Liver Transcriptome Profiling Identifies Key Genes Related to Lipid Metabolism in Yili Geese
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
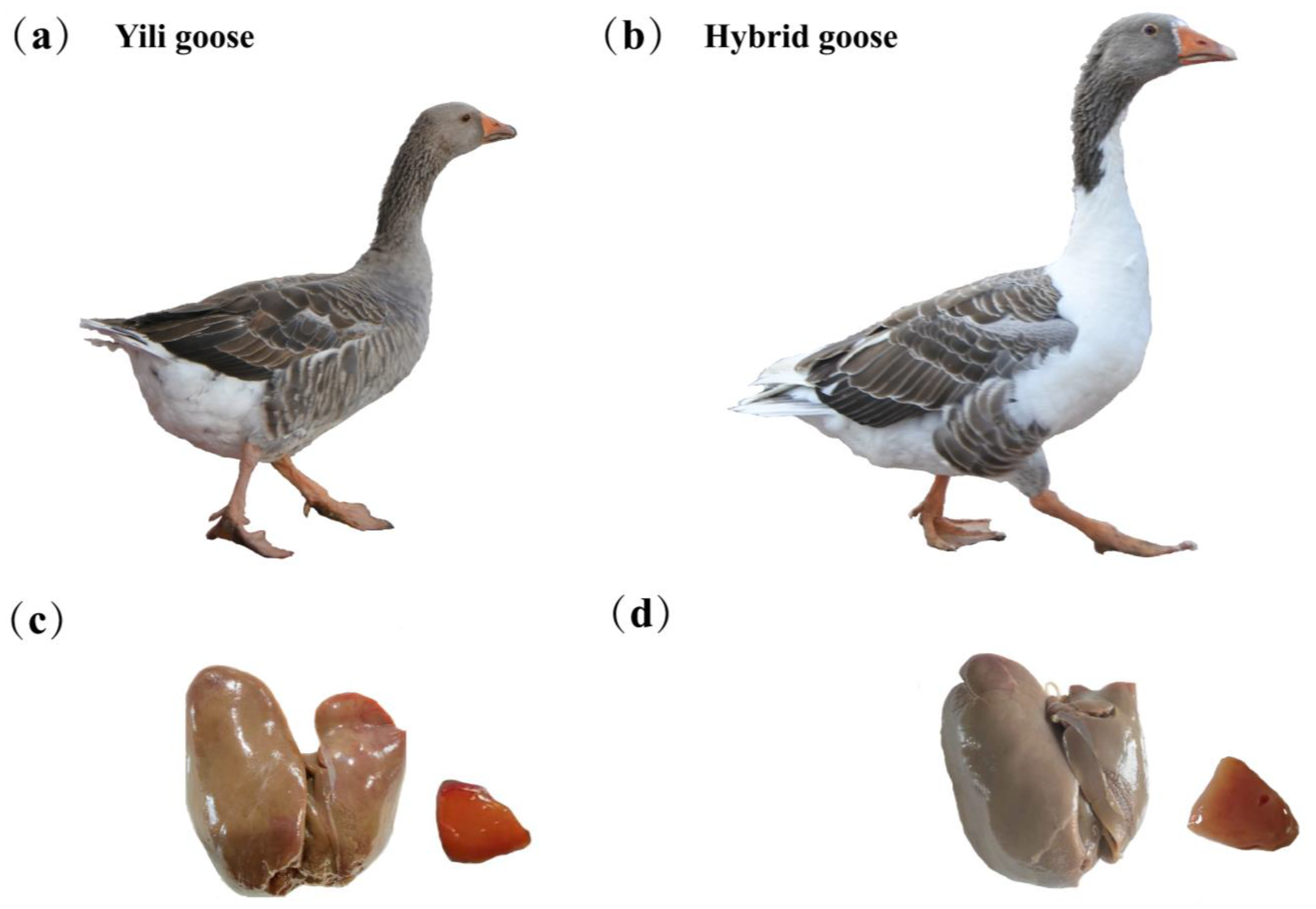
2.2. Animals and Sample Collection
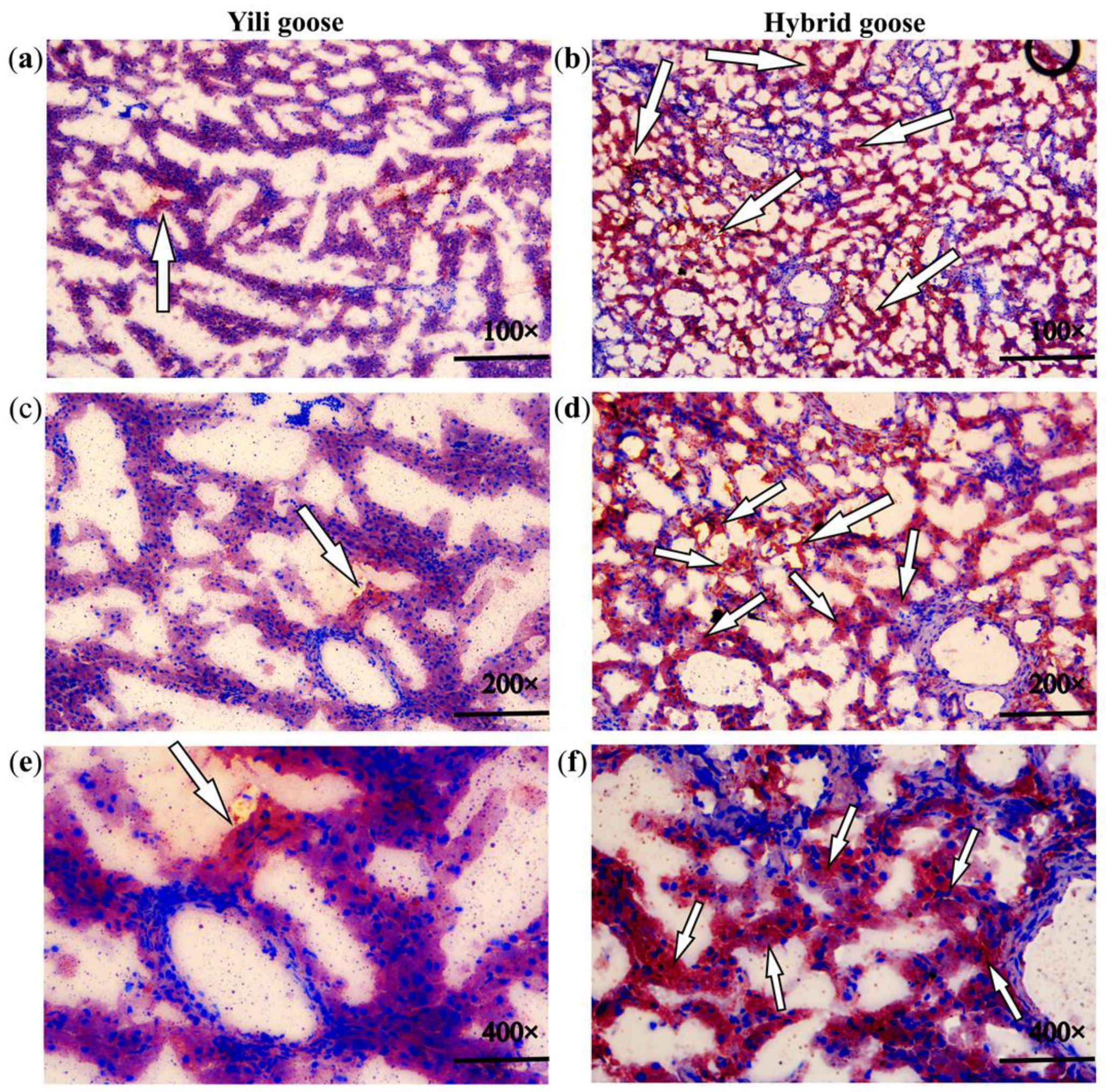
2.3. Histological Staining
2.4. RNA Extraction, Library Preparation, and Sequencing
2.5. Quality Control, Comparison, Assembly, and Quantification of Sequencing Data
2.6. Identification of DEGs and Homologous Search of Unknown Genes
2.7. Functional Annotation and Pathway Enrichment Analysis of DEGs
2.8. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) for DEGs Verification
2.9. Statistical Analysis
3. Results
3.1. Lipid-Related Phenotypes of Geese
3.2. Summary of RNA-Seq Data
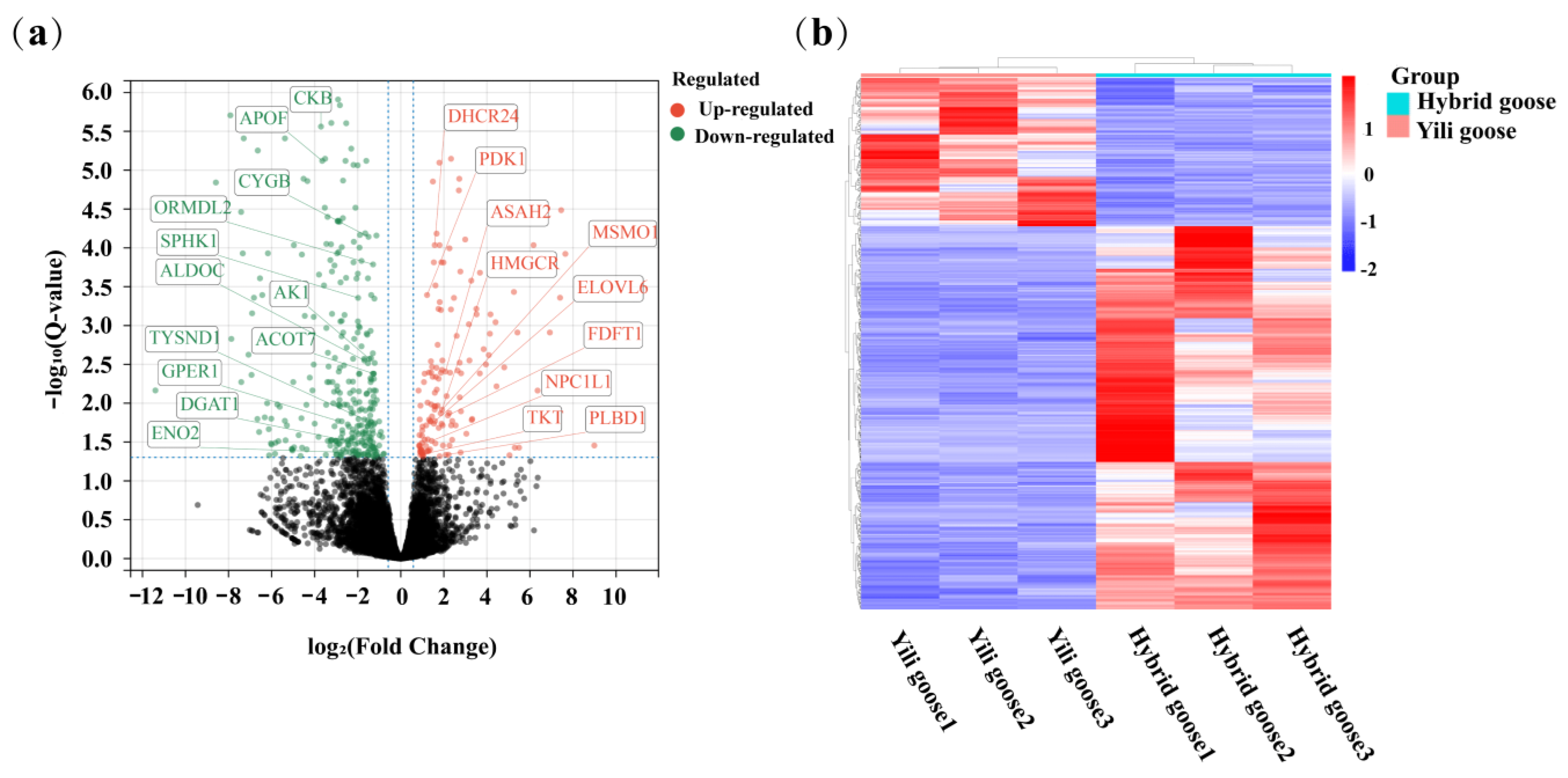
3.3. Differentially Expressed Genes Analysis
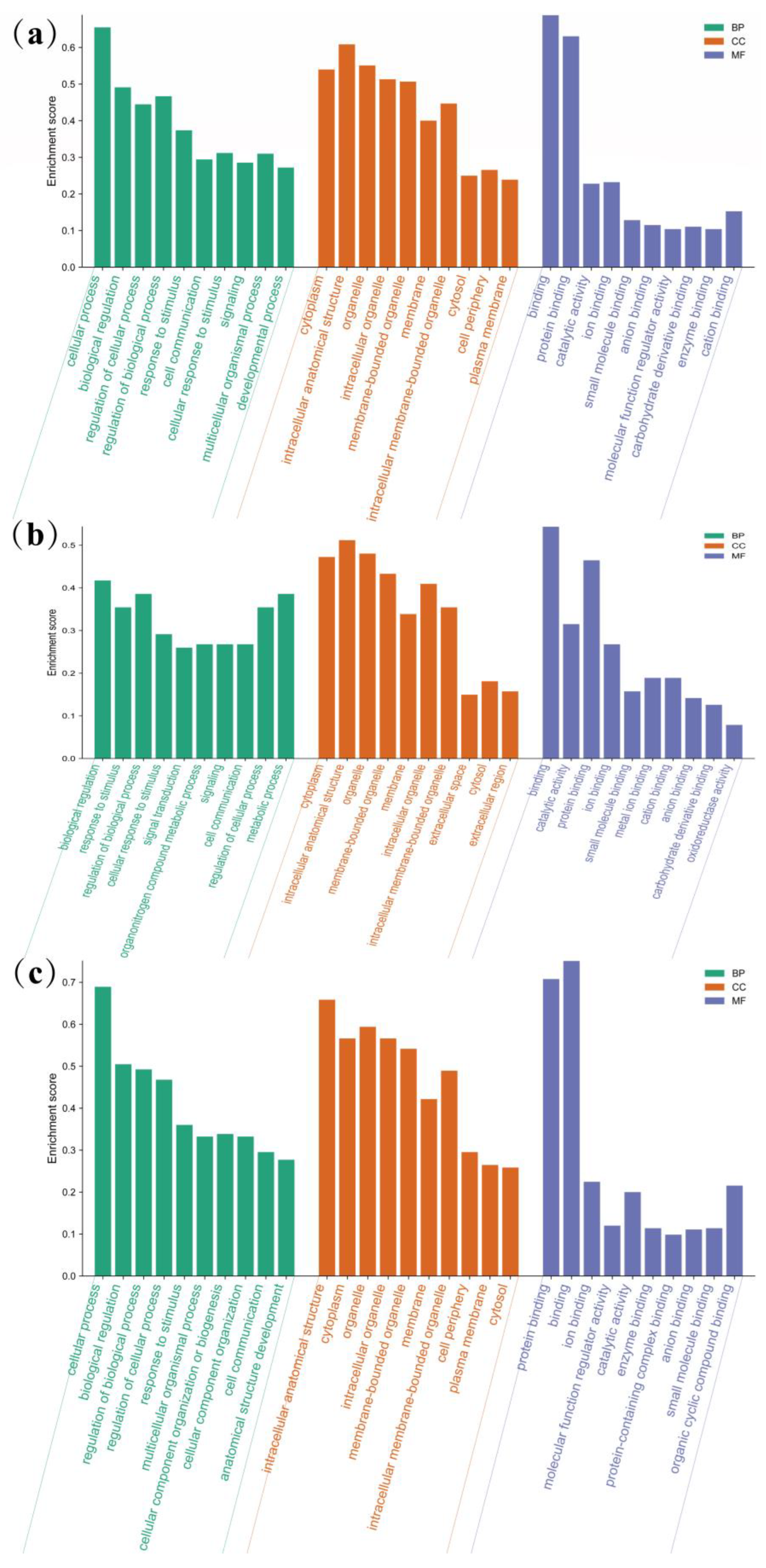
3.4. GO Annotation and Enrichment Analysis of Differentially Expressed Genes
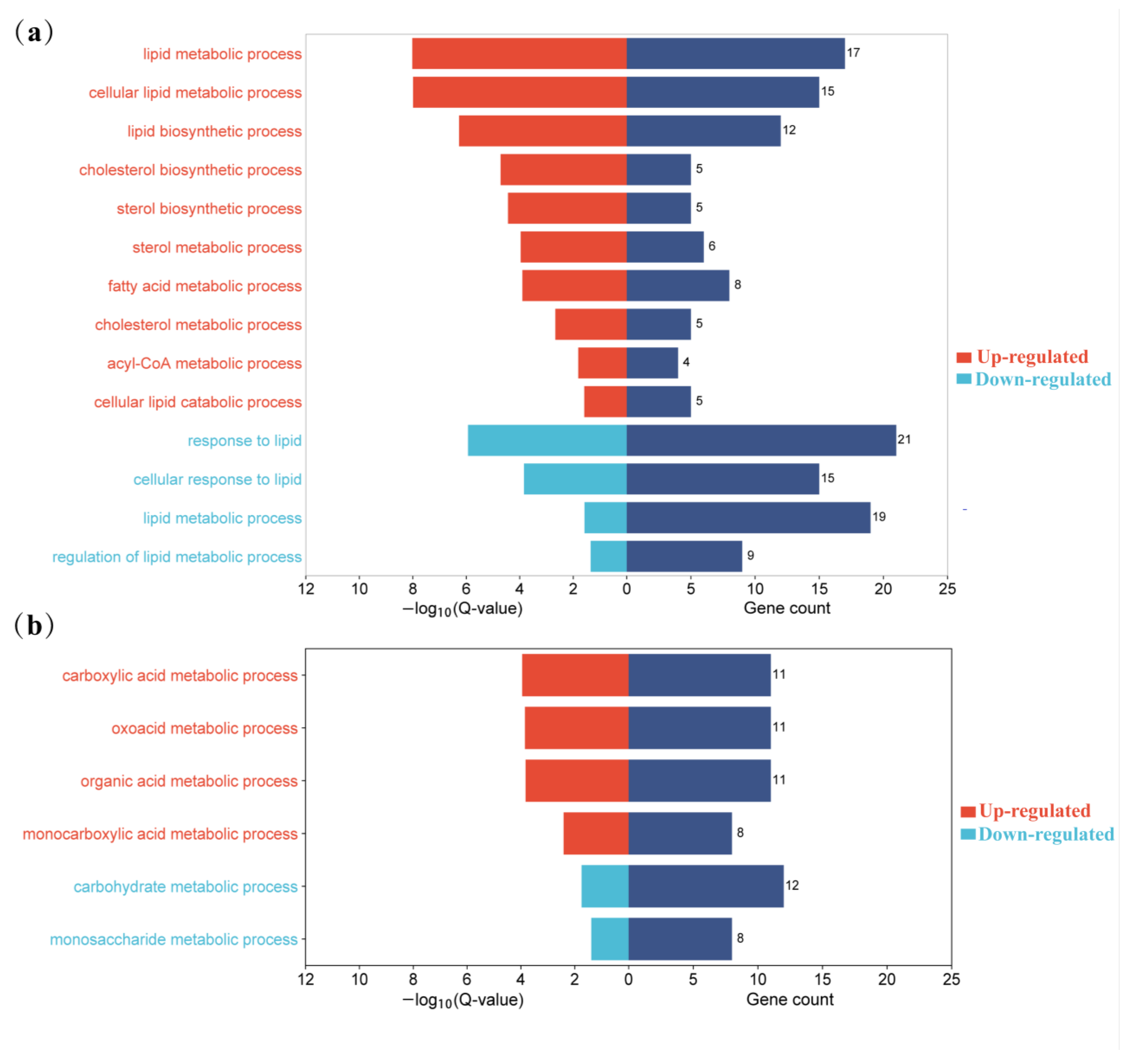
3.5. KEGG Pathway Analysis of DEG
3.6. Gene Screening and Protein Interaction Analysis
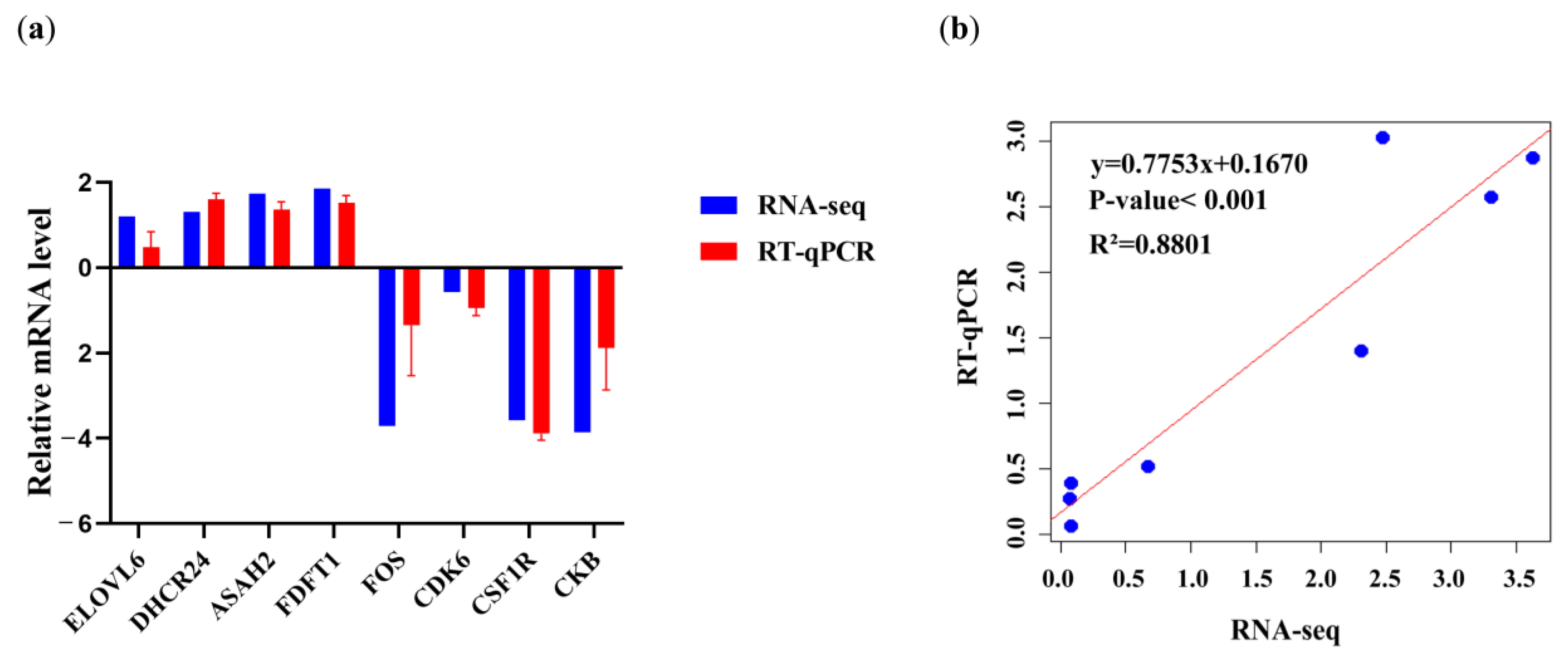
3.7. RNA-Seq Data Validation by RT-qPCR
4. Discussion

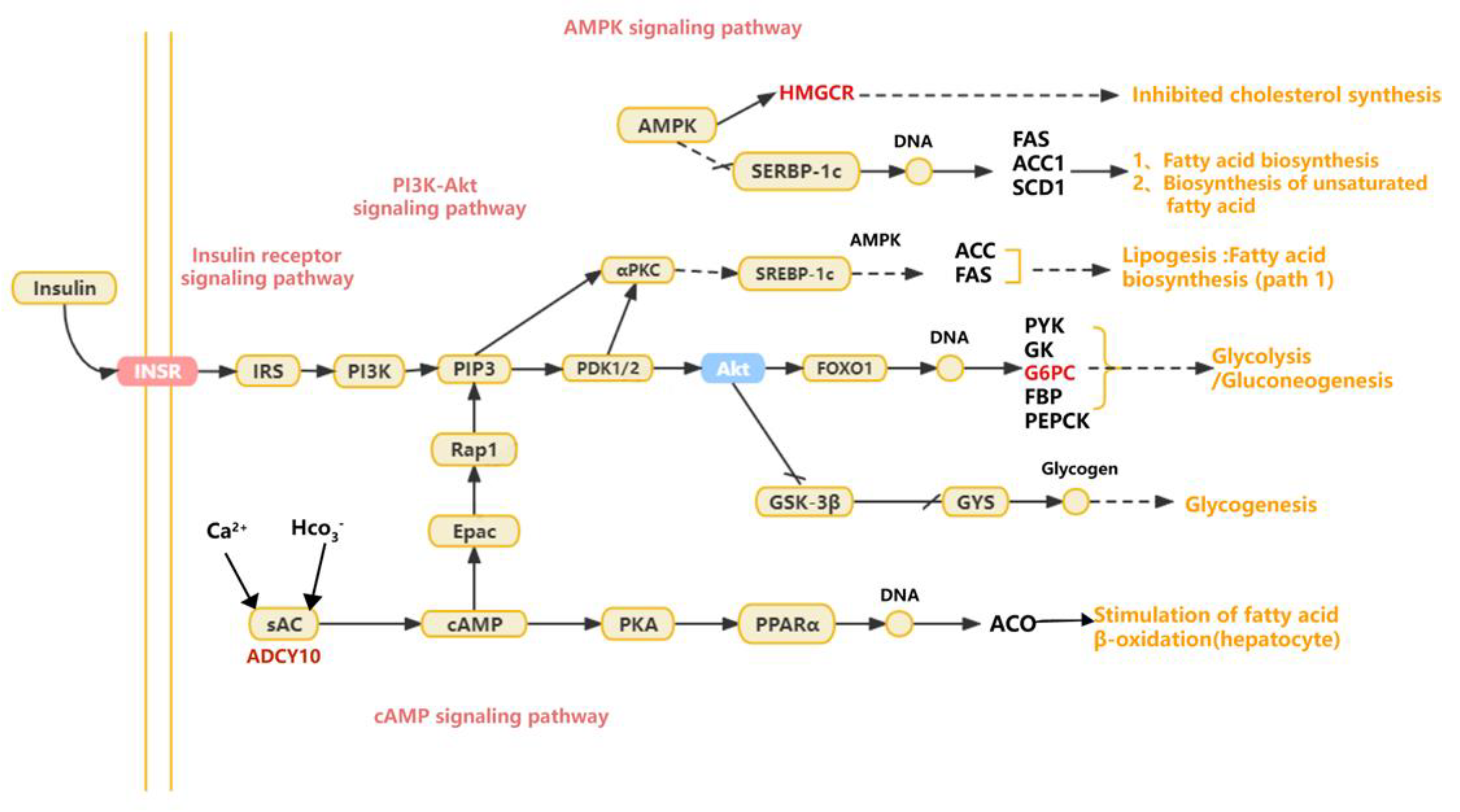
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, C.; Yang, F.; Wang, Q.; Zhou, X.; Xie, M.; Kang, P.; Wang, Y.; Peng, X. Nutritive Value of Mulberry Leaf Meal and its Effect on the Performance of 35–70-Day-Old Geese. J. Poult. Sci. 2017, 54, 41–46. [Google Scholar] [CrossRef][Green Version]
- Wu, Y.; Zhao, X.; Chen, L.; Wang, J.; Duan, Y.; Li, H.; Lu, L. Transcriptomic Analyses of the Hypothalamic-Pituitary-Gonadal Axis Identify Candidate Genes Related to Egg Production in Xinjiang Yili Geese. Animals 2020, 10, 90. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M. A Comparative Study on Growth Performance, Slaughtering Performance, Serum Parameters and Intestinal Development of Yili Goose and Its Hybrid Goose. Master’s Thesis, Xinjiang Agricultural University, Urumqi, China, 2021. [Google Scholar]
