Ferrer, M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 2016, 24, 402–413. [Google Scholar] [CrossRef]
- Viera-Silva, S.; Falony, G.; Darzi, Y.; Lima-Mendez, G.; Yunta, R.G.; Okuda, S.; Vandeputte, D.; Valles-Colomer, M.; Hildebrand, F.; Chaffron, S.; et al. Species-function relationships shape ecological properties of the human gut microbiome. Nat. Microbiol. 2016, 1, 16088. [Google Scholar] [CrossRef]
- Tian, L.; Wang, X.-W.; Wu, A.-K.; Fan, Y.; Friedman, J.; Dahlin, A.; Waldor, M.K.; Weinstock, G.M.; Weiss, S.T.; Liu, Y.-Y.; et al. Deciphering functional redundancy in the human microbiome. Nat. Commun. 2020, 11, 6217. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, M.; Pang, X.; Zhao, Y.; Wang, L.; Zhao, L. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. 2012, 6, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- Fei, N.; Zhao, L. An opportunistic pathogen isolated from the gut of an obese man causes obesity in germfree mice. ISME J. 2013, 7, 880–884. [Google Scholar] [CrossRef]
- Fernandes, K.A.; Kittelmann, S.; Rogers, C.W.; Gee, E.K.; Bolwell, C.F.; Bermingham, E.N.; Thomas, D.G. Faecal microbiota of forage-fed horses in New Zealand and the population dynamics of microbial communities following dietary change. PLoS ONE 2014, 9, e112846. [Google Scholar] [CrossRef]
- Williams, C.A.; Kronfeld, D.S.; Staniar, W.B.; Harris, P.A. Plasma glucose and insulin responses of Thoroughbred mares fed a meal high in starch and sugar or fat and fiber. J. Anim. Sci. 2001, 79, 2196–2201. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Kronfeld, D.S.; Cooper, W.L.; Harris, P.A. Glucose clearance in grazing mares is affected by diet, pregnancy, and lactation. J. Anim. Sci. 2003, 81, 1764–1771. [Google Scholar] [CrossRef]
- Salem, S.E.; Maddox, T.W.; Berg, A.; Antczak, P.; Ketley, J.M.; Williams, N.J.; Archer, D.C. Variation in faecal microbiota in a group of horses managed at pasture over a 12-month period. Sci Rep. 2018, 8, 8510. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, K.A.; Gee, E.K.; Rogers, C.W.; Kittelmann, S.; Biggs, P.J.; Bermingham, E.N.; Bolwell, C.F.; Thomas, D.G. Seasonal variation in the faecal microbiota of mature adult horses maintained on pasture in New Zealand. Animals 2021, 11, 2300. [Google Scholar] [CrossRef]
- Theelen, M.J.; Luiken, R.E.; Wagenaar, J.A.; Sloet van Oldruitenborgh-Oosterbaan, M.M.; Rossen, J.W.; Zomer, A.L. The Equine Faecal Microbiota of Healthy Horses and Ponies in The Netherlands: Impact of Host and Environmental Factors. Animals 2021, 11, 1762. [Google Scholar] [CrossRef] [PubMed]
- Selim, K.E.; Jakkola, S.; Karikoski, N.; Boston, R.; Reilas, T.; Särkijärvi, S.; Saastamoinen, M.; Kokkonen, T. Relationships among body condition, insulin resistance and subcutaneous adipose tissue gene expression during the grazing season in mares. PLoS ONE 2015, 10, e0125968. [Google Scholar] [CrossRef] [PubMed]
- Place, N.J.; McGowan, C.M.; Lamb, S.V.; Schanbacher, B.J.; McGowan, T.; Walsh, D.M. Seasonal variation in serum concentrations of selected metabolic hormones in horses. J. Vet. Intern. Med. 2010, 24, 650–654. [Google Scholar] [CrossRef]
- Funk, R.A.; Wooldridge, A.A.; Stewart, A.J.; Behrend, E.N.; Kemppainen, R.J.; Zhong, Q.; Johnson, A.K. Seasonal changes in the combined glucose-insulin tolerance test in normal aged horses. J. Vet. Intern. Med. 2012, 26, 1035–1041. [Google Scholar] [CrossRef]
