Simple Summary
In this study, we conducted a comprehensive investigation of the fatty acid composition in Ningxiang pigs using a genome-wide association study. Our findings revealed a combination of previously reported and novel candidate genes associated with saturated fatty acids (SFAs), monounsaturated fatty acids (MUFAs), and polyunsaturated fatty acids (PUFAs). Notably, we identified significant single-nucleotide polymorphisms (SNPs) that are closely linked to specific fatty acids, and some of these genes explained substantial phenotypic variance. These noteworthy discoveries have the potential to significantly improve meat quality and fat deposition in Ningxiang pigs through targeted breeding approaches. Our research provides valuable insights into the intricate composition of fatty acids, thus offering practical implications for elevating meat quality and ultimately benefiting both the pig industry and consumers. The significance of this study is underscored by its potential to drive positive changes in society by promoting healthier and superior-quality pork products.
Abstract
Ningxiang pigs exhibit a diverse array of fatty acids, making them an intriguing model for exploring the genetic underpinnings of fatty acid metabolism. We conducted a genome-wide association study using a dataset comprising 50,697 single-nucleotide polymorphisms (SNPs) and samples from over 600 Ningxiang pigs. Our investigation yielded novel candidate genes linked to five saturated fatty acids (SFAs), four monounsaturated fatty acids (MUFAs), and five polyunsaturated fatty acids (PUFAs). Significant associations with SFAs, MUFAs, and PUFAs were found for 37, 21, and 16 SNPs, respectively. Notably, some SNPs have significant PVE, such as ALGA0047587, which can explain 89.85% variation in Arachidic acid (C20:0); H3GA0046208 and DRGA0016063 can explain a total of 76.76% variation in Elaidic Acid (C18:1n-9(t)), and the significant SNP ALGA0031262 of Arachidonic acid (C20:4n-6) can explain 31.76% of the variation. Several significant SNPs were positioned proximally to previously reported genes. In total, we identified 11 candidate genes (hnRNPU, CEPT1, ATP1B1, DPT, DKK1, PRKG1, EXT2, MEF2C, IL17RA, ITGA1 and ALOX5), six candidate genes (ALOX5AP, MEDAG, ISL1, RXRB, CRY1, and CDKAL1), and five candidate genes (NDUFA4L2, SLC16A7, OTUB1, EIF4E and ROBO2) associated with SFAs, MUFAs, and PUFAs, respectively. These findings hold great promise for advancing breeding strategies aimed at optimizing meat quality and enhancing lipid metabolism within the intramuscular fat (IMF) of Ningxiang pigs.
1. Introduction
Ningxiang is a famous breed of pig in China, with a history spanning more than 1000 years. As an obese breed, Ningxiang is superior to lean meat breeds in terms of intramuscular fat (IMF). Its market weight is approximately 74 kg, and the carcass slaughter rate is approximately 74 percent [1]. Ningxiang pork products are popular due to their unique flavor and nutritional value. Research has shown that the flavor and nutritional value of meat are closely related to the IMF content and fatty acid composition [2,3,4].
Fatty acids play a crucial role in the flavor of pork as fat-soluble flavor precursors [5,6]. Oleic acid (C18:1) is a major monounsaturated fatty acid (MUFA) in the fatty acid composition and is the most abundant, accounting for approximately 40% of the total fatty acid content. C18:1 in beef fat provides the meat with good tenderness, flavor, and antioxidant capacity [7,8]. Moreover, the type and content of fatty acids in meat diets can impact human health. For example, linoleic acid aids in slowing the progression of atherosclerosis [9]. Furthermore, the ratios of n-3, n-6, and other polyunsaturated fatty acids (PUFAs) are closely related to human diseases, such as cardiovascular disease and depression, as well as growth and development [10,11,12,13]. In summary, the fatty acid profile of pork is a crucial evaluation criterion for meat quality.
