Mitochondrial Diversity and Phylogenetic Relationship of Eight Native Bulgarian Sheep Breeds
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Welfare and Ethical Statement
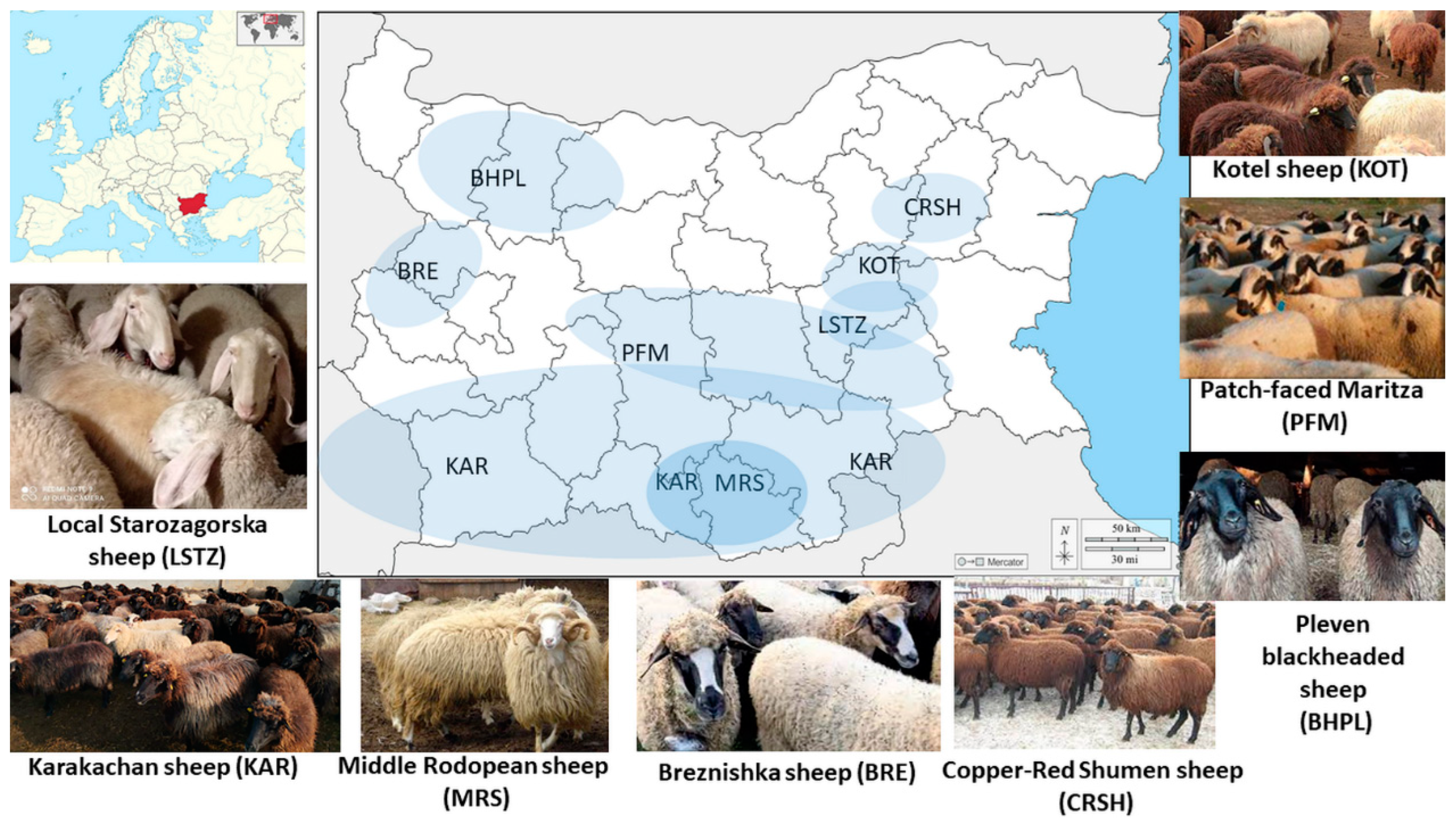
2.2. Sample Collection
2.3. DNA Extraction, PCR Amplification, and Sequencing
2.4. Bioinformatics and Data Analysis
3. Results
3.1. mtDNA Sequence Polymorphis in Native Bulgarian Sheep Breeds
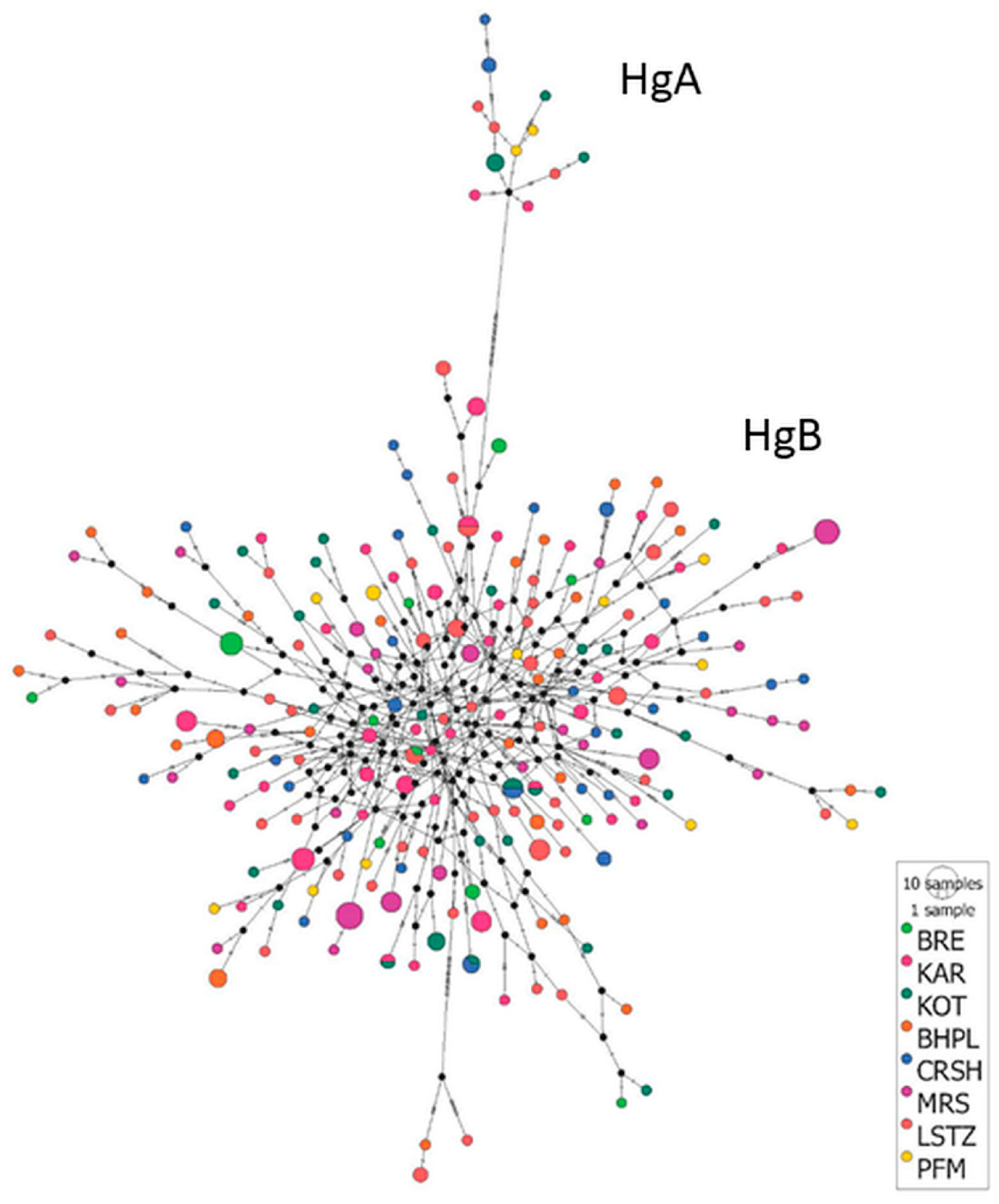
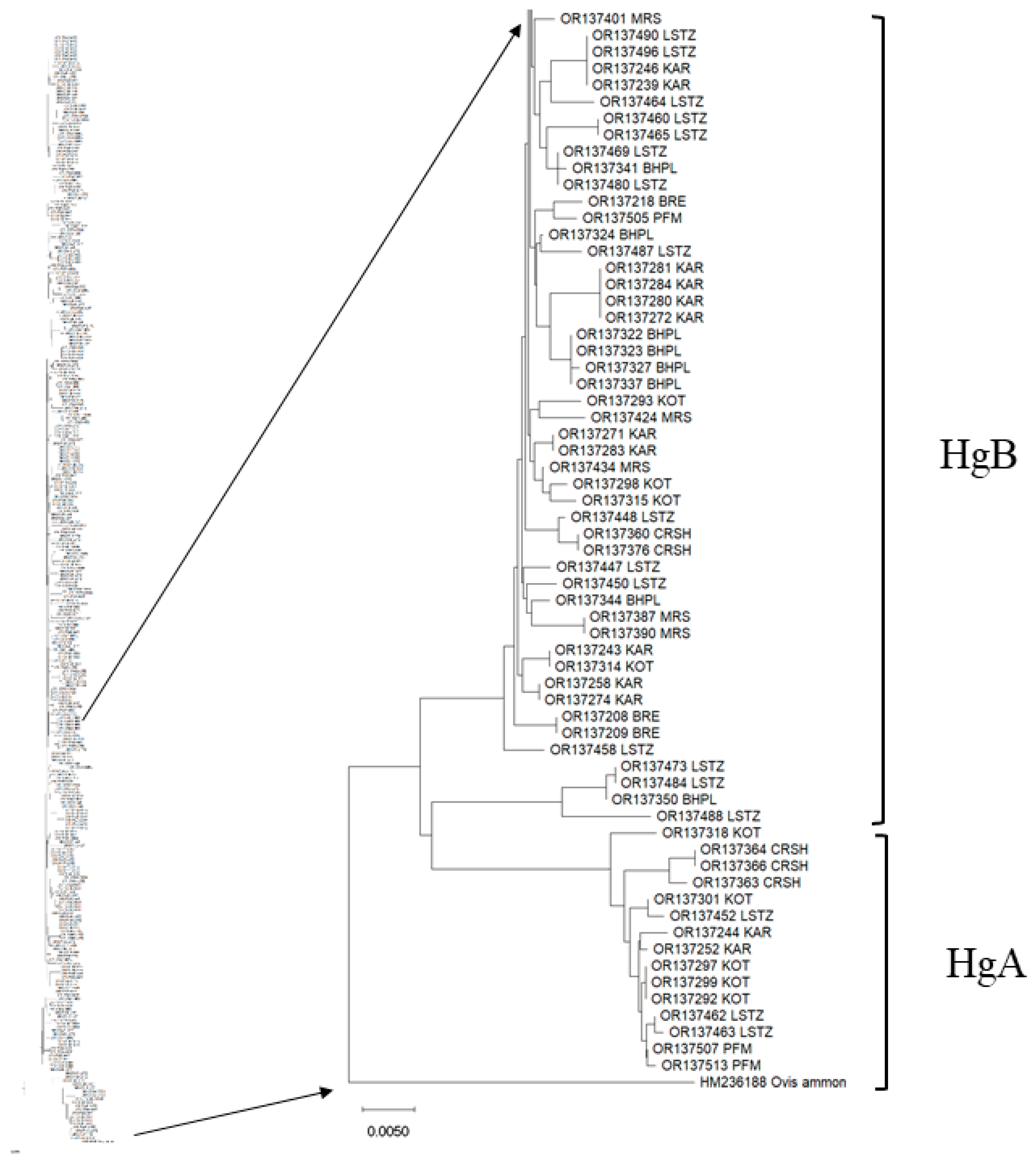
3.2. Haplotype Distribution among Breeds, Haplogroup Identification and Haplotype Network
3.3. Genetic Differentiation between Bulgarian Native Sheep Breeds
4. Discussion
4.1. mtDNA Diversity in Bulgarian Native Sheep Breeds
4.2. Genetic Differentiation and Similarities between Native Bulgarian Sheep Breeds
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alberto, F.J.; Boyer, F.; Orozco-terWengel, P.; Streeter, I.; Servin, B.; De Villemereuil, P.; Benjelloun, B.; Librado, P.; Biscarini, F.; Colli, L.; et al. Convergent genomic signatures of domestication in sheep and goats. Nat. Commun. 2018, 9, 813. [Google Scholar] [CrossRef]
- Ben Sassi-Zaidy, Y.; Mohamed-Brahmi, A.; Chaouch, M.; Maretto, F.; Cendron, F.; Charfi-Cheikhrouha, F.; Ben Abderrazak, S.; Djemali, M.; Cassandro, M. Historical westward migration phases of Ovis aries inferred from the population structure and the phylogeography of occidental Mediterranean native sheep breeds. Genes 2022, 13, 1421. [Google Scholar] [CrossRef]
- Ciani, E.; Mastrangelo, S.; Da Silva, A.; Marroni, F.; Ferenčaković, M.; Ajmone-Marsan, P.; Baird, H.; Barbato, M.; Colli, L.; Delvento, C.; et al. On the origin of European sheep as revealed by the diversity of the Balkan breeds and by optimizing population-genetic analysis tools. Genet. Sel. Evol. 2020, 52, 25. [Google Scholar] [CrossRef] [PubMed]
- Portanier, E.; Chevret, P.; Gélin, P.; Benedetti, P.; Sanchis, F.; Barbanera, F.; Kaerle, C.; Queney, G.; Bourgoin, G.; Devillard, S.; et al. New insights into the past and recent evolutionary history of the Corsican mouflon (Ovis gmelini musimon) to inform its conservation. Conserv. Genet. 2022, 23, 91–107. [Google Scholar] [CrossRef]
- Barbato, M.; Hailer, F.; Orozco-terWengel, P.; Kijas, J.; Mereu, P.; Cabras, P.; Mazza, R.; Pirastru, M.; Bruford, M.W. Genomic signatures of adaptive introgression from European mouflon into domestic sheep. Sci. Rep. 2017, 7, 7623. [Google Scholar] [CrossRef] [PubMed]
- Taylor, W.T.T.; Pruvost, M.; Posth, C.; Rendu, W.; Krajcarz, M.T.; Abdykanova, A.; Brancaleoni, G.; Spengler, R.; Hermes, T.; Schiavinato, S.; et al. Evidence for early dispersal of domestic sheep into Central Asia. Nat. Hum. Behav. 2021, 5, 1169–1179. [Google Scholar] [CrossRef]
