Genome-Wide Association and Pathway Analysis of Carcass and Meat Quality Traits in Karachai Young Goats
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Phenotypic Measurements
2.2. Statistical Analysis
2.3. Genotyping and Quality Control of Data
2.4. Genome-Wide Association Studies
2.5. Gene Analysis
2.6. Development of a Multiplex Test System
2.7. Meat Productivity and Meat Quality Analysis
3. Results
3.1. Population Stratification
3.2. Descriptive Statistics
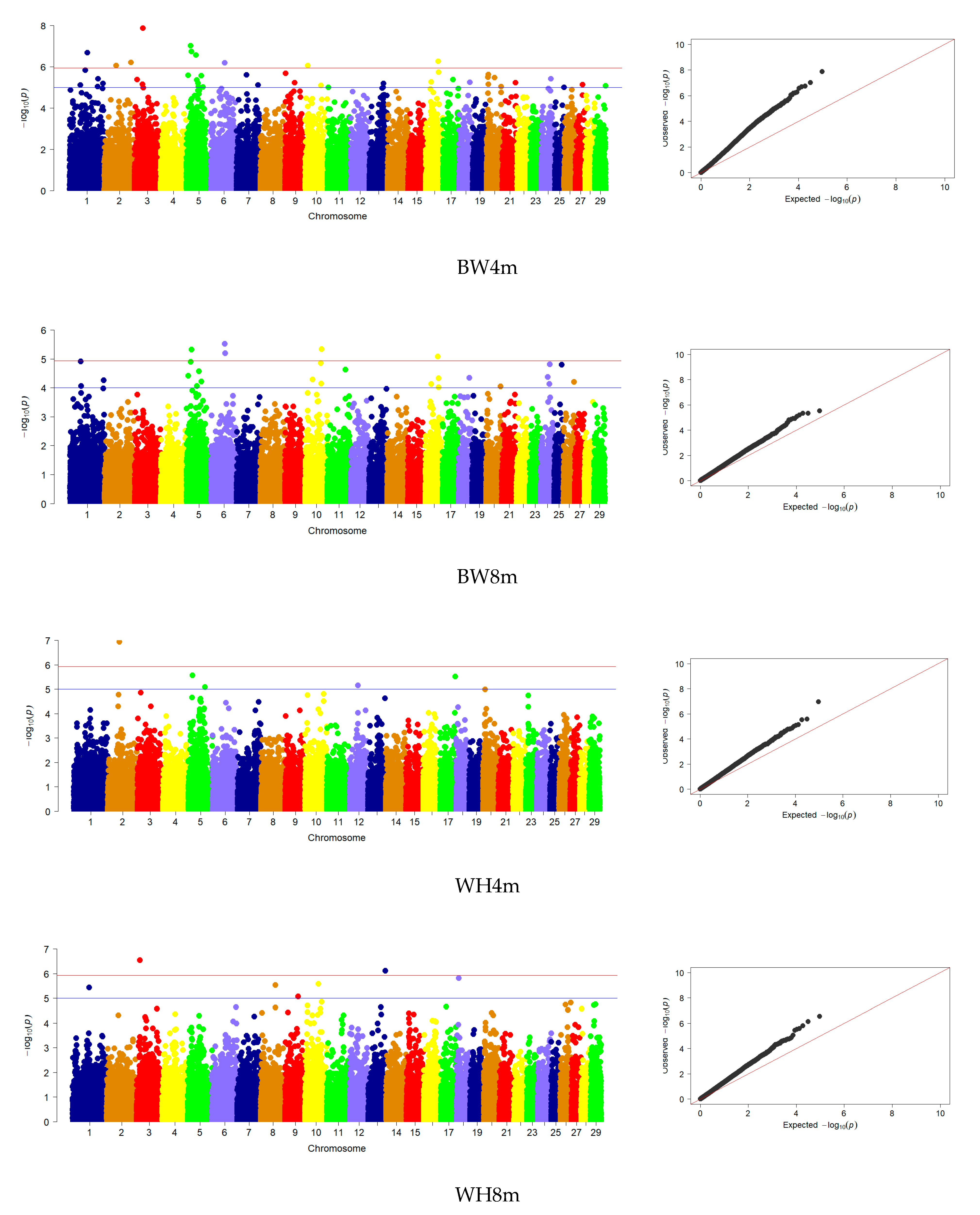
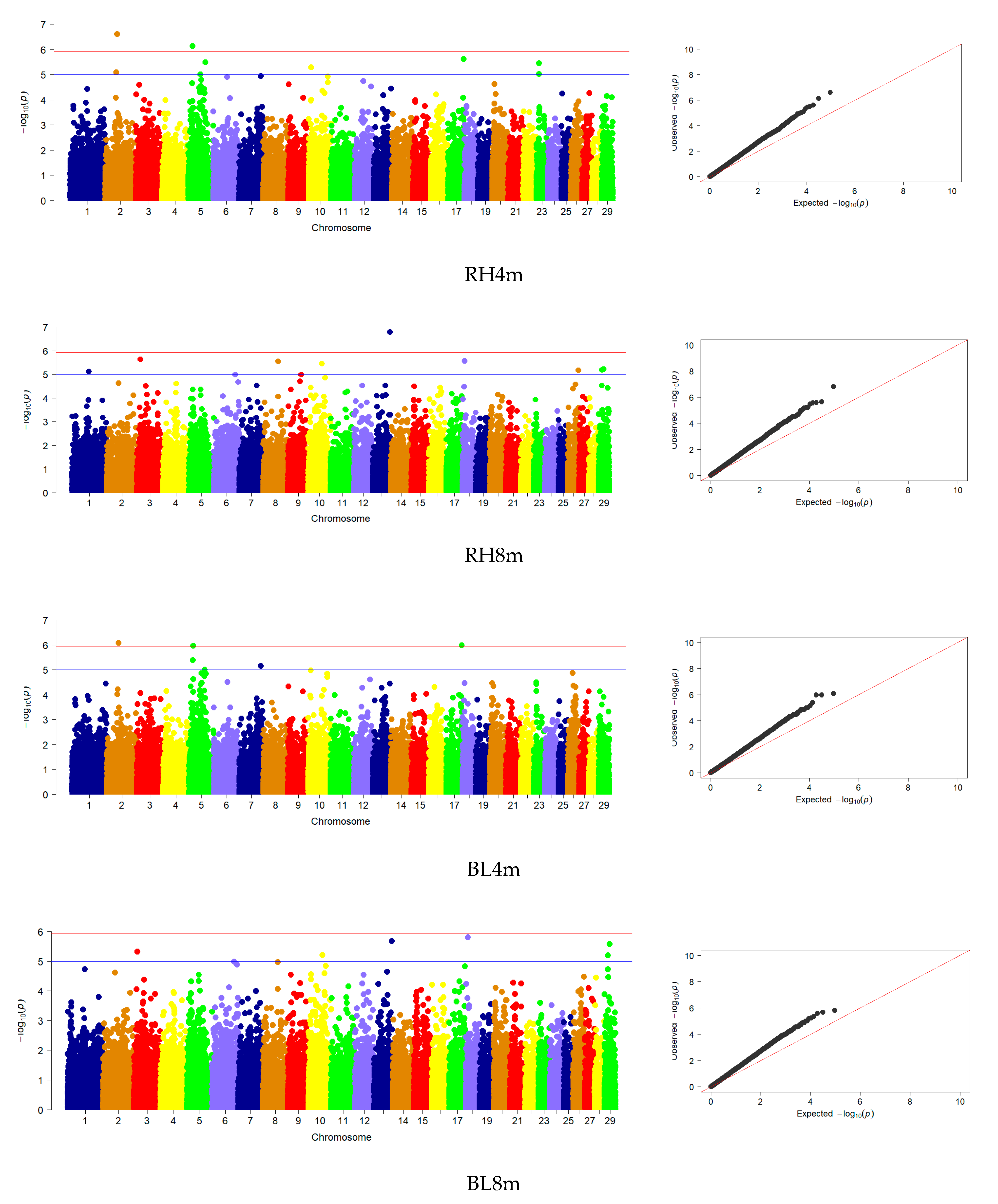
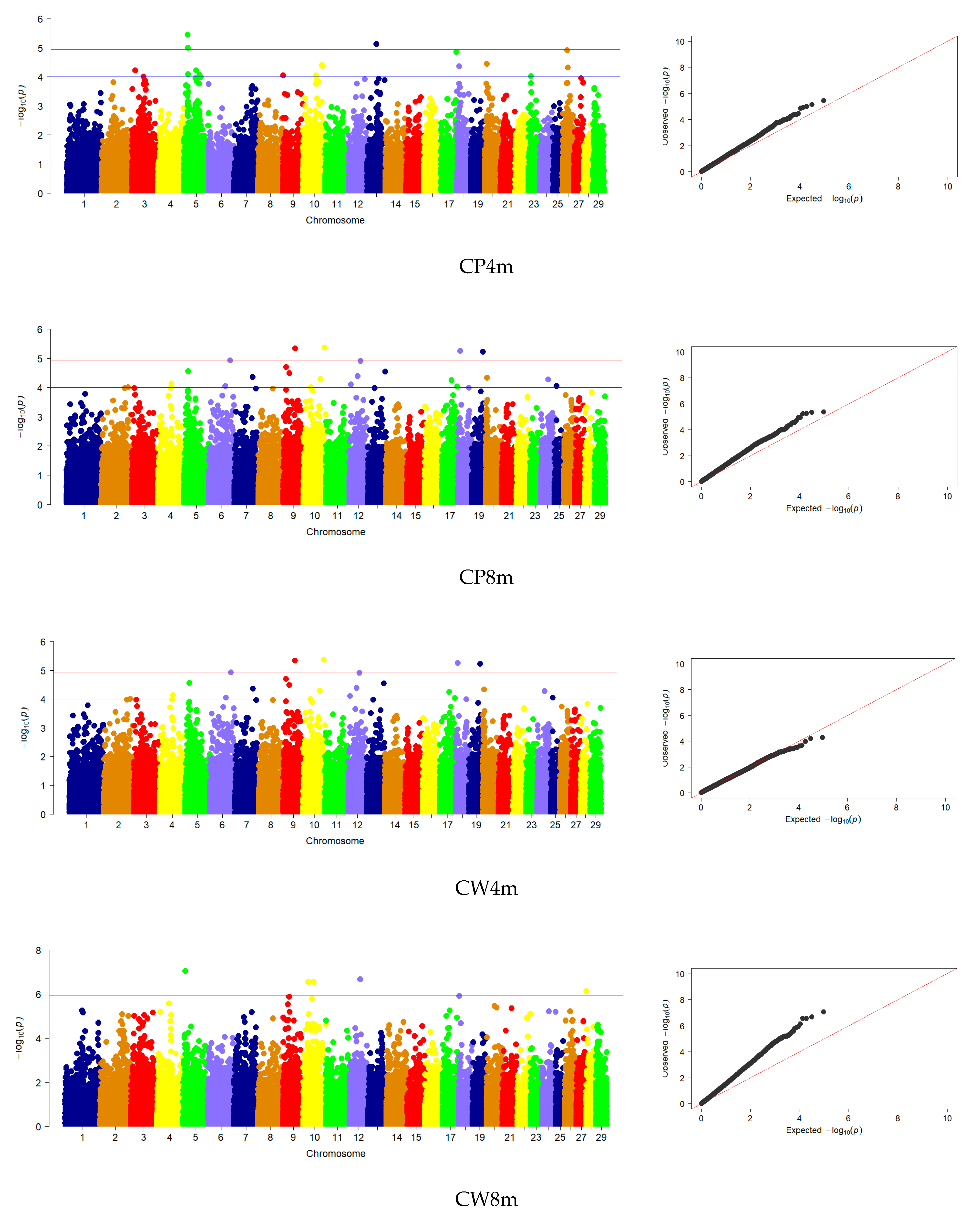
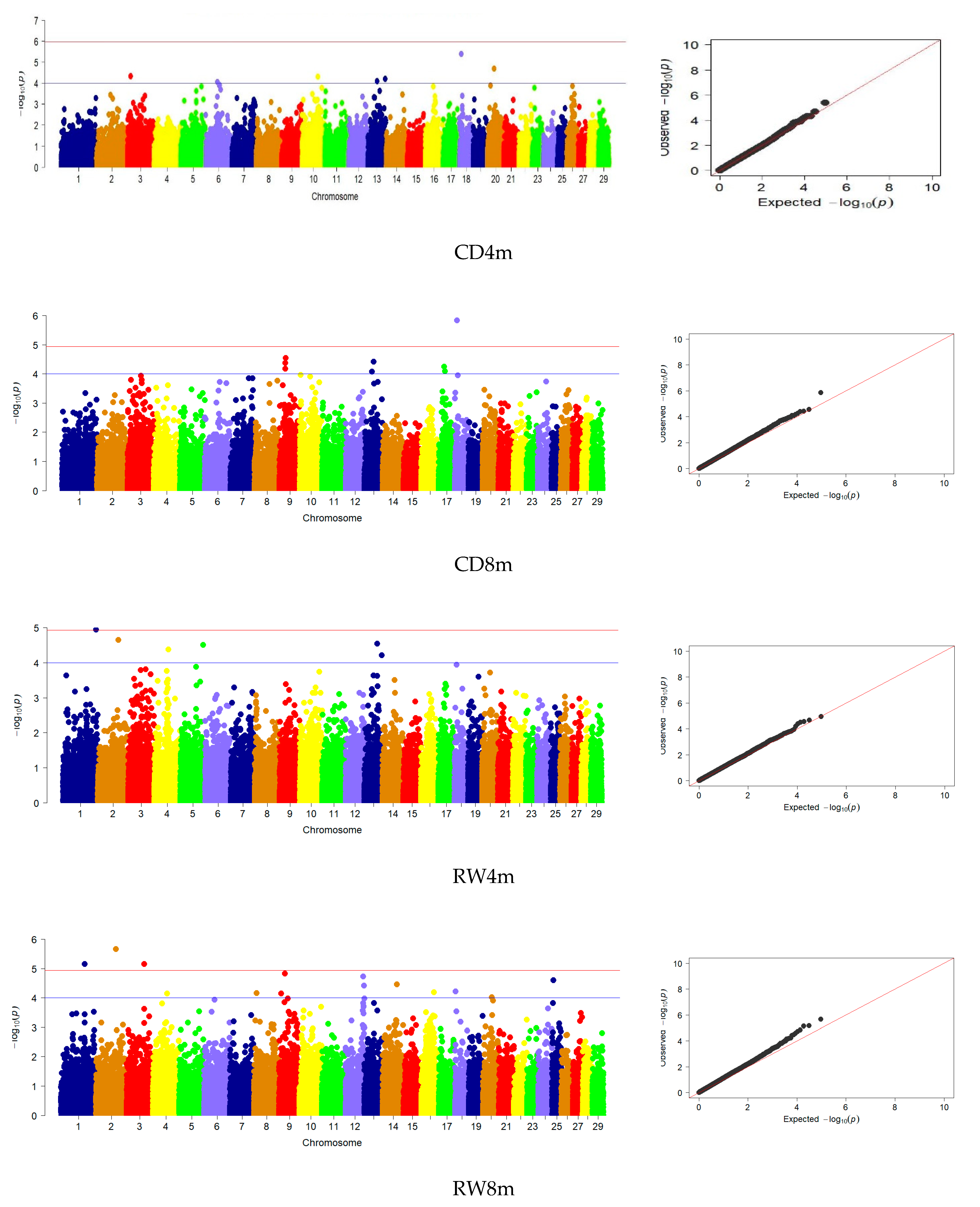
3.3. Genome-Wide Association Studies
3.4. Candidate Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alberto, F.J.; Boyer, F.; Orozco-Terwengel, P.; Streeter, I.; Servin, B.; de Villemereuil, P.; Benjelloun, B.; Librado, P.; Biscarini, F.; Colli, L.; et al. Convergent genomic signatures of domestication in sheep and goats. Nat. Commun. 2018, 9, 813. [Google Scholar] [CrossRef]
- Miller, B.A.; Lu, C.D. Current status of global dairy goat production: An overview. Asian-Australas. J. Anim. Sci. 2019, 32, 1219–1232. [Google Scholar] [CrossRef]
- Pulina, G.; Milán, M.J.; Lavín, M.P.; Theodoridis, A.; Morin, E.; Capote, J.; Thomas, D.L.; Francesconi, A.H.D.; Caja, G. Invited review: Current production trends, farm structures, and economics of the dairy sheep and goat sectors. J. Dairy Sci. 2018, 101, 6715–6729. [Google Scholar] [CrossRef]
- Reshma Nair, M.R.; Sejian, V.; Silpa, M.V.; Fonsêca, V.F.C.; de Melo Costa, C.C.; Devaraj, C.; Krishnan, G.; Bagath, M.; Nameer, P.O.; Bhatta, R. Goat as the ideal climate-resilient animal model in tropical environment: Revisiting advantages over other live-stock species. Int. J. Biometeorol. 2021, 65, 2229–2240. [Google Scholar] [CrossRef] [PubMed]
