The Two-Component System CpxRA Affects Antibiotic Susceptibility and Biofilm Formation in Avian Pathogenic Escherichia coli
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Culture Conditions
2.2. Genetic and Molecular Biology Techniques
2.3. Construction of the CpxRA-Deficient Mutant and Complemented Strains
2.4. Bacterial Growth Curves
2.5. Biofilm Formation Assays
2.6. Antibiotic Susceptibility Tests
2.7. Antibacterial Activity Assays
2.8. Total RNA Isolation, cDNA Generation, and Real-Time PCR Processing
2.9. His6-CpxR Protein Purification
2.10. Electrophoretic Mobility Shift Assays (EMSAs)
2.11. Statistical Analyses
3. Results
3.1. Identification of cpxRA Mutant and Complementary Strains of APEC40
3.2. cpxRA Deletion Did Not Affect Strain Growth
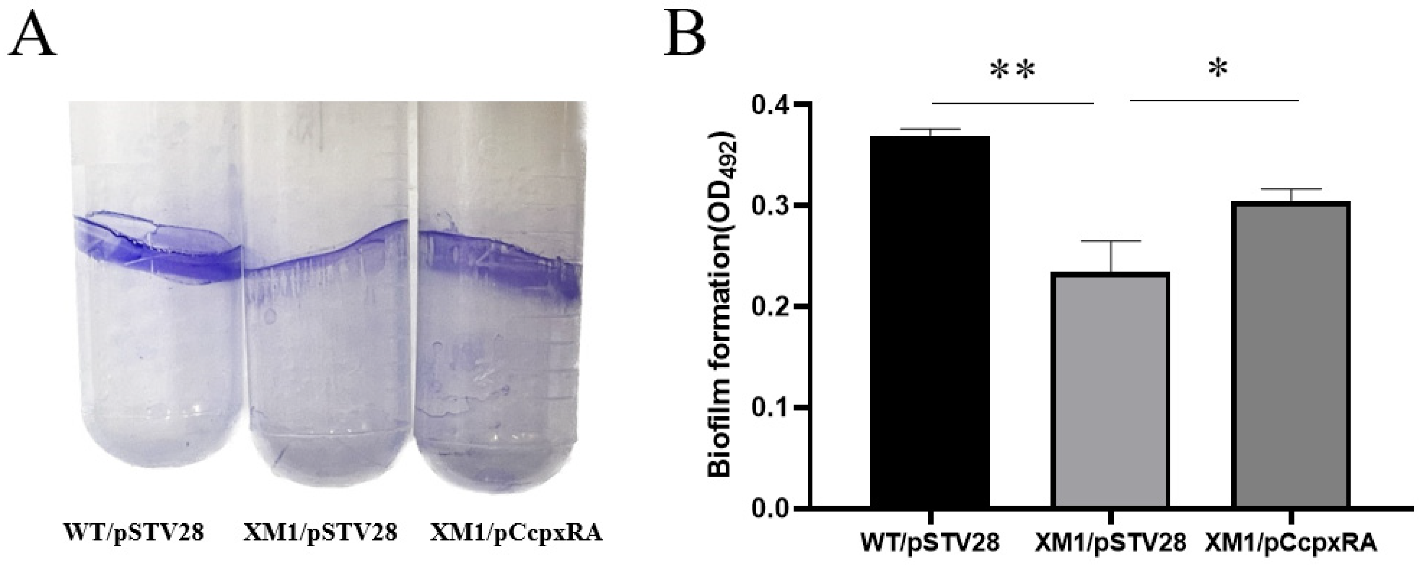
3.3. Deletion of cpxRA Inhibited Biofilm Formation
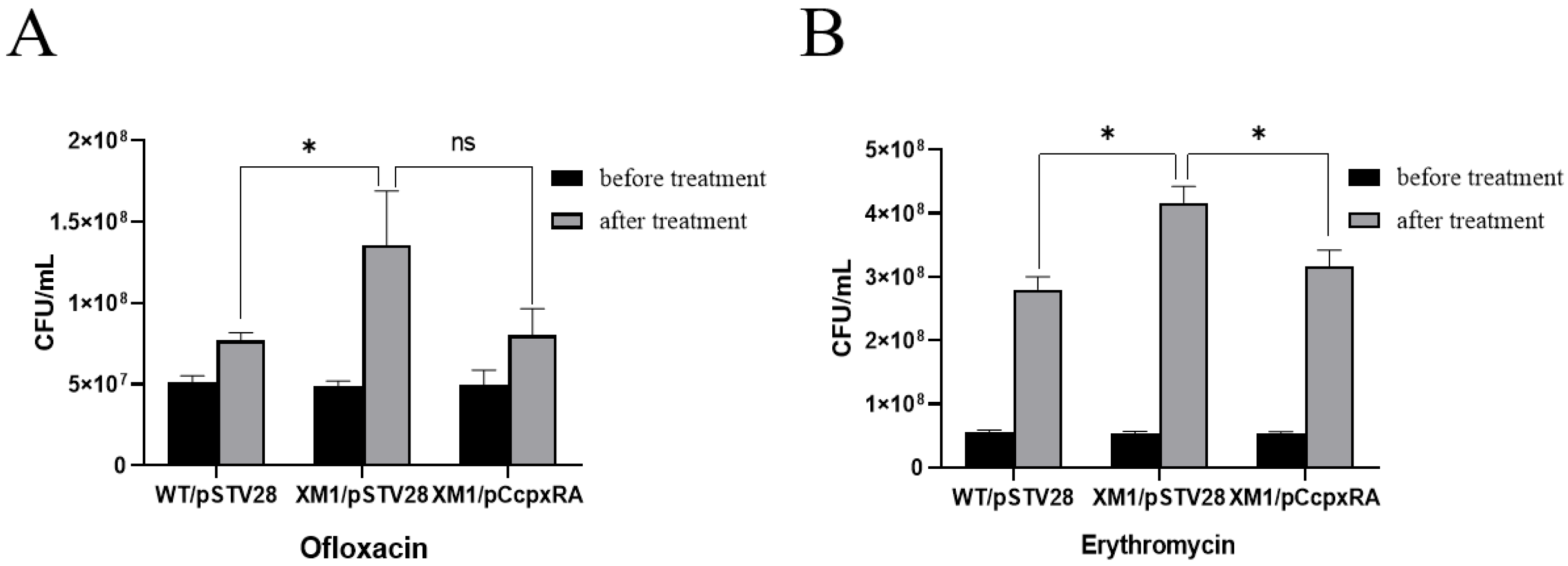
3.4. Deletion of the cpxRA Gene Decreased Bacterial Antibiotic Susceptibility
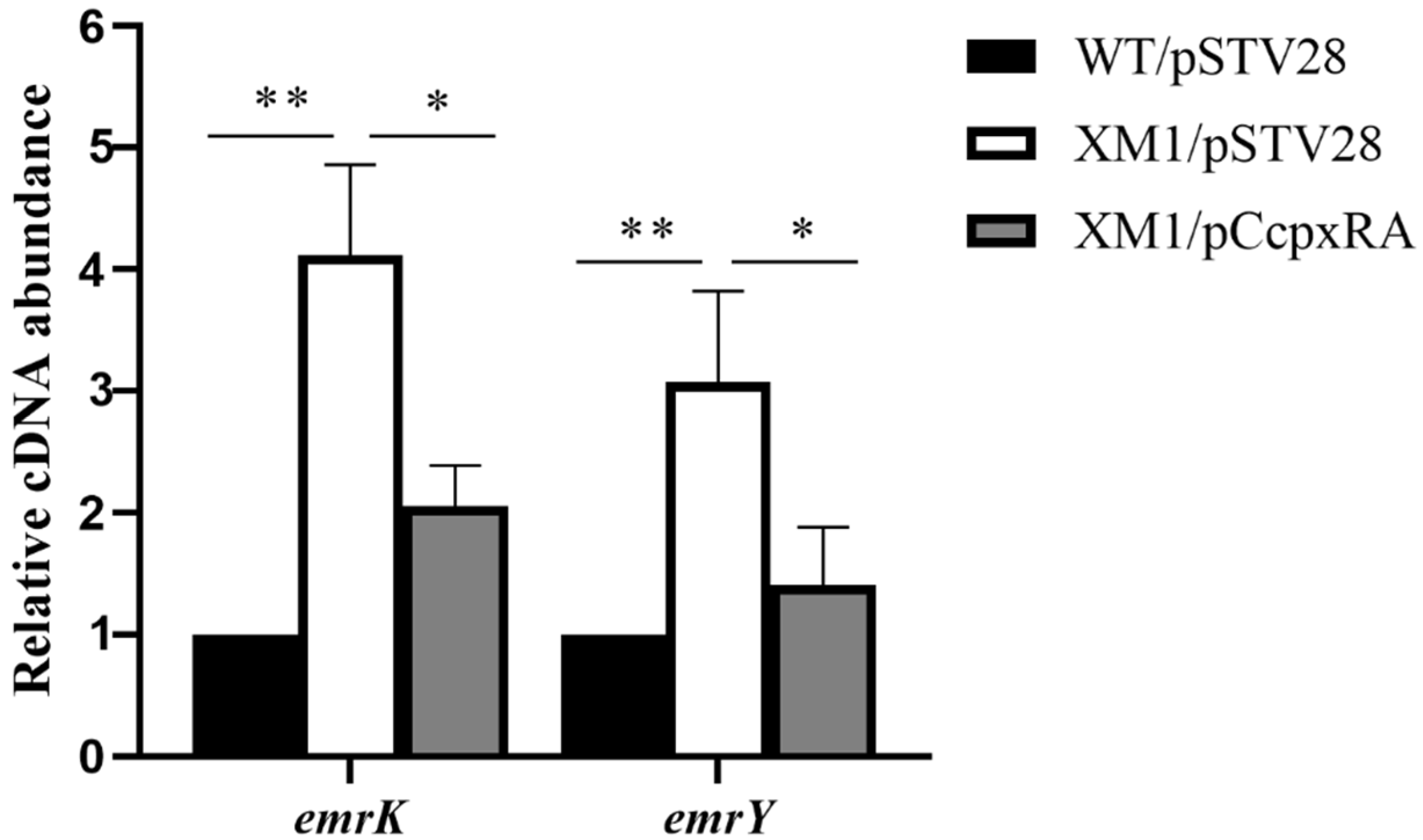
3.5. CpxRA Down-Regulated the Transcription Level of emrKY
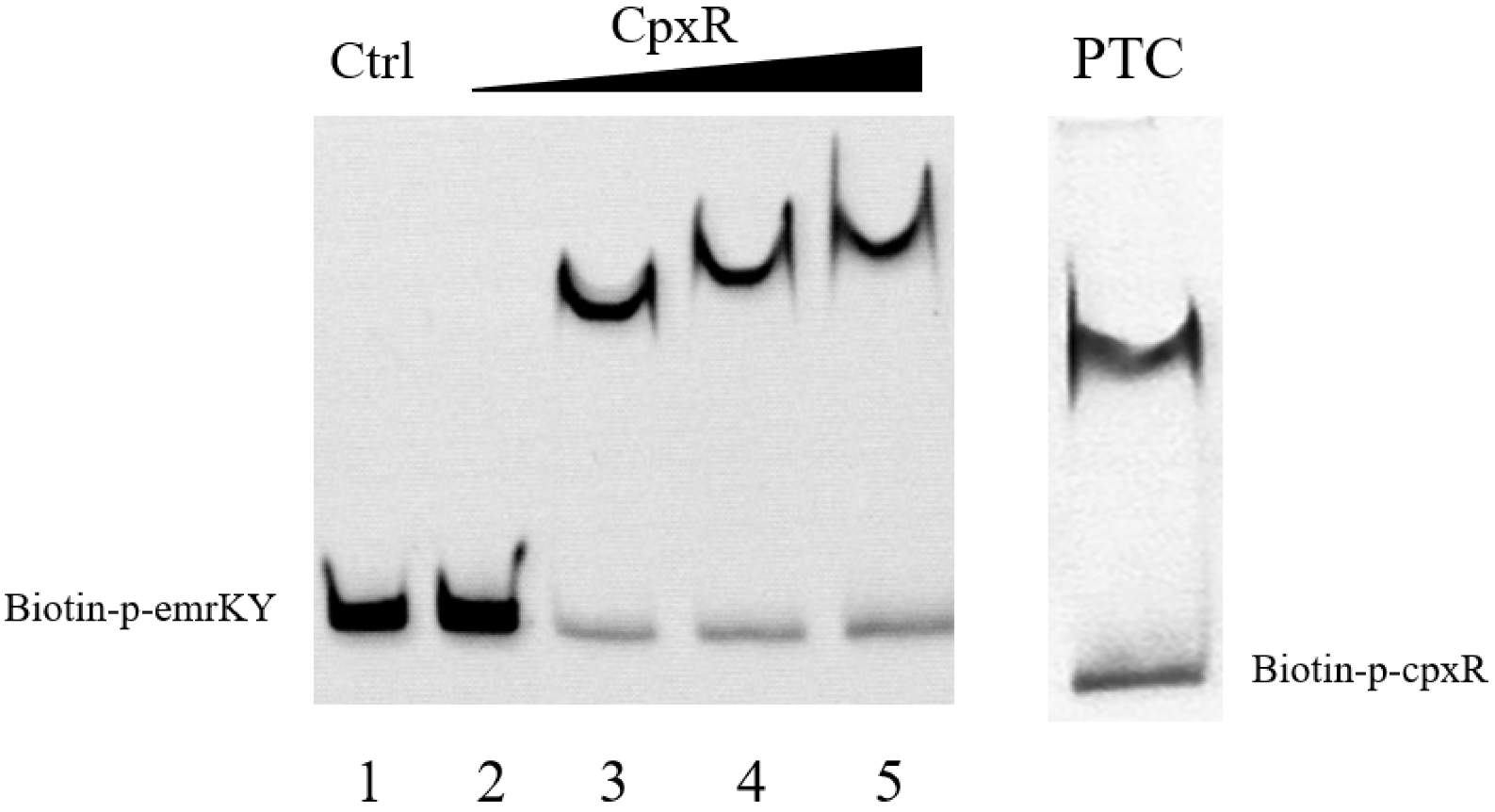
3.6. CpxR Binding to EmrKY Promoters
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Han, X.; Bai, H.; Tu, J.; Yang, L.; Xu, D.; Wang, S.; Qi, K.; Fan, G.; Zhang, Y.; Zuo, J.; et al. Deletion of luxS further attenuates the virulence of the avian pathogenic Escherichia coli aroA mutant. Microb. Pathog. 2015, 88, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Giovanardi, D.; Lupini, C.; Pesente, P.; Rossi, G.; Ortali, G.; Catelli, E. Characterization and antimicrobial resistance analysis of avian pathogenic Escherichia coli isolated from Italian turkey flocks. Poult. Sci. 2013, 92, 2661–2667. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Janßen, T.; Kießling, S.; Philipp, H.-C.; Wieler, L.H. Molecular epidemiology of avian pathogenic Escherichia coli (APEC) isolated from colisepticemia in poultry. Vet. Microbiol. 2004, 104, 91–101. [Google Scholar] [CrossRef]