- Pedrosa, S.; Uzun, M.; Arranz, J.-J.; Gutiérrez-Gil, B.; San Primitivo, F.; Bayón, Y. Evidence of three maternal lineages in near Eastern sheep supporting multiple domestication events. Proc. R. Soc. B. 2005, 272, 2211–2217. [Google Scholar] [CrossRef]
- Vigne, J.-D.; Carrère, I.; Briois, F.; Guilaine, J. The early process of mammal domestication in the Near East: New Evidence from the Pre-Neolithic and Pre-Pottery Neolithic in Cyprus. Curr. Anthropol. 2011, 52, S255–S271. [Google Scholar] [CrossRef]
- Vigne, J.-D. Zooarcheology and the biogeographical history of the mammals of Corsica and Sardinia since the last ice age. Mamm. Rev. 1992, 22, 87–96. [Google Scholar] [CrossRef]
- Robb, J. Material culture, landscapes of action, and emergent causation: A new model for the origins of the European Neolithic. Curr. Anthropol. 2013, 54, 657–683. [Google Scholar] [CrossRef]
- Becker, C.; Benecke, N.; Grabundžija, A.; Küchelmann, H.C.; Pollock, S.; Schier, W.; Schoch, C.; Schrakamp, I.; Schütt, B.; Schumacher, M. The textile revolution: Research into the origin and spread of wool production between the Near East and Central Europe. eTopoi J. Anc. Stud. 2016, 6, 102–151. [Google Scholar]
- Kreuz, A.; Marinova, E.; Schäfer, E.; Wiethold, J. A Comparison of Early Neolithic crop and weed assemblages from the Linearbandkeramik and the Bulgarian Neolithic Cultures: Differences and Similarities. Veget. Hist. Archaeobot. 2005, 14, 237–258. [Google Scholar] [CrossRef]
- Vitezović, S. Neolithisation of technology: Innovation and tradition in the Starcevo culture osseous industry. Doc. Praehist. 2016, 43, 123–137. [Google Scholar] [CrossRef]
- Bouby, L.; Marinval, P.; Durand, F.; Figueiral, I.; Briois, F.; Martzluff, M.; Perrin, T.; Valdeyron, N.; Vaquer, J.; Guilaine, J.; et al. Early Neolithic (ca. 5850-4500 cal BC) agricultural diffusion in the Western Mediterranean: An update of archaeobotanical data in SW France. PLoS ONE 2020, 15, e0230731. [Google Scholar] [CrossRef]
- Perrin, T.; Manen, C. Potential Interactions between Mesolithic hunter-gatherers and neolithic farmers in the Western Mediterranean: The geochronological data revisited. PLoS ONE 2021, 16, e0246964. [Google Scholar] [CrossRef] [PubMed]
- Martins, H.; Oms, F.X.; Pereira, L.; Pike, A.W.G.; Rowsell, K.; Zilhão, J. Radiocarbon dating the beginning of the Neolithic in Iberia: New results, new problems. J. Mediterr. Archaeol. 2015, 28, 105–131. [Google Scholar] [CrossRef]