- Lam, Y.Y.; Zhang, C.; Zhao, L. Causality in dietary interventions—Building a case for gut microbiota. Genome Med. 2018, 10, 62. [Google Scholar] [CrossRef]
- Chen, T.; Liu, A.B.; Sun, S.; Ajami, N.J.; Ross, M.C.; Wang, H.; Zhang, L.; Reuhl, K.; Kobayashi, K.; Onishi, J.C.; et al. Green tea polyphenols modify the gut microbiome in db/db mice as co-abundance grouips correlating with the blood glucose lowering effect. Mol. Nutr. Food Res. 2019, 63, 180164. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, L. Strain-level dissection of the contribution of the gut microbiome to human metabolic disease. Genome Med. 2016, 8, 41. [Google Scholar] [CrossRef] [PubMed]
| Nutrients 2,3 | Forage 4 | |||||||
|---|---|---|---|---|---|---|---|---|
| HAY-SP | CSG-SP | WSG | CSG-FA | HAY-FA | ||||
| BRS | CRS | BRS | CRS | BRS | CRS | |||
| DE, Mcal/kg | 2.02 | 2.18 | 2.06 | 2.15 | 2.03 | 2.34 | 2.22 | 2.00 |
| CP, % | 5.6 | 13.1 | 16.5 | 21.4 | 22.7 | 23.7 | 21.5 | 10.2 |
| ADF, % | 40.9 | 35.3 | 36.4 | 31.4 | 36.9 | 30.5 | 30.0 | 42.2 |
| NDF, % | 66.1 | 59.4 | 62.1 | 58.7 | 61.8 | 50.5 | 55.4 | 65.8 |
| NSC, % | 18.4 | 12.4 | 8.5 | 7.1 | 2.6 | 11.3 | 11.5 | 9.1 |
| WSC, % | 18.0 | 10.3 | 7.4 | 4.8 | 2.3 | 10.2 | 10.2 | 8.1 |
| ESC, % | 7.0 | 6.6 | 3.7 | 4.0 | 1.9 | 8.3 | 6.9 | 6.9 |
| Starch, % | 0.40 | 2.07 | 1.17 | 2.30 | 1.37 | 1.13 | 1.23 | 1.00 |
| Step | Statistic | Count |
|---|---|---|
| Initial | Total | 881,189 |
| Quality and Chimera Filtering 2 | Total | 730,065 |
| Minimum sample−1 | 12,460 | |
| Maximum sample−1 | 30,170 | |
| Mean sample−1 | 18,252 | |
| Median sample−1 | 18,890 | |
| Removal of Low Abundance Features | Total | 648,797 |
| Minimum sample−1 | 10,989 | |
| Maximum sample−1 | 26,948 | |
| Mean sample−1 | 16,220 | |
| Median sample−1 | 15,587 |
| Forage 2 | BCG 3 | LDA 4 | ASV Taxonomic Lineage 5 |
|---|---|---|---|
| HAY-SP | BCG_9 | 4.55 | Firmicutes | Clostridia | Oscillospirales | Oscillospiraceae | Papillibacter | uncultured_Clostridiales Bacteroidota | Bacteroidia | Bacteroidales | Rikenellaceae | Rikenellaceae_RC9_gut_group | uncultured_bacterium Firmicutes | Clostridia | Christensenellales | Christensenellaceae | Christensenellaceae_R-7_group | uncultured_Clostridia Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae (2 ASV) Firmicutes | Clostridia | Oscillospirales | Oscillospiraceae | NK4A214_group | uncultured_rumen |
| BCG_18 | 5.03 | Firmicutes | Clostridia | Oscillospirales | Ruminococcaceae | Ruminococcus | Ruminococcus_sp. Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae (3 ASV) Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Lachnospiraceae_UCG-009 | uncultured_bacterium Firmicutes | Clostridia | Christensenellales | Christensenellaceae | Christensenellaceae_R-7_group | uncultured_prokaryote | |
| BCG_93 | 4.55 | Bacteroidota | Bacteroidia | Bacteroidales | p-251-o5 | p-251-o5 | uncultured_bacterium Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | probable_genus_10 | uncultured_bacterium Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Pseudobutyrivibrio | |