Genome-wide association studies (GWAS) can efficiently and accurately identify candidate genes related to target traits using single nucleotide polymorphisms (SNPs) as genetic molecular markers. In recent years, researchers have discovered numerous candidate genes associated with porcine IMF deposition and fatty acid composition through GWAS. A GWAS of the IMF in the Italian White breed by Davoli et al. revealed seven new SNPs, and three new candidate genes were annotated. These genes were not related to the IMF content [14]. Van et al. [15] used data from the Axiom pig 660K array to conduct a GWAS on the IMF of 454 Duroc and 659 Landrace boars and identified two quantitative trait locus (QTL) regions for newly synthesized fatty acid traits on SSC4 and SSC14 in Duroc pigs. Viterbo et al. [16] performed a GWAS on the fatty acid composition of 480 purebred Duroc pigs, identifying 25, 29, and 16 SNP loci significantly associated with stearic acid, oleic acid, and saturated fatty acids (SFAs), respectively. The genetic factors contributing to the fatty acid composition of Ningxiang pigs remain unknown. This study aimed to examine significant candidate genes for specific fatty acid compositions and explore potential biological pathways by conducting a GWAS on the fatty acid composition of Ningxiang pigs.
2. Materials and Methods
2.1. Animal Harvest and Sample Collection
The feeding and dietary conditions of all Ningxiang pigs were the same, and samples were collected in two batches (July 2019 and August 2020). Longissimus dorsi samples (taken from the 12th ribs) were collected from 691 Ningxiang pigs that were slaughtered at a predetermined age (180 ± 5 days age) at the Ningxiang Chu Weixiang Slaughterhouse and Meat Processing, LLC (Ningxiang City, Hunan Province, China). A sample of approximately 2 × 1 × 1 cubic centimeters was quickly taken and stored in a self-sealing bag with dry ice for IMF measurement. Additionally, the samples used for DNA extraction were simultaneously stored in a liquid nitrogen tank.
2.2. Determining Intramuscular Fat and Fatty Acids
In this study, we used Soxhlet extraction to measure the IMF contents of the 691 longissimus dorsi samples according to the standard “Meat and Meat Products-Determination of Free Fat Content” (GB/T 9695.1-2008) [17]. The composition of fatty acids was determined via gas chromatography (GC), with the specific procedure detailed below. First, 0.5 g of sample powder was accurately weighed, dried to a constant weight and placed in a 10 mL centrifuge tube. Next, 2 mL of a benzene and petroleum ether mixture (1:1 by volume) was added, and the tube was wrapped in tin foil and left in a dark place for 24 h. Then, 2 mL of a 0.4 mol/L KOH methanol solution was added for methylation. The sample was shaken well and left to stand for 15 min. Double-distilled water was added to a volume of 10 mL, and the sample was centrifuged at 10,000 rpm for 10 min to obtain 100 µL of supernatant. The supernatant was finally diluted with hexane. A gas chromatograph (Agilent 7890A, Santa Clara, CA, USA) was used to determine the content of medium- to long-chain fatty acids. The GC analysis conditions were as follows: the chromatographic column was an SP-2560 (100 m × 0.25 mm, 0.20 µm) capillary column, and high-purity nitrogen was used as the carrier gas. The heating program was as follows: initial temperature of 140 °C maintained for 5 min, then increased to 240 °C at 4 °C/min; sample inlet temperature of 260 °C; flame ionization detector (FID) temperature of 260 °C; split ratio of 100:1; and injection volume of 1 µL. A total of 25 fatty acids were detected (Table S1), and only those present in more than 80% of individuals (N ≥ 533) were retained (Table 1).

Table 1.
Detailed information on 14 fatty acids present in the IMF of the longissimus dorsi of Ningxiang pigs.
2.3. DNA Extraction, Genotyping and Quality Control
2.3.1. DNA Extraction and Genotyping
Genomic DNA was extracted from muscle tissue using the standard phenol-chloroform method, and the DNA was dissolved in TE buffer. The concentration and purity of the DNA samples were measured using a Nanodrop One spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Samples with an A260/280 ratio of 1.7~2.0 were genotyped using the GeneSeek Genomic Profiling (GGP) version 2 Porcine 50K SNP chip (Neogen Corporation, Lincoln, NE, USA), which comprises 50,697 SNP loci.