- Silanikove, N. The physiological basis of adaptation in goats to harsh environments. Small Rumin.Res. 2000, 35, 181–193. [Google Scholar] [CrossRef]
- Mazinani, M.; Rude, B. Population, World Production and Quality of Sheep and Goat Products. Am. J. Anim. Veter-Sci. 2020, 15, 291–299. [Google Scholar] [CrossRef]
- Daly, K.G.; Delser, P.M.; Mullin, V.E.; Scheu, A.; Mattiangeli, V.; Teasdale, M.D.; Hare, A.J.; Burger, J.; Verdugo, M.P.; Collins, M.J.; et al. Ancient goat genomes reveal mosaic domestication in the Fertile Crescent. Science 2018, 361, 85–88. [Google Scholar] [CrossRef]
- Zheng, Z.; Wang, X.; Li, M.; Li, Y.; Yang, Z.; Wang, X.; Pan, X.; Gong, M.; Zhang, Y.; Guo, Y.; et al. The origin of domestication genes in goats. Sci. Adv. 2020, 6, eaaz5216. [Google Scholar] [CrossRef]
- Dong, Y.; Xie, M.; Jiang, Y.; Xiao, N.; Du, X.; Zhang, W.; Tosser-Klopp, G.; Wang, J.; Yang, S.; Liang, J.; et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat. Biotechnol. 2012, 31, 135–141. [Google Scholar] [CrossRef]
- Tosser-Klopp, G.; Bardou, P.; Bouchez, O.; Cabau, C.; Crooijmans, R.; Dong, Y.; Donnadieu-Tonon, C.; Eggen, A.; Heuven, H.C.M.; Jamli, S.; et al. Design and Characterization of a 52K SNP Chip for Goats. PLoS ONE 2014, 9, e86227. [Google Scholar] [CrossRef]
- Selionova, M.I.; Trukhachev, V.I.; Aybazov, A.-M.M.; Stolpovsky, Y.A.; Zinovieva, N.A. Genetic markers of goats (Review). Agric. Biol. 2021, 56, 1031–1048. [Google Scholar] [CrossRef]
- Siddiki, A.M.A.M.Z.; Miah, G.; Islam, S.; Kumkum, M.; Rumi, M.H.; Baten, A.; Hossain, M.A. Goat Genomic Resources: The Search for Genes Associated with Its Economic Traits. Int. J. Genom. 2020, 2020, 5940205. [Google Scholar] [CrossRef]