- Hofmanová, Z.; Kreutzer, S.; Hellenthal, G.; Sell, C.; Diekmann, Y.; Díez-del-Molino, D.; Van Dorp, L.; López, S.; Kousathanas, A.; Link, V.; et al. Early farmers from across Europe directly descended from Neolithic Aegeans. Proc. Natl. Acad. Sci. USA 2016, 113, 6886–6891. [Google Scholar] [CrossRef]
- Shennan, S. The First Farmers of Europe. In The First Farmers of Europe: An Evolutionary Perspective; Cambridge World Archaeology; Cambridge University Press: Cambridge, UK, 2018; p. 253. [Google Scholar]
- Rivollat, M.; Réveillas, H.; Mendisco, F.; Pemonge, M.H.; Justeau, P.; Couture, C.; Lefranc, P.; Féliu, C.; Deguilloux, M.F. Ancient mitochondrial DNA from the Middle Neolithic necropolis of Obernai extends the genetic influence of the LBK to west of the Rhine. Am. J. Phys. Anthropol. 2016, 161, 522–529. [Google Scholar] [CrossRef]
- Liu, J.; Ding, X.; Zeng, Y.; Yue, Y.; Guo, X.; Guo, T.; Chu, M.; Wang, F.; Han, J.; Feng, R.; et al. Genetic diversity and phylogenetic evolution of Tibetan sheep based on mtDNA D-Loop Sequences. PLoS ONE 2016, 11, e0159308. [Google Scholar] [CrossRef] [PubMed]
- Gáspárdy, A.; Berger, B.; Zabavnik-Piano, J.; Kovács, E.; Annus, K.; Zenke, P.; Sáfár, L.; Maróti-Agóts, Á. Comparison of mtDNA control region among descendant breeds of the extinct Zaupel sheep revealed haplogroup C and D in Central Europe. Vet. Med. Sci. 2021, 7, 2330–2338. [Google Scholar] [CrossRef] [PubMed]
- Wanjala, G.; Bagi, Z.; Kusza, S. Meta-analysis of mitochondrial DNA Control Region Diversity to shed light on phylogenetic relationship and demographic history of African Sheep (Ovis aries) breeds. Biology 2021, 10, 762. [Google Scholar] [CrossRef] [PubMed]
- Demirci, S.; Koban Baştanlar, E.; Dağtaş, N.D.; Pişkin, E.; Engin, A.; Özer, F.; Yüncü, E.; Doğan, S.A.; Togan, I. Mitochondrial DNA diversity of modern, ancient and wild sheep (Ovis gmelinii anatolica) from Turkey: New insights on the evolutionary history of sheep. PLoS ONE 2013, 8, e81952. [Google Scholar] [CrossRef] [PubMed]
- Rannamäe, E.; Lõugas, L.; Niemi, M.; Kantanen, J.; Maldre, L.; Kadõrova, N.; Saarma, U. Maternal and paternal genetic diversity of ancient sheep in Estonia from the Late Bronze Age to the post-medieval period and comparison with other regions in Eurasia. Anim. Genet. 2016, 47, 208–218. [Google Scholar] [CrossRef]
- Yurtman, E.; Özer, O.; Yüncü, E.; Dağtaş, N.D.; Koptekin, D.; Çakan, Y.G.; Özkan, M.; Akbaba, A.; Kaptan, D.; Atağ, G.; et al. Archaeogenetic analysis of Neolithic sheep from Anatolia suggests a complex demographic history since domestication. Commun. Biol. 2021, 4, 1279. [Google Scholar] [CrossRef] [PubMed]
- Dymova, M.A.; Zadorozhny, A.V.; Mishukova, O.V.; Khrapov, E.A.; Druzhkova, A.S.; Trifonov, V.A.; Kichigin, I.G.; Tishkin, A.A.; Grushin, S.P.; Filipenko, M.L. Mitochondrial DNA analysis of ancient sheep from Altai. Anim. Genet. 2017, 48, 615–618. [Google Scholar] [CrossRef]