2.3.2. Quality Control and Genotype Imputation
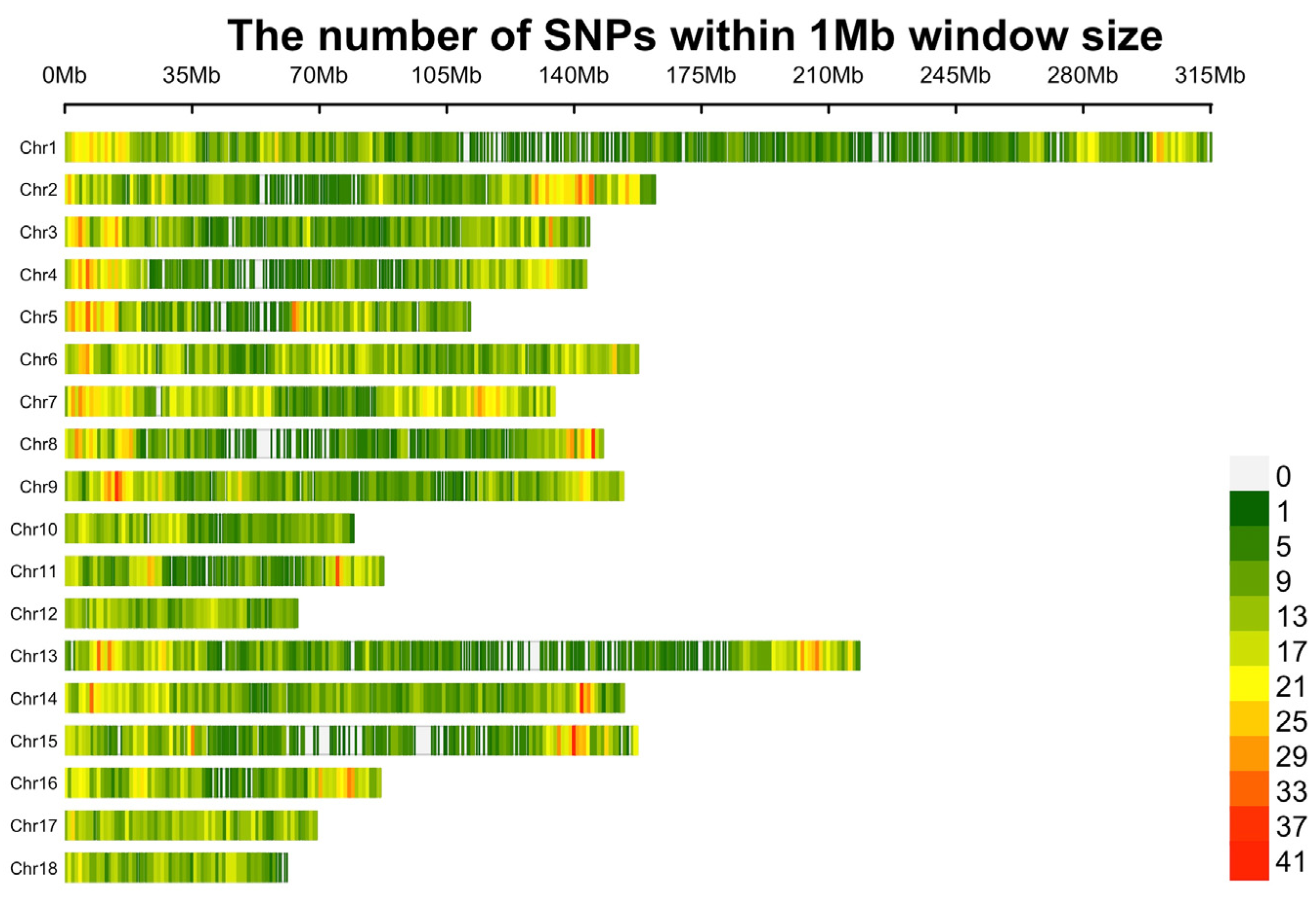
First, SNPs located on the X and Y chromosomes and unknown or duplicate locations were removed. To decrease the missing genotype rate, Beagle 5.1 software [18] was employed to impute the remaining 38,817 SNPs. Then, quality control was conducted using PLINK v1.9 [19] with the following criteria: (1) SNP call rate ≥ 90%; (2) minor allele frequency (MAF) ≥ 1%; and (3) Hardy–Weinberg equilibrium (HWE) testing p value ≥ 10−6. After quality control, 12,201 SNPs were removed due to missing genotype rates, MAFs, and HWE. Finally, 691 individuals and 25,809 SNPs remained for subsequent analysis (Figure 1).