- Wang, J.; Peng, X.; Wu, Y.; Zhao, X.; Duan, Y.; Liu, J.; Fang, S.; Li, H. Comparative Study on Slaughtering Performance, Meat Quality, and Antioxidant Performance of Yili Goose and Its Hybrids with Huoyan Goose. Heilongjiang Anim. Sci. Vet. Med. 2019, 102, 51–55+59. [Google Scholar] [CrossRef]
- Mancinelli, A.C.; Menchetti, L.; Birolo, M.; Bittante, G.; Chiattelli, D.; Castellini, C. Crossbreeding to improve local chicken breeds: Predicting growth performance of the crosses using the Gompertz model and estimated heterosis. Poult. Sci. 2023, 102, 102783. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Liu, B.; Lei, H.; Lin, Z.; Chen, J.; Zhu, Y.; Ye, H.; Yang, L.; Wang, W. Effects of Dietary Tryptophan on Growth Performance, Plasma Parameters, and Internal Organs of 1–28-Day-Old Sichuan White Geese. J. Poult. Sci. 2023, 60, 2023008. [Google Scholar] [CrossRef]
- He, H.; Chen, J.; Tang, Y.; Zhang, J.; Qiu, S.; Han, X.; Li, L.; Cao, T.; Xiang, B.; Liu, A. Effects of dietary crude fiber levels on growth performance, slaughter performance, and serum biochemical indicators of Sichuan white geese. J. Southwest Univ. 2021, 43, 44–51. [Google Scholar] [CrossRef]
- Li, Q.; Zhong, H.; Peng, X.; Wang, Q. Fitting and Analysis of Growth Law and Body Weight Curve for Sichuan White Goose. Chin. Poult. 2018, 40, 52–54. [Google Scholar] [CrossRef]
- Zhang, J.; Xiong, Z.; Li, L.; Tian, X.; Jie, X.; Cheng, Y.; Wan, K.; Liu, A.; Xiang, B.; HE, H. Functional Analysis of Liver Transcriptomes of Goose with Different Body Weight. Acta Vet. Zootech. Sin. 2020, 51, 987–996. [Google Scholar] [CrossRef]