- Schroeder, O.; Benecke, N.; Frölich, K.; Peng, Z.; Kaniuth, K.; Sverchkov, L.; Reinhold, S.; Belinskiy, A.; Ludwig, A. Endogenous retroviral insertions indicate a secondary introduction of domestic sheep lineages to the Caucasus and Central Asia between the Bronze and Iron Age. Genes 2017, 8, 165. [Google Scholar] [CrossRef]
- Lancioni, H.; Di Lorenzo, P.; Ceccobelli, S.; Perego, U.A.; Miglio, A.; Landi, V.; Antognoni, M.T.; Sarti, F.M.; Lasagna, E.; Achilli, A. Phylogenetic relationships of three Italian merino-derived sheep breeds evaluated through a complete mitogenome analysis. PLoS ONE 2013, 8, e73712. [Google Scholar] [CrossRef]
- Gabbianelli, F.; Gargani, M.; Pariset, L.; Mariotti, M.; Alhaique, F.; De Minicis, E.; Barelli, L.; Ciammetti, E.; Redi, F.; Valentini, A. Mitochondrial DNA analysis of medieval sheep (Ovis aries) in central Italy reveals the predominance of haplogroup B already in the Middle Ages. Anim. Genet. 2015, 46, 329–332. [Google Scholar] [CrossRef]
- Machová, K.; Málková, A.; Vostrý, L. Sheep post-domestication expansion in the context of mitochondrial and Y chromosome haplogroups and haplotypes. Genes 2022, 13, 613. [Google Scholar] [CrossRef]
- Pedrosa, S.; Arranz, J.-J.; Brito, N.; Molina, A.; San Primitivo, F.; Bayón, Y. Mitochondrial diversity and the origin of Iberian sheep. Genet. Sel. Evol. 2007, 39, 91. [Google Scholar] [CrossRef]
- Pariset, L.; Mariotti, M.; Gargani, M.; Joost, S.; Negrini, R.; Perez, T.; Bruford, M.; Ajmone Marsan, P.; Valentini, A. Genetic diversity of sheep breeds from Albania, Greece, and Italy assessed by mitochondrial DNA and nuclear polymorphisms (SNPs). Sci. World J. 2011, 11, 1641–1659. [Google Scholar] [CrossRef]
- Li, J.; Song, F.; Lang, M.; Xie, M. Comprehensive Insights into the genetic background of Chinese populations using Y chromosome markers. R. Soc. Open Sci. 2023, 10, 230814. [Google Scholar] [CrossRef]
- Tapio, M.; Marzanov, N.; Ozerov, M.; Ćinkulov, M.; Gonzarenko, G.; Kiselyova, T.; Murawski, M.; Viinalass, H.; Kantanen, J. Sheep mitochondrial DNA variation in European, Caucasian, and Central Asian areas. Mol. Biol. Evol. 2006, 23, 1776–1783. [Google Scholar] [CrossRef] [PubMed]
- Zhao, E.; Yu, Q.; Zhang, N.; Kong, D.; Zhao, Y. Mitochondrial DNA diversity and the origin of Chinese indigenous sheep. Trop. Anim. Health Prod. 2013, 45, 1715–1722. [Google Scholar] [CrossRef] [PubMed]
- Meadows, J.R.S.; Cemal, I.; Karaca, O.; Gootwine, E.; Kijas, J.W. Five ovine mitochondrial lineages identified from sheep breeds of the Near East. Genetics 2007, 175, 1371–1379. [Google Scholar] [CrossRef] [PubMed]