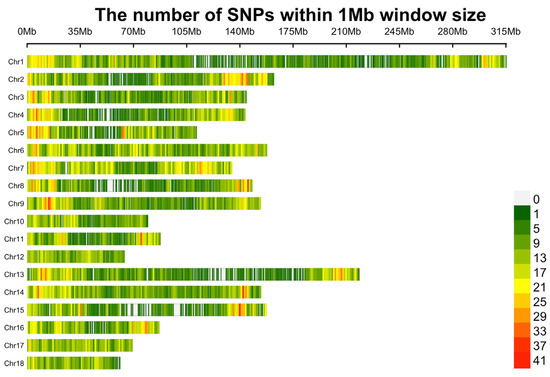
Figure 1.
Distribution plot of SNPs after quality control.
2.4. Population Stratification
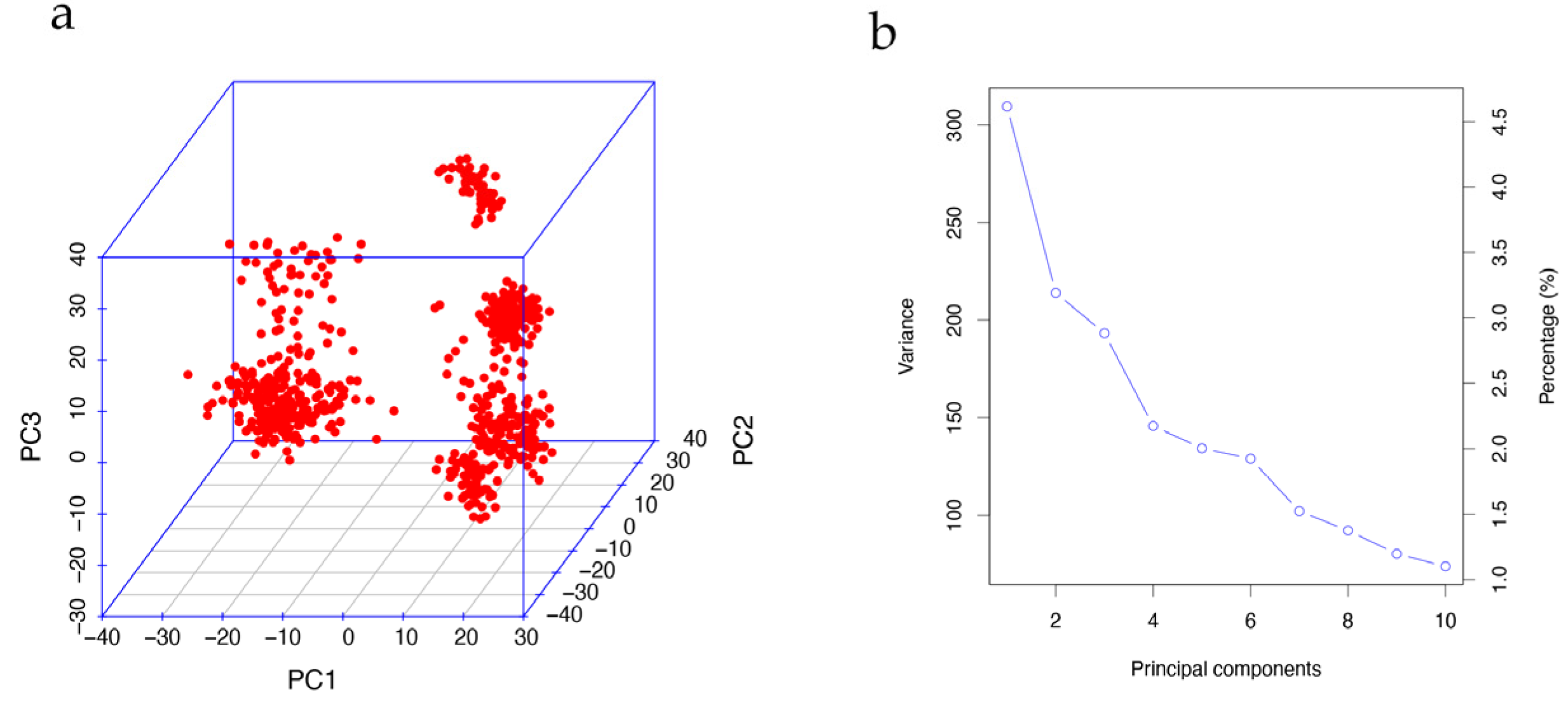
To mitigate the risk of concealed population stratification leading to spurious results in the GWAS, we conducted principal component analysis (PCA) using imputed genotypes (25,809 SNPs) with PLINK v1.9 (command: --pca). As depicted in Figure 2a, the population exhibited significant population stratification, necessitating the incorporation of the first principal components (PCs) for correction.
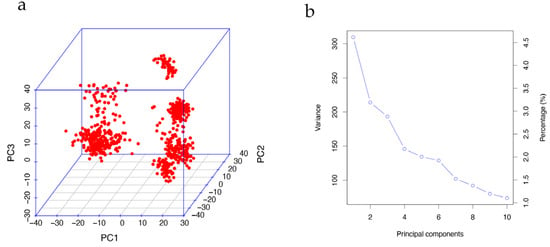
Figure 2.
Principal component analysis. (a) Visualization of the first three PC values showing the existence of population stratification. (b) Screen plot of the first 10 PC values. Decreasing trends indicate that the first five PCs can be appropriately used to correct the population stratification.
2.5. Genome-Wide Association Study
The association between SNPs and fatty acids was examined using the BLINK model (Bayesian-information and Linkage-disequilibrium Iteratively Nested Keyway) using the GAPIT 3.0 package [20]. BLINK, an enhanced version of FarmCPU, enhances statistical power by relaxing the assumption of even distribution of trait-related genes across the genome and incorporates the Bayesian information criterion (BIC) in fixed effects models to improve computational efficiency [21]. The model integrates Equations (1)–(3) according to the BIC strategy, iteratively calculating and excluding all pseudo-quantitative trait nucleotides (QTNs) to identify significant loci.