- Zhao, X.; Xuan, R.; Wang, A.; Li, Q.; Zhao, Y.; Du, S.; Duan, Q.; Wang, Y.; Ji, Z.; Guo, Y.; et al. High-Throughput Sequencing Reveals Transcriptome Signature of Early Liver Development in Goat Kids. Genes 2022, 13, 833. [Google Scholar] [CrossRef]
- Liu, X.; Guo, C.; Liu, Y.; Qu, X. Research progress on molecular protective mechanisms of goose liver steatosis and goose fatty liver formation. Chin. J. Anim. Nutr. 2018, 30, 2453–2458. [Google Scholar] [CrossRef]
- Jiang, K. RNA-seq and ChIP-seq Integrative Analysis reveals the Molecular Mechanism of Estrogen Regulation Liver Lipid Metabolism in Chicken. Ph.D. Thesis, Henan Agricultural University, Zhengzhou, China, 2021. [Google Scholar]
- Lyu, W.; Xiang, Y.; Wang, X.; Li, J.; Yang, C.; Yang, H.; Xiao, Y. Differentially Expressed Hepatic Genes Revealed by Transcriptomics in Pigs with Different Liver Lipid Contents. Oxid. Med. Cell. Longev. 2022, 2022, 2315575. [Google Scholar] [CrossRef]
- Ge, K.; Chen, X.; Kuang, J.; Yang, L.; Geng, Z. Comparison of liver transcriptome from high- and low-intramuscular fat Chaohu ducks provided additional candidate genes for lipid selection. 3 Biotech 2019, 9, 251. [Google Scholar] [CrossRef]
- Gunawan, A.; Listyarini, K.; Harahap, R.S.; Jakaria; Roosita, K.; Sumantri, C.; Inounu, I.; Akter, S.H.; Islam, M.A.; Uddin, M.J. Hepatic transcriptome analysis identifies genes, polymorphisms and pathways involved in the fatty acids metabolism in sheep. PLoS ONE 2021, 16, e0260514. [Google Scholar] [CrossRef]
- Cao, H.Y.; Xu, F.; Chen, Z.L.; Lin, B.S.; Zheng, X.B.; Yuan, S.H.; Liang, H.; Weng, J.P. Effect of exendin-4 on lipid deposition in skeletal muscle of diet-induced obese mice and its underlying mechanism. Zhonghua Yi Xue Za Zhi 2017, 97, 131–136. [Google Scholar] [CrossRef]