- Dimov, D.; Vuchkov, A. Sheep genetic resources in Bulgaria with focus on breeds with coloured wool. Genet. Resour. 2020, 2, 11–24. [Google Scholar] [CrossRef]
- FAO. The Second Report on the State of the World’s Animal Genetic Resources for Food and Agriculture; Scherf, B.D., Pilling, D., Eds.; FAO Commission on Genetic Resources for Food and Agriculture Assessments: Rome, Italy, 2015; Available online: http://www.fao.org/3/a-i4787e/index.html (accessed on 13 June 2022).
- Kusza, S.; Dimov, D.; Nagy, I.; Bõsze, Z.; Jávor, A.; Kukovics, S. Microsatellite analysis to estimate genetic relationships among five Bulgarian sheep breeds. Genet. Mol. Biol. 2010, 33, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Hristova, D.; Metodiev, S.; Nikolov, V.; Vassilev, D.; Todorovska, E. Genetic variation of Bulgarian autochthonous sheep breeds using microsatellite markers. Genetika 2017, 49, 247–258. [Google Scholar] [CrossRef]
- Mihailova, Y. Genetic diversity and structure of 2 indigenous sheep breeds (Kotel and Teteven) in Bulgaria using microsatellite markers. Biotechnol. Biotechnol. Equip. 2021, 35, 576–585. [Google Scholar] [CrossRef]
- Mihailova, Y.; Rusanov, K.; Rusanova, M.; Vassileva, P.; Atanassov, I.; Nikolov, V.; Todorovska, E.G. Genetic diversity and population structure of Bulgarian autochthonous sheep breeds revealed by microsatellite analysis. Animals 2023, 13, 1878. [Google Scholar] [CrossRef]
- Odjakova, T.; Todorov, P.; Radoslavov, G.; Hristov, P. Microsatellite genotyping of two Bulgarian sheep breeds. Diversity 2022, 14, 210. [Google Scholar] [CrossRef]
- Odjakova, T.; Todorov, P.; Kalaydzhiev, G.; Salkova, D.; Dundarova, H.; Radoslavov, G.; Hristov, P. A study on the genetic diversity and subpopulation structure of three Bulgarian mountainous sheep breeds, based on genotyping of microsatellite markers. Small Rumin. Res. 2023, 226, 107034. [Google Scholar] [CrossRef]
- Hiendleder, S.; Lewalski, H.; Wassmuth, R.; Janke, A. The complete mitochondrial DNA sequence of the domestic sheep (Ovis aries) and comparison with the other major ovine haplotype. J. Mol. Evol. 1998, 47, 441–448. [Google Scholar] [CrossRef]
- Revelo, H.A.; López-Alvarez, D.; Landi, V.; Rizzo, L.; Alvarez, L.A. Mitochondrial DNA variations in Colombian creole sheep confirm an Iberian origin and shed light on the dynamics of introduction events of African genotypes. Animals 2020, 10, 1594. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X.; Li, W.H. Statistical tests of neutrality of mutations. Genetics 1993, 133, 693–709. [Google Scholar] [CrossRef]