- Dho-Moulin, M.; Fairbrother, J.M. Avian pathogenic Escherichia coli (APEC). Vet. Res. 1999, 30, 299–316. [Google Scholar] [PubMed]
- Mellata, M. Human and Avian Extraintestinal Pathogenic Escherichia coli: Infections, Zoonotic Risks, and Antibiotic Resistance Trends. Foodborne Pathog. Dis. 2013, 10, 916–932. [Google Scholar] [CrossRef]
- Guabiraba, R.; Schouler, C. Avian colibacillosis: Still many black holes. FEMS Microbiol. Lett. 2015, 362, fnv118. [Google Scholar] [CrossRef]
- Sadeyen, J.-R.; Kaiser, P.; Stevens, M.P.; Dziva, F. Analysis of immune responses induced by avian pathogenic Escherichia coli infection in turkeys and their association with resistance to homologous re-challenge. Vet. Res. 2014, 45, 19. [Google Scholar] [CrossRef]
- Kathayat, D.; Lokesh, D.; Ranjit, S.; Rajashekara, G. Avian Pathogenic Escherichia coli (APEC): An Overview of Virulence and Pathogenesis Factors, Zoonotic Potential, and Control Strategies. Pathogens 2021, 10, 467. [Google Scholar] [CrossRef]
- Agunos, A.; Leger, D.; Carson, C. Review of antimicrobial therapy of selected bacterial diseases in broiler chickens in Canada. Can. Vet. J. 2012, 53, 1289–1300. [Google Scholar]
- Johnson, T.J.; Wannemuehler, Y.; Doetkott, C.; Johnson, S.J.; Rosenberger, S.C.; Nolan, L.K. Identification of Minimal Predictors of Avian Pathogenic Escherichia coli Virulence for Use as a Rapid Diagnostic Tool. J. Clin. Microbiol. 2008, 46, 3987–3996. [Google Scholar] [CrossRef]
- Johnson, T.J.; Kariyawasam, S.; Wannemuehler, Y.; Mangiamele, P.; Johnson, S.J.; Doetkott, C.; Skyberg, J.A.; Lynne, A.M.; Johnson, J.R.; Nolan, L.K. The Genome Sequence of Avian Pathogenic Escherichia coli Strain O1:K1:H7 Shares Strong Similarities with Human Extraintestinal Pathogenic E. coli Genomes. J. Bacteriol. 2007, 189, 3228–3236. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Li, G.; Wilking, H.; Kiebling, S.; Alt, K.; Antáo, E.-M.; Laturnus, C.; Diehl, I.; Glodde, S.; Homeier, T. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: How closely related are they? Int. J. Med. Microbiol. 2007, 297, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.M.; Stegger, M.; Aziz, M.; Johnson, T.J.; Waits, K.; Nordstrom, L.; Gauld, L.; Weaver, B.; Rolland, D.; Statham, S.; et al. Escherichia coli ST131- H 22 as a Foodborne Uropathogen. mBio 2018, 9, e00470-18. [Google Scholar] [CrossRef] [PubMed]
- Moulin-Schouleur, M.; Répérant, M.; Laurent, S.; Brée, A.; Mignon-Grasteau, S.; Germon, P.; Rasschaert, D.; Schouler, C. Extraintestinal Pathogenic Escherichia coli Strains of Avian and Human Origin: Link between Phylogenetic Relationships and Common Virulence Patterns. J. Clin. Microbiol. 2007, 45, 3366–3376. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Camp, J.; Schuster, S.; Vavra, M.; Schweigger, T.; Rossen, J.W.A.; Reuter, S.; Kern, W.V. Limited Multidrug Resistance Efflux Pump Overexpression among Multidrug-Resistant Escherichia coli Strains of ST131. Antimicrob. Agents Chemother. 2021, 65, e01735-20. [Google Scholar] [CrossRef]
- Kim, H.S.; Nagore, D.; Nikaido, H. Multidrug Efflux Pump MdtBC of Escherichia coli Is Active Only as a B2C Heterotrimer. J. Bacteriol. 2010, 192, 1377–1386. [Google Scholar] [CrossRef]
- Saier, M.H., Jr. A Functional-Phylogenetic Classification System for Transmembrane Solute Transporters. Microbiol. Mol. Biol. Rev. 2000, 64, 354–411. [Google Scholar] [CrossRef]
- Kumar, S.; Mukherjee, M.M.; Varela, M.F. Modulation of Bacterial Multidrug Resistance Efflux Pumps of the Major Facilitator Superfamily. Int. J. Bacteriol. 2013, 2013, 204141. [Google Scholar] [CrossRef]
- Teelucksingh, T.; Thompson, L.K.; Cox, G. The Evolutionary Conservation of Escherichia coli Drug Efflux Pumps Supports Physiological Functions. J. Bacteriol. 2020, 202, e00367-20. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the Natural environment to infectious diseases. Nat. Rev. Genet. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Labbate, M.; Queck, S.Y.; Koh, K.S.; Rice, S.A.; Givskov, M.; Kjelleberg, S. Quorum Sensing-Controlled Biofilm Development in Serratia liquefaciens MG1. J. Bacteriol. 2004, 186, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef]