- Getaneh, M.; Alemayehu, K. Candidate genes associated with economically important traits in dairy goats. Cogent Food Agric. 2022, 8, 2149131. [Google Scholar] [CrossRef]
- Bagatoli, A.; de Melo, A.L.P.; Gasparino, E.; Rodrigues, M.T.; Ferreira, L.; Garcia, O.S.R.; Soares, M.A.M. Association between polymorphisms of APOB, SLC27A6, AGPAT6 and PRLR genes and milk production and quality traits in goat. Small Rumin. Res. 2021, 203, 106484. [Google Scholar] [CrossRef]
- Kang, X.; Li, M.; Liu, M.; Liu, S.; Pan, M.G.; Wiggans, G.R.; Rosen, B.D.; Liu, G.E. Genomics Copy number variation analysis reveals variants associated with milk production traits in dairy goats. Genomics 2020, 112, 4934–4937. [Google Scholar] [CrossRef]
- Scholtens, M.; Jiang, A.; Smith, A.; Littlejohn, M.; Lehnert, K.; Snell, R.; Lopez-Villalobos, N.; Garrick, D.; Blair, H. Genome-wide association studies of lactation yields of milk, fat, protein and somatic cell score in New Zealand dairy goats. J. Anim. Sci. Biotechnol. 2020, 11, 55. [Google Scholar] [CrossRef]
- Wang, X.; Cai, B.; Zhou, J.; Zhu, H.; Niu, Y.; Ma, B.; Yu, H.; Lei, A.; Yan, H.; Shen, Y.; et al. Disruption of FGF5 in cashmere goats using CRISPR/Cas9 results in more secondary hair follicles and longer fibers. PLoS ONE 2016, 11, e0164640. [Google Scholar] [CrossRef]
- Wang, J.; Hao, Z.; Zhou, H.; Luo, Y.; Hu, J.; Liu, X.; Li, S.; Hickford, J.G. A keratin-associated protein (KAP) gene that is associated with variation in cashmere goat fleece weight. Small Rumin. Res. 2018, 167, 104–109. [Google Scholar] [CrossRef]