- Xiaoling, L.I.; Fengxia, S.; Zimeng, S.; Yingxue, Z.; Jie, L.I.; Qiuxiang, Z. Effect of Dangfei Liganning capsule on liver X receptor α/steroid regulatory element binding protein-1/fatty acid synthase signal pathway in rats with metabolic- associated fatty liver disease. J. Tradit. Chin. Med. 2022, 42, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Jehle, J.; Schöne, B.; Bagheri, S.; Avraamidou, E.; Danisch, M.; Frank, I.; Pfeifer, P.; Bindila, L.; Lutz, B.; Lütjohann, D.; et al. Elevated levels of 2-arachidonoylglycerol promote atherogenesis in ApoE-/- mice. PLoS ONE 2018, 13, e0197751. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Wang, J.; Han, C.; Zhai, N.; Lv, J.; Zhou, Z.; Tang, H.; Xiang, S.; Li, L. Identification of differentially expressed genes between hepatocytes of Landes geese (Anser anser) and Sichuan White geese (Anser cygnoides). Mol. Biol. Rep. 2010, 37, 4059–4066. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xing, Y.; Fan, X.; Liu, T.; Zhao, M.; Liu, L.; Hu, X.; Cui, H.; Geng, T.; Gong, D. Fasting and Refeeding Affect the Goose Liver Transcriptome Mainly Through the PPAR Signaling Pathway. J. Poult. Sci. 2021, 58, 245–257. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Belinky, F.; Nativ, N.; Stelzer, G.; Zimmerman, S.; Iny Stein, T.; Safran, M.; Lancet, D. PathCards: Multi-source consolidation of human biological pathways. Database 2015, 2015, 1–13. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Liu, Y.; He, D. Research Progress of Steatosis Mechanism in Goose Liver. Chin. Poutry 2020, 42, 1. [Google Scholar] [CrossRef]
- Wang, G.; Jin, L.; Li, Y.; Tang, Q.; Hu, S.; Xu, H.; Gill, C.A.; Li, M.; Wang, J. Transcriptomic analysis between Normal and high-intake feeding geese provides insight into adipose deposition and susceptibility to fatty liver in migratory birds. BMC Genom. 2019, 20, 372. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wu, Y.-m.; Zhang, Y.; Tang, S.-y.; Han, S.-s.; Cui, C.; Tan, B.; Yu, J.; Kang, H.-y.; Chen, G.-d.; et al. miR-27b-5p regulates chicken liver disease via targeting IRS2 to suppress the PI3K/AKT signal pathway. J. Integr. Agric. 2023. [Google Scholar] [CrossRef]