- Capra, E.J.; Laub, M.T. Evolution of Two-Component Signal Transduction Systems. Annu. Rev. Microbiol. 2012, 66, 325–347. [Google Scholar] [CrossRef]
- Eguchi, Y.; Utsumi, R. Introduction to bacterial signal transduction networks. Adv. Exp. Med. Biol. 2008, 631, 1–6. [Google Scholar] [PubMed]
- Li, H.; Liu, F.; Peng, W.; Yan, K.; Zhao, H.; Liu, T.; Cheng, H.; Chang, P.; Yuan, F.; Chen, H.; et al. The CpxA/CpxR Two-Component System Affects Biofilm Formation and Virulence in Actinobacillus pleuropneumoniae. Front. Cell. Infect. Microbiol. 2018, 8, 72. [Google Scholar] [CrossRef] [PubMed]
- Weatherspoon-Griffin, N.; Yang, D.; Kong, W.; Hua, Z.; Shi, Y. The CpxR/CpxA Two-component Regulatory System Up-regulates the Multidrug Resistance Cascade to Facilitate Escherichia coli Resistance to a Model Antimicrobial Peptide. J. Biol. Chem. 2014, 289, 32571–32582. [Google Scholar] [CrossRef]
- Zhao, Z.; Xu, Y.; Jiang, B.; Qi, Q.; Tang, Y.J.; Xian, M.; Wang, J.; Zhao, G. Systematic Identification of CpxRA-Regulated Genes and Their Roles in Escherichia coli Stress Response. mSystems 2022, 7, e0041922. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Yu, L.M.; Li, W.C.; Qi, K.Z.; Wang, S.Y.; Chen, X.L.; Ni, J.T.; Deng, R.N.; Shang, F.; Xue, T. McbR is involved in biofilm formation and H2O2 stress response in avian pathogenic Escherichia coli X40. Poult. Sci. 2019, 98, 4094–4103. [Google Scholar] [CrossRef]
- Yu, L.; Li, W.; Liu, Z.; Yu, J.; Wang, W.; Shang, F.; Xue, T. Role of McbR in the regulation of antibiotic susceptibility in avian pathogenic Escherichia coli. Poult. Sci. 2020, 99, 6390–6401. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shang, F.; Shen, J.; Xu, J.; Chen, X.; Ni, J.; Yu, L.; Xue, T. LsrR, the effector of AI-2 quorum sensing, is vital for the H2O2 stress response in mammary pathogenic Escherichia coli. Vet. Res. 2021, 52, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Pogliano, J.; Lynch, A.S.; Belin, D.; Lin, E.C.; Beckwith, J. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 1997, 11, 1169–1182. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Sharma, S.; Sharma, P.; Chandola, D.; Dang, S.; Gupta, S.; Gabrani, R. Escherichia coli biofilm: Development and therapeutic strategies. J. Appl. Microbiol. 2016, 121, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Beloin, C.; Roux, A.; Ghigo, J.M. Escherichia coli biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 249–289. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Wood, T.K. OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ. Microbiol. 2009, 11, 2735–2746. [Google Scholar] [CrossRef] [PubMed]
- Raivio, T.L.; Leblanc, S.K.; Price, N.L. The Escherichia coli Cpx envelope stress response regulates genes of diverse function that impact antibiotic resistance and membrane integrity. J. Bacteriol. 2013, 195, 2755–2767. [Google Scholar] [CrossRef] [PubMed]
- Raivio, T.L.; Silhavy, T.J. The sigmaE and Cpx regulatory pathways: Overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 1999, 2, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Otto, K.; Silhavy, T.J. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 2002, 99, 2287–2292. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Richmond, G.E.; Piddock, L.J.V. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 2014, 9, 1165–1177. [Google Scholar] [CrossRef]
- Frawley, E.R.; Crouch, M.L.; Bingham-Ramos, L.K.; Robbins, H.F.; Wang, W.; Wright, G.D.; Fang, F.C. Iron and citrate export by a major facilitator superfamily pump regulates metabolism and stress resistance in Salmonella Typhimurium. Proc. Natl. Acad. Sci. USA 2013, 110, 12054–12059. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J. Multidrug-resistance efflux pumps—Not just for resistance. Nat. Rev. Genet. 2006, 4, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Wei, Y.; Li, H.; Shi, D.; Yang, D.; Yin, J.; Zhou, S.; Chen, T.; Li, J.; Jin, M. Emerging pollutant metformin in water promotes the development of multiple-antibiotic resistance in Escherichia coli via chromosome mutagenesis. J. Hazard. Mater. 2022, 430, 128474. [Google Scholar] [CrossRef] [PubMed]
- De Wulf, P.; McGuire, A.M.; Liu, X.; Lin, E.C. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 2002, 277, 26652–26661. [Google Scholar] [CrossRef]

| Strains or Plasmids | Description | Reference or Source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | Clone host strain | Invitrogen |
| BL21(DE3) | Expression strain | Invitrogen |
| WT | Avian pathogenic E. coli (APEC) 40, wild-type | Laboratory stock |
| XM1 | APECX40 cpxAR-deletion mutant | This study |
| WT/pSTV28 | WT with the empty vector pSTV28, Cmr | This study |
| XM1/pSTV28 | XM with the empty vector pSTV28, Cmr | This study |
| XM1/pCcpxRA | XM with the complement plasmid pCcpxAR, Cmr | This study |
| Plasmids | ||
| pKD3 | cat gene, template plasmid, Cmr Ampr | [29] |
| pKD46 | Expresses λ Red recombinase Exo, Bet, and Gam, temperature sensitive, Ampr | [29] |
| pCP20 | FLP+λcI857+ λpRRep(Ts), temperature sensitive, Cmr Ampr | [29] |
| pSTV28 | Low copy number cloning vector, Cmr | Takara |
| pCcpxRA | pSTV28 with cpxAR gene, Cmr | This study |
| pET28a(+) | Expression vector, Kanr | Novagen |
| pET-cpxR | pET28a(+) with cpxR gene, Kanr | This study |
| Primer Name | Oligonucleotide (5′-3′) |
|---|---|
| APEC40-cpxRA-f | CGAGATGGAAGGCTTCAACGTGATTGTTGCCCACGATGGGTGTAGGCTGGAGCTGCTT |
| APEC40-cpxRA-r | CACCCAGCCACGATGCTGCTGAATGGCGGTTTCAACAATCTGAATATCCTCCTTAGTTC |
| CM-f | TGTAGGCTGGAGCTGCTT |
| CM-r | CATATGAATATCCTCCTTAGTTC |
| Check-cpxRA-in-f | TTTGACCGCCCGCTATTA |
| Check-cpxRA-in-r | CTTGCTTTCACCGCTACGAC |
| Check-cpxRA-out-f | ACTGACTGCCAGCGTTGA |