- Dangar, N.S.; Pandya, G.M.; Ramani, U.V.; Kharadi, V.B.; Brahmkshtri, B.P. Association study of fecundity gene BMP 15 with prolificacy in surti goats under farm and field condition of South Gujarat Region. Ind. J. Vet. Sci. Biotech. 2022, 18, 100–104. [Google Scholar]
- Wang, J.-J.; Li, Z.-D.; Zheng, L.-Q.; Zhang, T.; Shen, W.; Lei, C.-Z. Genome-wide detection of selective signals for fecundity traits in goats (Capra hircus). Gene 2022, 818, 146221. [Google Scholar] [CrossRef]
- Islam, R.; Liu, X.; Gebreselassie, G.; Abied, A.; Ma, Q.; Ma, Y. Genome-wide association analysis reveals the genetic locus for high reproduction trait in Chinese Arbas Cashmere goat. Genes Genom. 2020, 42, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, X.; Qi, T.; Hui, Y.; Yan, H.; Qu, L.; Lan, X.; Pan, C. Whole-genome sequencing to identify candidate genes for litter size and to uncover the variant function in goats (Capra hircus). Genomics 2020, 113, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Jiang, J.; Wang, G.; Zhou, P.; Li, J.; Chen, C.; Liu, L.; Li, N.; Xia, Y.; Ren, H. Genome-wide association analysis of nine reproduction and morphological traits in three goat breeds from Southern China. Anim. Biosci. 2023, 36, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Yao, N.; Yang, M.; Liu, X.; Dong, K.; Zhao, Q.; Pu, Y.; He, X.; Guan, W.; Yang, N.; et al. Exome sequencing reveals genetic differentiation due to high-altitude adaptation in the Tibetan cashmere goat (Capra hircus). BMC Genom. 2016, 17, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, C.; Guo, Y.; She, S.; Wang, B.; Jiang, Y.; Bai, Y.; Song, X.; Li, L.; Shi, L.; et al. Screening of Deletion Variants within the Goat PRDM6 Gene and Its Effects on Growth Traits. Animals 2020, 10, 208. [Google Scholar] [CrossRef]