where y is a vector of phenotypic data; a is the vector of fixed effects or covariates, including IMF content and the first five PCs; b is a vector of marker effects; p is the effect of pseudo-QTNs; X, Z, and Q are the incidence matrices corresponding to a, b, and p, respectively; and e is the vector of residual errors. The BLINK model uses Equation (1) to define pseudo-QTNs as a covariate for Equation (2). The SNP obtained from Equation (2) determines the information of QTNs according to linkage disequilibrium (LD) and then employs Equation (3) to perform accuracy detection of QTNs using the BIC strategy. The false discovery rate (FDR) method of multiple testing, as described by Benjamini-Hochberg, was utilized to measure the statistical significance of association studies at a genome-wide level. The cut-off for considering SNPs as significant was set at FDR ≤ 0.1. The phenotypic variance explained (PVE) by genetic effects was calculated as follows [22]:
where MAF is the minor allele frequency for the SNP, is the effect of the SNP marker, N is the sample size, and is the standard error of .
2.6. Estimation of Heritability and Genetic Correlation
Heritability (h2) was estimated using the following Formula (5) in HIBLUP [23]:
where and are the additive genetic variance and the residual variance, respectively. The phenotypic and genetic correlation (rp and rg) between IMF and FAs were estimated using HIBLUP software [23].
where and represent the phenotypic and genetic covariance, respectively. and are the phenotypic and genetic variance, respectively.