| Check-cpxRA-out-r | TGCCGGGTTATCGAAAAG |
| pET-cpxR-f | ACTTTAAGAAGGAGATATACCATGAATAAAATCCTGTTA |
| pET-cpxR-r | GTGGTGGTGGTGGTGCTCGAGTGAAGCAGAAACCATCAGAT |
| pCcpxRA-kpnⅠ-f | GCGGGTACCTGACGGCAGCGGTAACTA |
| pCcpxRA-hindⅢ-r | GCGAAGCTTAACTCCGCTTATACAGCG |
| M13-f | TGTAAAACGACGGCCAGT |
| M13-r | CAGGAAACAGCTATGACC |
| T7-f | TAATACGACTCACTATAGGG |
| T7-r | TGCTAGTTATTGCTCAGCGG |
| rt-16s-f | TTTGAGTTCCCGGCC |
| rt-16s-r | CGGCCGCAAGGTTAA |
| rt-emrY-f | CGATATTGGGCGGTTATA |
| rt-emrY-r | GGGTCAGTCCTGGTAGATT |
| rt-emrK-f | TGCCACTATCGCACTCAA |
| rt-emrK-r | GGTATGCTCCAGCGTTTC |
| p-emrKY-biotin-f | TCTCCCTTCTCCCTGTAGTAA |
| p-emrKY-r | TATTATCTCTCATTTCTCATAGATG |
| Antibiotics | MIC (μg/mL) of Three E. coli Strains | ||
|---|---|---|---|
| WT/pSTV28 | XM1/pSTV28 | XM1/pCcpxRA | |
| Erythromycin | 15 | 15 | 15 |
| Ofloxacin | 0.25 | 0.25 | 0.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, K.; Wang, H.; Lv, Z.; Hu, Y.; Wang, H.; Shu, F.; Zhu, C.; Xue, T. The Two-Component System CpxRA Affects Antibiotic Susceptibility and Biofilm Formation in Avian Pathogenic Escherichia coli. Animals 2023, 13, 383. https://doi.org/10.3390/ani13030383
Ma K, Wang H, Lv Z, Hu Y, Wang H, Shu F, Zhu C, Xue T. The Two-Component System CpxRA Affects Antibiotic Susceptibility and Biofilm Formation in Avian Pathogenic Escherichia coli. Animals. 2023; 13(3):383. https://doi.org/10.3390/ani13030383
Chicago/Turabian StyleMa, Kai, Hui Wang, Zhenfei Lv, Yutong Hu, Hongli Wang, Fang Shu, Chengfeng Zhu, and Ting Xue. 2023. "The Two-Component System CpxRA Affects Antibiotic Susceptibility and Biofilm Formation in Avian Pathogenic Escherichia coli" Animals 13, no. 3: 383. https://doi.org/10.3390/ani13030383
APA StyleMa, K., Wang, H., Lv, Z., Hu, Y., Wang, H., Shu, F., Zhu, C., & Xue, T. (2023). The Two-Component System CpxRA Affects Antibiotic Susceptibility and Biofilm Formation in Avian Pathogenic Escherichia coli. Animals, 13(3), 383. https://doi.org/10.3390/ani13030383