- Achilli, A.; Olivieri, A.; Pala, M.; Metspalu, E.; Fornarino, S.; Battaglia, V.; Accetturo, M.; Kutuev, I.; Khusnutdinova, E.; Pennarun, E.; et al. Mitochondrial DNA variation of modern Tuscans supports the Near Eastern origin of Etruscans. Am. J. Hum. Genet. 2007, 80, 759–768. [Google Scholar] [CrossRef]
- Koseniuk, A.; Słota, E. Mitochondrial control region diversity in Polish sheep breeds. Arch. Anim. Breed. 2016, 59, 227–233. [Google Scholar] [CrossRef]
- Koshkina, O.; Deniskova, T.; Dotsev, A.; Kunz, E.; Selionova, M.; Medugorac, I.; Zinovieva, N. Phylogenetic analysis of Russian native sheep breeds based on mtDNA sequences. Genes 2023, 14, 1701. [Google Scholar] [CrossRef] [PubMed]
- Cinkulov, M.; Popovski, Z.; Porcu, K.; Tanaskovska, B.; Hodzić, A.; Bytyqi, H.; Mehmeti, H.; Margeta, V.; Djedović, R.; Hoda, A.; et al. Genetic diversity and structure of the West Balkan Pramenka sheep types as revealed by microsatellite and mitochondrial DNA analysis. J. Anim. Breed. Genet. 2008, 125, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F.; Davis, S.J.; Pereira, L.; McEvoy, B.; Bradley, D.G.; Amorim, A. Genetic signatures of a Mediterranean influence in Iberian Peninsula sheep husbandry. Mol. Biol. Evol. 2006, 23, 1420–1426. [Google Scholar] [CrossRef]
- Peng, M.; Fan, L.; Shi, N.; Ning, T.; Yao, Y.; Murphy, R.W.; Wang, W.; Zhang, Y. DomeTree: A canonical toolkit for mitochondrial DNA analyses in domesticated animals. Mol. Ecol. Resour. 2015, 15, 1238–1242. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the Neighbor-Joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Yarkov, D.; Stankov, K.; Stankov, I. Historical review of the development of Bulgarian livestock production. Bulg. J. Agric. Sci. 2022, 28, 564–578. [Google Scholar]
- Zhelyazkova, P.; Dimov, D.; Andonov, S. Genetic-parameter estimation of milk yield in White Maritza sheep breed using different test day models. Arch. Anim. Breed. 2023, 66, 253–263. [Google Scholar] [CrossRef]
- Borychowski, M.; Stępień, S.; Polcyn, J.; Tošović-Stevanović, A.; Ćalović, D.; Lalić, G.; Žuža, M. Socio-economic determinants of small family farms’ resilience in selected Central and Eastern European countries. Sustainability 2020, 12, 10362. [Google Scholar] [CrossRef]
- Stuart, D.; Gunderson, R. Nonhuman animals as fictitious commodities: Exploitation and consequences in industrial agriculture. Soc. Anim. 2020, 28, 291–310. [Google Scholar] [CrossRef]
- Greenfield, H.J. The secondary products revolution: The past, the present and the future. World Archaeol. 2010, 42, 29–54. [Google Scholar] [CrossRef]