- Gu, B.; Sun, R.; Fang, X.; Zhang, J.; Zhao, Z.; Huang, D.; Zhao, Y.; Zhao, Y. Genome-Wide Association Study of Body Conformation Traits by Whole Genome Sequencing in Dazu Black Goats. Animals 2022, 12, 548. [Google Scholar] [CrossRef]
- Wang, K.; Cui, Y.; Wang, Z.; Yan, H.; Meng, Z.; Zhu, H.; Qu, L.; Lan, X.; Pan, C. One 16 bp insertion/deletion (indel) within the KDM6A gene revealing strong associations with growth traits in goat. Gene 2018, 686, 16–20. [Google Scholar] [CrossRef]
- Bai, Y.; Yuan, R.; Luo, Y.; Kang, Z.; Zhu, H.; Qu, L.; Lan, X.; Song, X. Exploration of Genetic Variants within the Goat A-Kinase Anchoring Protein 12 (AKAP12) Gene and Their Effects on Growth Traits. Animals 2021, 11, 2090. [Google Scholar] [CrossRef]
- Bi, Y.; Chen, Y.; Xin, D.; Liu, T.; He, L.; Kang, Y.; Pan, C.; Shen, W.; Lan, X.; Liu, M. Effect of indel variants within the sorting nexin 29 (SNX29) gene on growth traits of goats. Anim. Biotechnol. 2020, 33, 914–919. [Google Scholar] [CrossRef]
- Moaeen-Ud-Din, M.; Muner, R.D.; Khan, M.S. Genome wide association study identifies novel candidate genes for growth and body conformation traits in goats. Sci. Rep. 2022, 12, 9891. [Google Scholar] [CrossRef]
- Luigi-Sierra, M.G.; Landi, V.; Guan, D.; Delgado, J.V.; Castelló, A.; Cabrera, B.; Mármol-Sánchez, E.; Alvarez, J.F.; Gómez-Carpio, M.; Martínez, A.; et al. A genome-wide association analysis for body, udder, and leg conformation traits recorded in Murciano-Granadina goats. J. Dairy Sci. 2020, 103, 11605–11617. [Google Scholar] [CrossRef] [PubMed]
- Moaeen-ud-Din, M.; Khan, M.S.; Muner, R.D.; Reecy, J.M. Genome Wide Association Study in Goat Identified Novel SNPs and Genes for Growth. 2023. Available online: https://doi.org/10.21203/rs.3.rs-2966814/v1 (accessed on 9 June 2022).