| BCG_116 | 4.52 | Bacteroidota | Bacteroidia | Bacteroidales | p-251-o5 | p-251-o5 | uncultured_bacterium Firmicutes | Negativicutes | Veillonellales-Selenomonadales | Selenomonadaceae | Anaerovibrio | uncultured_bacterium Firmicutes | Clostridia | Oscillospirales | Ruminococcaceae | Ruminococcus | |
| BCG_201 | 4.28 | Firmicutes | Clostridia | Oscillospirales | Oscillospiraceae | NK4A214_group Bacteroidota | Bacteroidia | Bacteroidales | p-251-o5 | p-251-o5 | uncultured_bacterium | |
| BCG_305 | 4.27 | Firmicutes | Bacilli | Erysipelotrichales | Erysipelotrichaceae | Catenisphaera | uncultured_bacterium Fibrobacterota | Fibrobacteria | Fibrobacterales | Fibrobacteraceae | Fibrobacter | uncultured_bacterium | |
| HAY-FA | BCG_22 | 4.67 | Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | Lactobacillus | Lactobacillus_equigenerosi Firmicutes | Clostridia | Oscillospirales | [Eubacterium]_coprostanoligenes_group | [Eubacterium]_coprostanoligenes_group Firmicutes | Clostridia | Oscillospirales | Oscillospiraceae | uncultured |
| BCG_118 | 4.56 | Firmicutes | Bacilli | Erysipelotrichales | Erysipelatoclostridiaceae | UCG-004 | uncultured_rumen Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Marvinbryantia Firmicutes | Clostridia | Oscillospirales | Oscillospiraceae | UCG-005 | uncultured_bacterium | |
| BCG_173 | 4.20 | Spirochaetota | Spirochaetia | Spirochaetales | Spirochaetaceae | Treponema | uncultured_bacterium Bacteroidota | Bacteroidia | Bacteroidales | F082 | F082 | uncultured_bacterium Bacteroidota | Bacteroidia | Bacteroidales | Rikenellaceae | Rikenellaceae_RC9_gut_group | uncultured_bacterium | |
| BCG_300 | 4.87 | Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Lachnospiraceae_XPB1014_group | uncultured_bacterium Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus | |
| BCG_309 | 4.25 | Firmicutes | Clostridia | Oscillospirales | Ethanoligenenaceae | Incertae_Sedis | uncultured_bacterium Firmicutes | Clostridia | Christensenellales | Christensenellaceae | Christensenellaceae_R-7_group | uncultured_rumen |
| Forage 2 | BCG 3 | LDA 4 | ASV Taxonomic Lineage 5 |
|---|---|---|---|
| CSG-SP | BCG_43 | 4.98 | Spirochaetota | Spirochaetia | Spirochaetales | Spirochaetaceae | Sphaerochaeta | uncultured_rumen Firmicutes | Negativicutes | Acidaminococcales | Acidaminococcaceae | Phascolarctobacterium Firmicutes | Clostridia | Oscillospirales | Oscillospiraceae | UCG-002 | uncultured_bacterium Firmicutes | Clostridia | Oscillospirales | UCG-010 | UCG-010 | uncultured_bacterium Firmicutes | Clostridia | Christensenellales | Christensenellaceae | Christensenellaceae_R-7_group |
| BCG_94 | 4.70 | Bacteroidota | Bacteroidia | Bacteroidales | Prevotellaceae | Prevotella | uncultured_Prevotellaceae Firmicutes | Clostridia | Clostridiales | Clostridiaceae | Sarcina | uncultured_bacterium Firmicutes | Clostridia | Clostridiales | Clostridiaceae | Clostridium_sensu_stricto_1 | |
| BCG_233 | 4.80 | Firmicutes | Clostridia | Clostridiales | Clostridiaceae | Clostridium_sensu_stricto_1 Bacteroidota | Bacteroidia | Bacteroidales | Bacteroidales_BS11_gut_group | Bacteroidales_BS11_gut_group | uncultured_bacterium | |