- Carotti, S.; Aquilano, K.; Valentini, F.; Ruggiero, S.; Alletto, F.; Morini, S.; Picardi, A.; Antonelli-Incalzi, R.; Lettieri-Barbato, D.; Vespasiani-Gentilucci, U. An overview of deregulated lipid metabolism in nonalcoholic fatty liver disease with special focus on lysosomal acid lipase. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 319, G469–G480. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z. Effects of Oleic Oil on Exoression of Genes Related TO Lipid Metabolic Homeostasis in Goose Primary Hepatocytes. Master’s Thesis, Sichuan Agricultural University, Ya’an, China, 2009. [Google Scholar]
- Loh, K.; Tam, S.; Murray-Segal, L.; Huynh, K.; Meikle, P.J.; Scott, J.W.; van Denderen, B.; Chen, Z.; Steel, R.; LeBlond, N.D.; et al. Inhibition of Adenosine Monophosphate-Activated Protein Kinase-3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase Signaling Leads to Hypercholesterolemia and Promotes Hepatic Steatosis and Insulin Resistance. Hepatol. Commun. 2019, 3, 84–98. [Google Scholar] [CrossRef]
- Luu, W.; Zerenturk, E.J.; Kristiana, I.; Bucknall, M.P.; Sharpe, L.J.; Brown, A.J. Signaling regulates activity of DHCR24, the final enzyme in cholesterol synthesis. J. Lipid Res. 2014, 55, 410–420. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, H.; Li, G.; Liu, Y.; Wang, C.; He, D. Exploring the genetic basis of fatty liver development in geese. Sci. Rep. 2020, 10, 14279. [Google Scholar] [CrossRef]
- McWilliams, S.R.; Karasov, W.H. Phenotypic flexibility in digestive system structure and function in migratory birds and its ecological significance. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2001, 128, 579–593. [Google Scholar] [CrossRef]