- Liu, X.; Ma, L.; Wang, M.; Wang, K.; Li, J.; Yan, H.; Zhu, H.; Lan, X. Two indel variants of prolactin receptor (PRLR) gene are associated with growth traits in goat. Anim. Biotechnol. 2019, 31, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, X.; Zhang, Z.; An, Q.; Wen, Y.; Wang, D.; Liu, X.; Li, Z.; Lyu, S.; Li, L.; et al. Copy number variation of CADM2 gene revealed its association with growth traits across Chinese Capra hircus (goat) populations. Gene 2020, 741, 144519. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.-Y.; Li, L.-J.; Zhang, Z.-J.; Wang, E.-Y.; Wang, J.; Xu, J.-W.; Liu, H.-B.; Wen, Y.-F.; He, H.; Lei, C.-Z.; et al. Copy number variation of MYLK4 gene and its growth traits of Capra hircus (goat). Anim. Biotechnol. 2019, 31, 532–537. [Google Scholar] [CrossRef]
- Pardo, J.I.S.; Bermejo, J.V.D.; Ariza, A.G.; Jurado, J.M.L.; Navas, C.M.; Pastrana, C.I.; Martínez, M.d.A.M.; González, F.J.N. Candidate Genes and Their Expressions Involved in the Regulation of Milk and Meat Production and Quality in Goats (Capra hircus). Animals 2022, 12, 988. [Google Scholar] [CrossRef]
- Erokhin, A.; Russian State Agrarian University—Moscow Timiryazev Agricultural Academy; Karasev, E.; Erokhin, S. Dynamics of goat population and production of goat milk and meat in the world and in Russia. Sheep Goats Woolen Bus. 2020, 4, 22–25. (In Russian) [Google Scholar] [CrossRef]
- Aybazov, M.M.; Selionova, M.I.; Mamontova, T.V. Exterior and some biological indices of Karachai goats. Zootechniya 2019, 12, 5–9. (In Russian) [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- PLINK 1.9 and 2.0. Available online: https://zzz.bwh.harvard.edu/plink/plink2.shtml (accessed on 5 June 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2012; Available online: http://www.R-project.org (accessed on 9 June 2022).
- Kinsella, R.J.; Kähäri, A.; Haider, S.; Zamora, J.; Proctor, G.; Spudich, G.; Almeida-King, J.; Staines, D.; Derwent, P.; Kerhornou, A.; et al. Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database 2011, 2011, bar030. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Holland, P.M.; Abramson, R.D.; Watson, R.; Gelfand, D.H. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 1991, 88, 7276–7280. [Google Scholar] [CrossRef] [PubMed]
- The European Parliament and the Council of the European Union. Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the Protection of Animals Used for Scientific Purposes. Off. J. Eur. Union 2010, 276, 33–79. Available online: https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2010:276:0033:0079:EN:PDF (accessed on 2 February 2020).
- European Convention for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes. European Treaty Series—No. 123. Available online: https://rm.coe.int/168007a67b (accessed on 10 October 2022).
- GOST 9793-2016; Meat and Meat Products. Method for Determination of Moisture Content. Standartinform: Moscow, Russia, 2018. Available online: https://docs.cntd.ru/document/1200144231 (accessed on 16 September 2022). (In Russian)
- GOST 23042–2015; Meat and Meat Products. Methods of Fat Determination. Standartinform: Moscow, Russia, 2019. Available online: https://docs.cntd.ru/document/1200133107 (accessed on 16 September 2022). (In Russian)
- GOST 25011–2017; Meat and Meat Products. Protein Determination Methods. Standartinform: Moscow, Russia, 2018. Available online: https://docs.cntd.ru/document/1200146783 (accessed on 16 September 2022). (In Russian)
- Selionova, M.I.; Mamontova, T.V.; Aybazov, M.M.; Petrovic, V.C.; Petrovic, M.P. Quality of Aborigenous Karachay Goat Meat Under Different Conditions. In Proceedings of the 13th International Symposium Modern Trends in Livestock Production, Belgrade, Serbia, 6–8 October 2021; pp. 117–125. [Google Scholar]
- Saleh, A.A.; Rashad, A.M.; Hassanine, N.N.; Sharaby, M.A. Candidate genes and signature of selection associated with different biological aspects and general characteristics of goat. Emerg. Anim. Species 2022, 5, 100013. [Google Scholar] [CrossRef]
- Tilahun, Y.; Gipson, T.A.; Alexander, T.; McCallum, M.L.; Hoyt, P.R. Genome-Wide Association Study towards Genomic Predictive Power for High Production and Quality of Milk in American Alpine Goats. Int. J. Genom. 2020, 2020, 6035694. [Google Scholar] [CrossRef] [PubMed]