- Marciniak, A. The secondary products revolution: Empirical evidence and its current zooarchaeological critique. J. World Prehist. 2011, 24, 117–130. [Google Scholar] [CrossRef]
- Bondár, M. Prehistoric innovations: Wheels and wheeled vehicles. Acta Antiq. Hung. 2018, 69, 271–297. [Google Scholar] [CrossRef]
- Greenfield, H.; Marciniak, A. Retention of old technologies following the end of the Neolithic: Microscopic analysis of the butchering marks on animal bones from Çatalhöyük East. World Archaeol. 2019, 51, 76–103. [Google Scholar] [CrossRef]
- Sherratt, A. Plough and pastoralism: Aspects of the secondary products revolution. In Pattern of the Past; Hodder, I., Isaac, G., Hammond, N., Eds.; Cambridge University Press: Cambridge, UK, 1981; pp. 261–306. [Google Scholar]
- Sherratt, A. The secondary exploitation of animals in the Old World. World Archaeol. 1983, 15, 90–104. [Google Scholar] [CrossRef]
- Kandoussi, A.; Boujenane, I.; Piro, M.; Petit, D.P. Genetic diversity and population structure of Moroccan Beni Ahsen: Is this endangered ovine breed one of the ancestors of Merino? Ruminants 2022, 2, 201–211. [Google Scholar] [CrossRef]
- Breniquet, C.; Michel, C. Wool economy in the ancient Near East. In Wool Economy in the Ancient Near East and the Aegean: From the Beginnings of Sheep Husbandry to Institutional Textile Industry; Ancient Textile Series; Oxbow Books: Oxford, UK, 2014; Volume 17, pp. 1–11. [Google Scholar]
- Arbuckle, B.S.; Hammer, E.L. The rise of pastoralism in the ancient Near East. J. Archaeol. Res. 2019, 27, 391–449. [Google Scholar] [CrossRef]
- Rochus, C.M.; Tortereau, F.; Plisson-Petit, F.; Restoux, G.; Moreno-Romieux, C.; Tosser-Klopp, G.; Servin, B. Revealing the selection history of adaptive loci using genome-wide scans for selection: An example from domestic sheep. BMC Genom. 2018, 19, 71. [Google Scholar] [CrossRef]
- Osei-Amponsah, R.; Chauhan, S.S.; Leury, B.J.; Cheng, L.; Cullen, B.; Clarke, I.J.; Dunshea, F.R. Genetic selection for thermotolerance in ruminants. Animals 2019, 9, 948. [Google Scholar] [CrossRef] [PubMed]
- Abied, A.; Xu, L.; Sahlu, B.W.; Xing, F.; Ahbara, A.; Pu, Y.; Lin, J.; Berihulay, H.; Islam, R.; He, X.; et al. Genome-wide analysis revealed homozygosity and demographic history of five Chinese sheep breeds adapted to different environments. Genes 2020, 11, 1480. [Google Scholar] [CrossRef] [PubMed]

| Breed | No. of Sequences | h | π | Hd | Kt | S | Fu and Li’s D | Fu and Li’s F | Tajima’s D |
|---|---|---|---|---|---|---|---|---|---|
| BRE | 17 | 11 | 0.009 | 0.912 | 10.471 | 45 | −0.69502 p > 0.10 | −0.79361 p > 0.10 | −0.88686 p > 0.10 |
| KAR | 60 | 40 | 0.010 | 0.981 | 11.892 | 117 | −1.23094 p > 0.10 | −1.66000 p > 0.10 | −1.83455 p < 0.05 |