- Valsecchi, F.; Konrad, C.; Manfredi, G. Role of soluble adenylyl cyclase in mitochondria. Biochim. Biophys. Acta 2014, 1842, 2555–2560. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.-W.; Wei, J.; Liu, X.-J.; Zhang, N.-Y.; Sheng, L. HIIT and MICT attenuate high-fat diet-induced hepatic lipid accumulation and ER stress via the PERK-ATF4-CHOP signaling pathway. J. Physiol. Biochem. 2022, 78, 641–652. [Google Scholar] [CrossRef]
- Xie, Y.; Shi, X.; Sheng, K.; Han, G.; Li, W.; Zhao, Q.; Jiang, B.; Feng, J.; Li, J.; Gu, Y. PI3K/Akt signaling transduction pathway, erythropoiesis and glycolysis in hypoxia (Review). Mol. Med. Rep. 2019, 19, 783–791. [Google Scholar] [CrossRef]
- Taniguchi, C.M.; Kondo, T.; Sajan, M.; Luo, J.; Bronson, R.; Asano, T.; Farese, R.; Cantley, L.C.; Kahn, C.R. Divergent regulation of hepatic glucose and lipid metabolism by phosphoinositide 3-kinase via Akt and PKClambda/zeta. Cell Metab. 2006, 3, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, C.M.; Emanuelli, B.; Kahn, C.R. Critical nodes in signalling pathways: Insights into insulin action. Nat. Rev. Mol. Cell Biol. 2006, 7, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Saltiel, A.R.; Kahn, C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001, 414, 799–806. [Google Scholar] [CrossRef]
- Han, C.; Wei, S.; He, F.; Liu, D.; Wan, H.; Liu, H.; Li, L.; Xu, H.; Du, X.; Xu, F. The Regulation of Lipid Deposition by Insulin in Goose Liver Cells Is Mediated by the PI3K-AKT-mTOR Signaling Pathway. PLoS ONE 2015, 10, e0098759. [Google Scholar] [CrossRef] [PubMed]
- Bhatt-Wessel, B.; Jordan, T.W.; Miller, J.H.; Peng, L. Role of DGAT enzymes in triacylglycerol metabolism. Arch. Biochem. Biophys. 2018, 655, 1–11. [Google Scholar] [CrossRef]