- Talouarn, E.; The VarGoats Consortium; Bardou, P.; Palhière, I.; Oget, C.; Clément, V.; Tosser-Klopp, G.; Rupp, R.; Robert-Granié, C. Genome wide association analysis on semen volume and milk yield using different strategies of imputation to whole genome sequence in French dairy goats. BMC Genet. 2020, 21, 19. [Google Scholar] [CrossRef] [PubMed]
- Saif, R.; Mahmood, T.; Ejaz, A.; Fazlani, S.A.; Zia, S. Whole-genome selective sweeps analysis in Pakistani Kamori goat. Gene Rep. 2021, 26, 101429. [Google Scholar] [CrossRef]
- Mrode, R.; Tarekegn, G.M.; Mwacharo, J.M.; Djikeng, A. Invited review: Genomic selection for small ruminants in developed countries: How applicable for the rest of the world? Animal 2018, 12, 1333–1340. [Google Scholar] [CrossRef]
- Saravanan, K.; Panigrahi, M.; Kumar, H.; Nayak, S.S.; Rajawat, D.; Bhushan, B.; Dutt, T. Progress and future perspectives of livestock genomics in India: A mini review. Anim. Biotechnol. 2022, 34, 1979–1987. [Google Scholar] [CrossRef]
- Lu, Z.; Yue, Y.; Yuan, C.; Liu, J.; Chen, Z.; Niu, C.; Sun, X.; Zhu, S.; Zhao, H.; Guo, T.; et al. Genome-Wide Association Study of Body Weight Traits in Chinese Fine-Wool Sheep. Animals 2020, 10, 170. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, J.; Zhao, F.; Ren, H.; Xu, L.; Lu, J.; Zhang, S.; Zhang, X.; Wei, C.; Lu, G.; et al. Genome-Wide Association Studies for Growth and Meat Production Traits in Sheep. PLoS ONE 2013, 8, e66569. [Google Scholar] [CrossRef]
- Mancin, E.; Tuliozi, B.; Pegolo, S.; Sartori, C.; Mantovani, R. Genome Wide Association Study of Beef Traits in Local Alpine Breed Reveals the Diversity of the Pathways Involved and the Role of Time Stratification. Front. Genet. 2022, 12, 746665. [Google Scholar] [CrossRef]
- Wong, M.L.; Islas-Trejo, A.; Medrano, J.F. Structural characterization of the mouse high growth deletion and discovery of a novel fusion transcript between suppressor of cytokine signaling-2 (Socs-2) and viral encoded semaphorin receptor (Plexin C1). Gene 2002, 299, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.M.; Pita, R.H.; Malek, M.; Lopes, P.S.; Guimarães, S.E.F.; Rothschild, M.F. Analysis of the mouse high-growth region in pigs. J. Anim. Breed. Genet. 2009, 126, 404–412. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, X.; Benson, K.F.; Ashar, H.R.; Chada, K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 1995, 376, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.O.; Li, J.; Davis, B.W.; Upadhyay, S.; Al Muhisen, H.M.; Suva, L.J.; Clement, T.M.; Andersson, L. Hmga2 deficiency is associated with allometric growth retardation, infertility, and behavioral abnormalities in mice. G3 Genes|Genomes|Genet. 2021, 12, jkab417. [Google Scholar] [CrossRef] [PubMed]
- Xiaokai, L.; Quankui, H.; Lingki, F.; Yafeng, G.; Jing, L.; Ganqiu, L. Sequence and expression differences of BMP2 and, FGFR3 genes in Guangxi Bama mini pig and Landrace pig. Guangxi Agric. Sci. 2021, 52, 1709–1718. [Google Scholar]
- Quan, J.; Ding, R.; Wang, X.; Yang, M.; Yang, Y.; Zheng, E.; Gu, T.; Cai, G.; Wu, Z.; Liu, D.; et al. Genome-wide association study reveals genetic loci and candidate genes for average daily gain in Duroc pigs. Asian-Australas. J. Anim. Sci. 2018, 31, 480–488. [Google Scholar] [CrossRef]
- Pan, B.; Long, H.; Yuan, Y.; Zhang, H.; Peng, Y.; Zhou, D.; Liu, C.; Xiang, B.; Huang, Y.; Zhao, Y.; et al. Identification of Body Size Determination Related Candidate Genes in Domestic Pig Using Genome-Wide Selection Signal Analysis. Animals 2022, 12, 1839. [Google Scholar] [CrossRef]
- Rimbault, M.; Beale, H.C.; Schoenebeck, J.J.; Hoopes, B.C.; Allen, J.J.; Kilroy-Glynn, P.; Wayne, R.K.; Sutter, N.B.; Ostrander, E.A. Derived variants at six genes explain nearly half of size reduction in dog breeds. Genome Res. 2013, 23, 1985–1995. [Google Scholar] [CrossRef]
- Kwak, G.-H.; Kim, T.-H.; Kim, H.-Y. Down-regulation of MsrB3 induces cancer cell apoptosis through reactive oxygen species production and intrinsic mitochondrial pathway activation. Biochem. Biophys. Res. Commun. 2017, 483, 468–474. [Google Scholar] [CrossRef]