| KOT | 36 | 31 | 0.016 | 0.995 | 18.705 | 112 | −2.03216 p > 0.10 | −2.29412 p < 0.05 | −1.91532 p < 0.05 |
| BHPL | 32 | 27 | 0.011 | 0.986 | 12.617 | 97 | −1.68405 p > 0.10 | −1.89471 p > 0.10 | −1.80146 p > 0.10 |
| CRSH | 34 | 28 | 0.011 | 0.986 | 12.617 | 105 | −0.02132 p > 0.10 | −0.47016 p > 0.10 | −1.20019 p > 0.10 |
| MRS | 48 | 27 | 0.009 | 0.953 | 11.359 | 87 | −0.82732 p > 0.10 | −1.20424 p > 0.10 | −1.49593 p > 0.10 |
| LSTZ | 69 | 54 | 0.013 | 0.991 | 15.014 | 147 | −0.59446 p > 0.10 | −1.20335 p > 0.10 | −1.75938 p > 0.10 |
| PFM | 14 | 13 | 0.017 | 1.000 | 20.333 | 78 | −0.32847 p > 0.10 | −0.49186 p > 0.10 | −0.86217 p > 0.10 |
| Population. | KAR | PBH | CRSH | KOT | BRE | MRS | LZST | PFM |
|---|---|---|---|---|---|---|---|---|
| KAR | 0.000 | |||||||
| PBH | 0.01040 | 0.000 | ||||||
| CRSH | 0.01323 | 0.01410 | 0.000 | |||||
| KOT | 0.01348 | 0.01442 | 0.01619 | 0.000 | ||||
| BRE | 0.00979 | 0.01038 | 0.01326 | 0.01352 | 0.000 | |||
| MRS | 0.00992 | 0.01075 | 0.01352 | 0.01393 | 0.01002 | 0.000 | ||
| LZST | 0.01120 | 0.01196 | 0.01456 | 0.01473 | 0.01125 | 0.01163 | 0.000 | |
| PFM | 0.01509 | 0.01587 | 0.01770 | 0.01776 | 0.01522 | 0.01541 | 0.01623 | 0.000 |
| Source of Variation | Degrees of Freedom d.f. | Sum of Squares, SS | Variance Components, VC | Percentage of Variation V % |
|---|---|---|---|---|
| Among populations | 7 | 96.897 | 0.17608 | 2.42 |
| Within populations | 307 | 2180.386 | 7.10223 | 97.58 |
| Total | 314 | 2277.283 | 7.27832 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalaydzhiev, G.; Palova, N.; Dundarova, H.; Lozanova, L.; Mehandjyiski, I.; Radoslavov, G.; Hristov, P. Mitochondrial Diversity and Phylogenetic Relationship of Eight Native Bulgarian Sheep Breeds. Animals 2023, 13, 3655. https://doi.org/10.3390/ani13233655
Kalaydzhiev G, Palova N, Dundarova H, Lozanova L, Mehandjyiski I, Radoslavov G, Hristov P. Mitochondrial Diversity and Phylogenetic Relationship of Eight Native Bulgarian Sheep Breeds. Animals. 2023; 13(23):3655. https://doi.org/10.3390/ani13233655
Chicago/Turabian StyleKalaydzhiev, Georgi, Nadezhda Palova, Heliana Dundarova, Lyudmila Lozanova, Ivan Mehandjyiski, Georgi Radoslavov, and Peter Hristov. 2023. "Mitochondrial Diversity and Phylogenetic Relationship of Eight Native Bulgarian Sheep Breeds" Animals 13, no. 23: 3655. https://doi.org/10.3390/ani13233655
APA StyleKalaydzhiev, G., Palova, N., Dundarova, H., Lozanova, L., Mehandjyiski, I., Radoslavov, G., & Hristov, P. (2023). Mitochondrial Diversity and Phylogenetic Relationship of Eight Native Bulgarian Sheep Breeds. Animals, 13(23), 3655. https://doi.org/10.3390/ani13233655