- Yang, C.; Li, Q.; Huang, W.; Lin, Y.; Wang, Y.; Xiang, H.; Zhu, J. Cloning of Goat DGAT1 Gene and Its Regulation on Lipid Deposition in Preadipocytes. Acta Vet. Zootech. Sin. 2022, 53, 76–87. [Google Scholar] [CrossRef]
- Chen, H.C.; Stone, S.J.; Zhou, P.; Buhman, K.K.; Farese, R.V., Jr. Dissociation of obesity and impaired glucose disposal in mice overexpressing acyl coenzyme a:diacylglycerol acyltransferase 1 in white adipose tissue. Diabetes 2002, 51, 3189–3195. [Google Scholar] [CrossRef]


| Sample | Read Numbers (bp) | Clean Reads (bp) | Clean Ratio (%) | Clean Base | GC% | Q20 | Q30 | Mapped Ratio(%) |
|---|---|---|---|---|---|---|---|---|
| Yili goose1 | 40,245,484 | 40,008,006 | 99.41% | 6.00 G | 47.40 | 97.19% | 92.64% | 87.65% |
| Yili goose2 | 40,650,512 | 40,440,622 | 99.48% | 6.07 G | 47.29 | 96.98% | 92.11% | 87.74% |
| Yili goose3 | 43,524,680 | 43,270,170 | 99.42% | 6.49 G | 47.10 | 96.81% | 91.92% | 77.75% |
| Hybrid goose1 | 41,299,504 | 41,006,270 | 99.29% | 6.15 G | 46.91 | 94.10% | 86.81% | 80.06% |
| Hybrid goose2 | 40,032,290 | 39,803,768 | 99.43% | 5.97 G | 48.10 | 97.18% | 92.50% | 87.52% |
| Hybrid goose3 | 49,512,794 | 49,126,426 | 99.22% | 7.37 G | 47.84 | 96.94% | 92.12% | 82.73% |
| Term ID | Description | Q-Value | Gene Number |
|---|---|---|---|
| GO:0006629 | Lipid metabolic process | 0.010901014 | 38 |
| GO:0044255 | Cellular lipid metabolic process | 0.011382412 | 30 |
| GO:0008610 | Lipid biosynthetic process | 0.014621669 | 21 |
| GO:0006631 | Fatty acid metabolic process | 0.0189958 | 14 |
| GO:0016042 | Lipid catabolic process | 0.027932536 | 11 |
| GO:0035336 | Long-chain fatty-acyl-CoA metabolic process | 0.032135698 | 4 |
| GO:0006695 | Cholesterol biosynthetic process | 0.032585568 | 4 |
| GO:0006637 | Acyl-CoA metabolic process | 0.03457691 | 6 |
| GO:0016126 | Sterol biosynthetic process | 0.037132117 | 5 |