- Wu, M.; Li, S.; Zhang, G.; Fan, Y.; Gao, Y.; Huang, Y.; Lan, X.; Lei, C.; Ma, Y.; Dang, R. Exploring insertions and deletions (indels) of MSRB3 gene and their association with growth traits in four Chinese indigenous cattle breeds. Arch. Anim. Breed. 2019, 62, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gu, J.; Miyoshi, E.; Honke, K.; Taniguchi, N. Phenotype Changes of Fut8 Knockout Mouse: Core Fucosylation Is Crucial for the Function of Growth Factor Receptor(s). Methods Enzymol. 2006, 417, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Blaževitš, O.; Bolshette, N.; Vecchio, D.; Guijarro, A.; Croci, O.; Campaner, S.; Grimaldi, B. MYC-Associated Factor MAX is a Regulator of the Circadian Clock. Int. J. Mol. Sci. 2020, 21, 2294. [Google Scholar] [CrossRef] [PubMed]
- Silpa, M.; Naicy, T.; Aravindakshan, T.; Radhika, G.; Venkatachalapathy, R.; Kurian, E. Sirtuin3 gene tissue expression profiling, SNP detection and its association with body conformation traits in goats. Small Rumin. Res. 2019, 184, 106017. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Y.; Qin, Y.; Cai, W.; Zhang, X.; Xu, Y.; Dou, X.; Wang, Z.; Han, D.; Wang, J.; et al. Association analysis of single-nucleotide polymorphism in prolactin and its receptor with productive and body conformation traits in Liaoning cashmere goats. Arch. Anim. Breed. 2022, 65, 145–155. [Google Scholar] [CrossRef]
- Berihulay, H.; Li, Y.; Gebrekidan, B.; Gebreselassie, G.; Liu, X.; Jiang, L.; Ma, Y. Whole genome resequencing reveals selection signatures associated with important traits in Ethiopian indigenous goat populations. Front. Genet. 2019, 10, 1190. [Google Scholar] [CrossRef]


| Trait | Max | Min | Mean | Var | Std. Dev | CV % |
|---|---|---|---|---|---|---|
| Four months | ||||||
| BW | 36.5 kg | 19.1 kg | 24.75 kg | 15.58 | 3.94 | 14.54 |
| WH | 53.0 cm | 45.5 cm | 48.97 cm | 3.25 | 1.80 | 3.72 |
| RH | 54.5 cm | 46.5 cm | 49.85 cm | 3.49 | 1.87 | 3.81 |
| BL | 56.0 cm | 47.5 cm | 51.00 cm | 3.55 | 1.89 | 3.70 |
| CP | 57.5 cm | 49.5 cm | 53.12 cm | 2.71 | 1.65 | 3.13 |
| CW | 12.0 cm | 7.5 cm | 8.98 cm | 0.57 | 0.75 | 8.44 |
| CD | 23.5 cm | 17.5 cm | 19.78 cm | 1.53 | 1.23 | 6.35 |
| RW | 12.0 cm | 9.5 cm | 10,28 cm | 0.20 | 0.45 | 4.42 |
| Eight months | ||||||
| BW | 49.8 kg | 27.1 kg | 36.35 kg | 14.8 | 4.21 | 11.59 |
| WH | 60.5 cm | 47.5 cm | 52.78 cm | 8.11 | 2.84 | 5.38 |
| RH | 60.5 cm | 48.0 cm | 53.48 cm | 8.95 | 2.99 | 5.59 |
| BL | 61.0 cm | 48.1 cm | 54.12 cm | 9.15 | 3.02 | 5.58 |
| CP | 69.0 cm | 52.2 cm | 60.60 cm | 10.96 | 3.31 | 5.46 |
| CW | 14.0 cm | 8.0 cm | 10.21 cm | 1.64 | 1.28 | 12.55 |
| CD | 26.5 cm | 18.0 cm | 21.54 cm | 2.91 | 1.71 | 7.95 |
| RW | 13.9 cm | 9.5 cm | 11.30 cm | 0.67 | 0.82 | 6.86 |
| Trait | 8 Months | 4 Months | ||
|---|---|---|---|---|
| n | Chr | n | Chr | |
| BW | 5 | 5, 6, 10, 16 | 33 | 1, 2, 3, 5, 6, 7, 9, 10, 13, 16, 17, 20, 24, 26 |
| WH | 10 | 1, 3, 8, 9, 10, 13, 18 | 2 | 2, 20 |
| RH | 9 | 1, 3, 8, 10, 13, 18, 26, 29 | 4 | 2, 5, 20, 23 |
| BL | 6 | 3, 10, 13, 18, 29 | 2 | 2, 5 |
| CP | 4 | 9, 10, 18, 19 | - | - |
| CW | 30 | 1, 2, 3, 4, 5, 7, 9, 10, 12, 17, 18, 20, 21, 24, 26, 28 | - | - |
| CD | 8 | 9, 13, 17, 18 | 1 | 18 |
| RW | 14 | 1, 2, 3, 4, 8, 9, 12, 14, 16, 18, 20, 25 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Selionova, M.; Aibazov, M.; Sermyagin, A.; Belous, A.; Deniskova, T.; Mamontova, T.; Zharkova, E.; Zinovieva, N. Genome-Wide Association and Pathway Analysis of Carcass and Meat Quality Traits in Karachai Young Goats. Animals 2023, 13, 3237. https://doi.org/10.3390/ani13203237
Selionova M, Aibazov M, Sermyagin A, Belous A, Deniskova T, Mamontova T, Zharkova E, Zinovieva N. Genome-Wide Association and Pathway Analysis of Carcass and Meat Quality Traits in Karachai Young Goats. Animals. 2023; 13(20):3237. https://doi.org/10.3390/ani13203237
Chicago/Turabian StyleSelionova, Marina, Magomet Aibazov, Alexander Sermyagin, Anna Belous, Tatiana Deniskova, Tatiana Mamontova, Ekaterina Zharkova, and Natalia Zinovieva. 2023. "Genome-Wide Association and Pathway Analysis of Carcass and Meat Quality Traits in Karachai Young Goats" Animals 13, no. 20: 3237. https://doi.org/10.3390/ani13203237
APA StyleSelionova, M., Aibazov, M., Sermyagin, A., Belous, A., Deniskova, T., Mamontova, T., Zharkova, E., & Zinovieva, N. (2023). Genome-Wide Association and Pathway Analysis of Carcass and Meat Quality Traits in Karachai Young Goats. Animals, 13(20), 3237. https://doi.org/10.3390/ani13203237