| CSG-FA | BCG_45 | 4.35 | Firmicutes | Clostridia | Peptostreptococcales-Tissierellales | Anaerovoracaceae | Family_XIII_AD3011_group | uncultured_bacterium Firmicutes | Bacilli | Erysipelotrichales | Erysipelotrichaceae | Catenisphaera | uncultured_bacterium Actinobacteriota | Coriobacteriia | Coriobacteriales | Eggerthellaceae | Denitrobacterium | Denitrobacterium_detoxificans Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Marvinbryantia | uncultured_rumen Firmicutes | Clostridia | Peptostreptococcales-Tissierellales | Anaerovoracaceae |
| BCG_48 | 4.79 | Firmicutes | Clostridia | Oscillospirales | [Eubacterium]_coprostanoligenes_group | [Eubacterium]_coprostanoligenes_group | uncultured_bacterium Bacteroidota | Bacteroidia | Bacteroidales | Bacteroidales_UCG-001 | Bacteroidales_UCG-001 Bacteroidota | Bacteroidia | Bacteroidales | Rikenellaceae | Rikenellaceae_RC9_gut_group | uncultured_bacterium Firmicutes | Clostridia | Oscillospirales | Oscillospiraceae | NK4A214_group | |
| BCG_51 | 4.36 | Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Lachnospiraceae_XPB1014_group | uncultured_bacterium Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Lachnospiraceae_UCG-009 | uncultured_bacterium Firmicutes | Clostridia | Peptostreptococcales-Tissierellales | Anaerovoracaceae | uncultured | uncultured_bacterium Firmicutes | Bacilli | Erysipelotrichales | Erysipelatoclostridiaceae | Erysipelatoclostridium | uncultured_bacterium | |
| BCG_219 | 4.55 | Bacteroidota | Bacteroidia | Bacteroidales | Prevotellaceae | Alloprevotella | uncultured_bacterium Bacteroidota | Bacteroidia | Bacteroidales | Bacteroidales_RF16_group | Bacteroidales_RF16_group | uncultured_bacterium | |
| BCG_275 | 4.45 | Firmicutes | Clostridia | Oscillospirales | Oscillospiraceae | NK4A214_group | uncultured_bacterium Verrucomicrobiota | Kiritimatiellae | WCHB1-41 | WCHB1-41 | WCHB1-41 | uncultured_bacterium |
| Forage 2 | BCG 3 | LDA 4 | ASV Taxonomic Lineage 5 |
|---|---|---|---|
| WSG | BCG_35 | 4.68 | Verrucomicrobiota | Verrucomicrobiae | Verrucomicrobiales | Akkermansiaceae | Akkermansia | uncultured_bacterium Firmicutes | Clostridia | Christensenellales | Christensenellaceae | Christensenellaceae_R-7_group Fibrobacterota | Fibrobacteria | Fibrobacterales | Fibrobacteraceae | Fibrobacter | Fibrobacter_sp. Spirochaetota | Spirochaetia | Spirochaetales | Spirochaetaceae | Treponema | uncultured_bacterium Firmicutes | Clostridia | Peptostreptococcales-Tissierellales | Anaerovoracaceae | Family_XIII_AD3011_group | uncultured_bacterium |
| BCG_100 | 4.21 | Firmicutes | Clostridia | Oscillospirales | Oscillospiraceae | NK4A214_group Spirochaetota | Spirochaetia | Spirochaetales | Spirochaetaceae | Treponema Firmicutes | Clostridia | Oscillospirales | Oscillospiraceae | UCG-005 | metagenome | |
| BCG_113 | 4.85 | Bacteroidota | Bacteroidia | Bacteroidales | Bacteroidales_RF16_group | Bacteroidales_RF16_group | uncultured_bacterium Firmicutes | Clostridia | Clostridiales | Clostridiaceae | Clostridium_sensu_stricto_1 | Clostridium_butyricum Firmicutes | Clostridia | Oscillospirales | Oscillospiraceae | Papillibacter | uncultured_bacterium | |
| BCG_124 | 4.58 | Firmicutes | Clostridia | Christensenellales | Christensenellaceae | Christensenellaceae_R-7_group Synergistota | Synergistia | Synergistales | Synergistaceae | uncultured | uncultured_bacterium Firmicutes | Clostridia | Peptostreptococcales-Tissierellales | Anaerovoracaceae | Mogibacterium | |