| GO:0019216 | Regulation of lipid metabolic process | 0.037300739 | 10 |
| GO:0016125 | Sterol metabolic process | 0.038013044 | 7 |
| Term ID | Description | Q-Value | Gene Number |
|---|---|---|---|
| GO:0019752 | Carboxylic acid metabolic process | 0.015848476 | 23 |
| GO:0043436 | Oxoacid metabolic process | 0.016400085 | 23 |
| GO:0006082 | Organic acid metabolic process | 0.016554431 | 23 |
| GO:0032787 | Monocarboxylic acid metabolic process | 0.019241064 | 17 |
| GO:0005975 | Carbohydrate metabolic process | 0.02169968 | 16 |
| GO:0005996 | Monosaccharide metabolic process | 0.024555726 | 10 |
| GO:0019318 | Hexose metabolic process | 0.029380642 | 9 |
| GO:0006006 | Glucose metabolic process | 0.032569981 | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, H.; Zhang, J.; Li, Y.; Ahmad, H.I.; Li, T.; Liang, Q.; Li, Y.; Yang, M.; Han, J. Liver Transcriptome Profiling Identifies Key Genes Related to Lipid Metabolism in Yili Geese. Animals 2023, 13, 3473. https://doi.org/10.3390/ani13223473
Dong H, Zhang J, Li Y, Ahmad HI, Li T, Liang Q, Li Y, Yang M, Han J. Liver Transcriptome Profiling Identifies Key Genes Related to Lipid Metabolism in Yili Geese. Animals. 2023; 13(22):3473. https://doi.org/10.3390/ani13223473
Chicago/Turabian StyleDong, Huajiao, Jie Zhang, Yingying Li, Hafiz Ishfaq Ahmad, Tiantian Li, Qianqian Liang, Yan Li, Min Yang, and Jilong Han. 2023. "Liver Transcriptome Profiling Identifies Key Genes Related to Lipid Metabolism in Yili Geese" Animals 13, no. 22: 3473. https://doi.org/10.3390/ani13223473
APA StyleDong, H., Zhang, J., Li, Y., Ahmad, H. I., Li, T., Liang, Q., Li, Y., Yang, M., & Han, J. (2023). Liver Transcriptome Profiling Identifies Key Genes Related to Lipid Metabolism in Yili Geese. Animals, 13(22), 3473. https://doi.org/10.3390/ani13223473