| BCG_259 | 4.46 | Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | [Eubacterium]_hallii_group | uncultured_bacterium |
| Fecal Metabolites 2, ug g Feces −1 | Forage 3 | ||||
|---|---|---|---|---|---|
| HAY-SP | CSG-SP | WSG | CSG-FA | HAY-FA | |
| Total SCFA | 674 ± 76 ab | 1071 ± 121 c | 860 ± 98 ac | 1134 ± 129 c | 437 ± 50 b |
| Total BCFA | 48 ± 6 a | 136 ± 16 b | 113 ± 13 b | 126 ± 15 b | 70 ± 8 a |
| Acetate | 323 ± 34 ab | 473 ± 50 ac | 423 ± 45 ac | 524 ± 55 c | 229 ± 24 b |
| Butyrate | 101 ± 22 ab | 174 ± 30 a | 128 ± 22 a | 170 ± 29 a | 62 ± 11 b |
| Propionate | 217 ± 25 a | 365 ± 42 b | 274 ± 31 ab | 390 ± 45 b | 110 ± 13 c |
| Valerate | 12.0 ± 2.1 ab | 40.4 ± 7.2 c | 24.2 ± 4.3 bc | 35.3 ± 6.3 c | 18.1 ± 3.2 b |
| Hexanoate | 14.2 ± 3.3 a | 3.4 ± 0.8 b | 5.2 ± 1.2 b | 3.6 ± 0.8 b | 11.6 ± 2.7 a |
| Heptanoate | 2.8 ± 0.5 a | 0.8 ± 0.1 b | 1.2 ± 0.2 b | 0.8 ± 0.1 b | 1.7 ± 0.3 ab |
| Isobutyrate | 34.4 ± 3.5 a | 81.9 ± 8.3 b | 76.9 ± 7.8 b | 77.7 ± 7.9 b | 47.7 ± 4.8 a |
| Isovalerate | 13.5 ± 2.1 a | 51.2 ± 7.9 b | 35.0 ± 5.4 bc | 47.1 ± 7.3 b | 21.6 ± 3.4 ac |
| Model Accuracy 2 | ||||
|---|---|---|---|---|
| Full BCG Dataset 2 | Reduced BCG Dataset 3 | |||
| R2 | p-Value | R2 | p-Value | |
| Fecal Variables 4 | ||||
| Total SCFA | 0.06 | 0.13 | 0.24 | 0.001 |
| Total BCFA | 0.08 | 0.07 | 0.28 | 0.0004 |
| Acetate | 0.05 | 0.19 | 0.29 | 0.0003 |
| Butyrate | 0.04 | 0.20 | 0.26 | 0.002 |
| Propionate | 0.03 | 0.31 | 0.17 | 0.008 |
| Valerate | 0.24 | 0.49 | 0.15 | 0.01 |
| Hexanoate | 0.26 | 0.0007 | 0.48 | <0.0001 |
| Heptanoate | 0.11 | 0.03 | 0.24 | 0.001 |
| Isobutyrate | 0.01 | 0.54 | 0.27 | 0.0006 |
| Isovalerate | 0.26 | 0.0007 | 0.26 | 0.0007 |
| Fecal pH | 0.06 | 0.13 | 0.17 | 0.008 |
| Forage Nutrients | ||||
| Digestible Energy | 0.26 | 0.0008 | 0.34 | <0.0001 |
| Crude Protein | 0.41 | <0.0001 | 0.62 | <0.0001 |
| Acid Detergent Fiber | 0.19 | 0.005 | 0.39 | <0.0001 |
| Neutral Detergent Fiber | 0.27 | 0.0005 | 0.35 | <0.0001 |
| Non-Structural Carbohydrate | 0.53 | <0.0001 | 0.61 | <0.0001 |
| Water-Soluble Carbohydrate | 0.52 | <0.0001 | 0.67 | <0.0001 |
| Ethanol-Soluble Carbohydrate | 0.10 | 0.04 | 0.18 | 0.006 |
| Starch | 0.21 | 0.003 | 0.40 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weinert-Nelson, J.R.; Biddle, A.S.; Sampath, H.; Williams, C.A. Fecal Microbiota, Forage Nutrients, and Metabolic Responses of Horses Grazing Warm- and Cool-Season Grass Pastures. Animals 2023, 13, 790. https://doi.org/10.3390/ani13050790
Weinert-Nelson JR, Biddle AS, Sampath H, Williams CA. Fecal Microbiota, Forage Nutrients, and Metabolic Responses of Horses Grazing Warm- and Cool-Season Grass Pastures. Animals. 2023; 13(5):790. https://doi.org/10.3390/ani13050790
Chicago/Turabian StyleWeinert-Nelson, Jennifer R., Amy S. Biddle, Harini Sampath, and Carey A. Williams. 2023. "Fecal Microbiota, Forage Nutrients, and Metabolic Responses of Horses Grazing Warm- and Cool-Season Grass Pastures" Animals 13, no. 5: 790. https://doi.org/10.3390/ani13050790
APA StyleWeinert-Nelson, J. R., Biddle, A. S., Sampath, H., & Williams, C. A. (2023). Fecal Microbiota, Forage Nutrients, and Metabolic Responses of Horses Grazing Warm- and Cool-Season Grass Pastures. Animals, 13(5), 790. https://doi.org/10.3390/ani13050790