2.7. Identification of Candidate Genes
Candidate genes were identified based on their physical positions and functions according to the Sus scrofa 10.2 reference genome assembly. The SNP-containing or nearest annotated genes for each potential SNP were obtained from the Sus scrofa (10.2) gtf file (http://ftp.ensembl.org/pub/release-80/gtf/sus_scrofa/Sus_scrofa.Sscrofa10.2.80.gtf.gz accessed on 1 May 2015) and taken as candidate genes.
2.8. Functional Enrichment Analysis
The g:Profiler website [24] was used for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. A tailor-made algorithm was chosen for multiple testing correction (adjusted p value < 0.05).
3. Results
3.1. Descriptive Statistics for IMF and Fatty Acid Composition
The statistical analysis results of IMF and 25 fatty acids are presented in Table S1. The IMF content in the longissimus dorsi of Ningxiang pigs was determined to be 3.65%. In the overall fatty acid distribution, MUFAs had the highest proportion at 41.88%, followed by SFAs at 39.35%, and PUFAs at 12.78%. Eleven fatty acids, including C22:6n-3, C17:1, and C15:0, were removed due to a sample size below 553 individuals (<80%). The remaining 14 fatty acids and IMF were used for subsequent analysis (Table 1). Among these fatty acids, oleic acid (C18:1n-9(c)) exhibited the highest content (40.28%), while elaidic acid (C18:1n-9(t)) had the lowest content (0.11%).
3.2. Estimation of Genetic Parameters
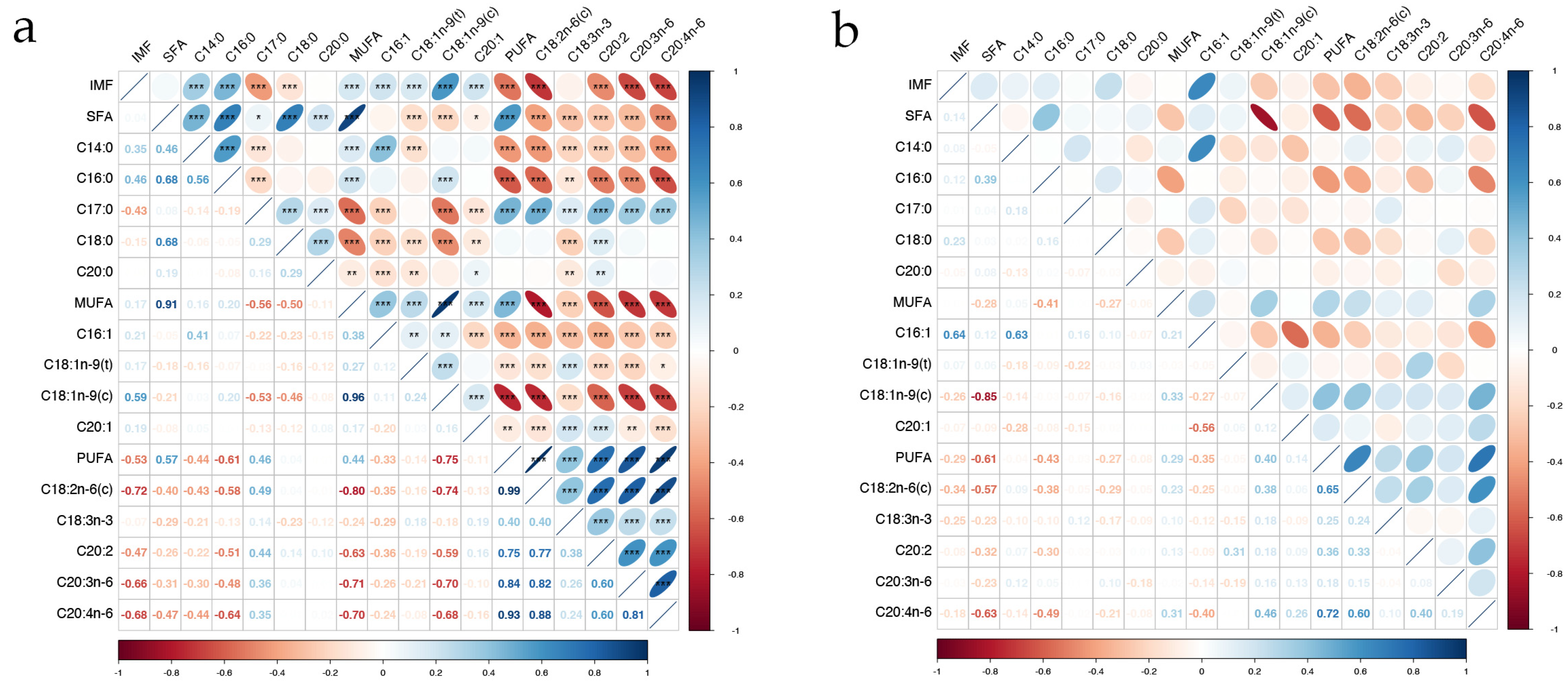
The results of the correlation analysis between fatty acids and IMF are presented in Figure 3a,b. In the phenotypic correlation analysis, the majority of fatty acids, except for SFA, C20:0, and C18:3n-3, exhibited a significant correlation with IMF (p < 0.001). SFA demonstrated significant positive phenotypic correlations with MUFAs and PUFAs (p < 0.001) but showed negative genetic correlations with four MUFAs and five PUFAs (p < 0.05). Furthermore, SFA, MUFA, and PUFA were significantly positively correlated with five SFAs, four MUFAs, and five PUFAs, respectively (p < 0.05). In the genetic correlation analysis, C16:1 had the highest correlation with IMF (rg = 0.64), and SFA was negatively correlated with most MUFAs and PUFAs, except C16:1 and C18:1n-9(t). All fatty acids had moderate to high heritabilities (0.37~0.89). Fatty acids C17:0 and C18:1n-9(t) had the lowest heritability estimates (0.45 and 0.37, respectively). Most fatty acids exhibited heritability estimates above 0.6. Notably, C18:0 exhibited the highest heritability estimate of 0.89 (Table 1).
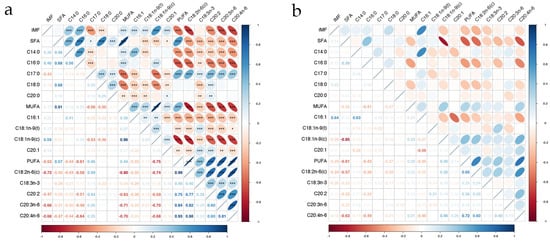
Figure 3.
Correlations among IMF and 14 fatty acids in the longissimus dorsi of Ningxiang pigs. “***” represents p value < 0.001, “**” represents p value < 0.01, “*” represents p value < 0.05. (a) Phenotypic correlation; (b) genetic correlation.
3.3. Genome-Wide Association Study Results for Fatty Acids
After quality control, 25,809 SNPs for 691 Ningxiang pigs were retained for the GWAS. In total, 74 genome-wide level SNPs were identified for 14 FAs in this study.
3.3.1. SFA
Thirty-seven genome-wide significant SNPs were identified for five SFAs (C14:0, C16:0, C17:0, C18:0, and C20:0). C20:0 had the most loci, which were located on SSC2, SSC3, SSC4, SSC5, SSC7, SSC8, SSC13, SSC14, and SSC16 (Figure S1). Moreover, some loci, such as ALGA0047587 (89.85%), ASGA0059505 (11.39%), and DRGA0011206 (6.85%) (Table S2), explained a large portion of the phenotypic variance.
3.3.2. MUFA
The Manhattan plots showed that 21 genome-wide significant loci were identified on 12 chromosomes (SSC1, SSC2, SSC3, SSC4, SSC5, SSC7, SSC9, SSC11, SSC13, SSC14, SSC16, and SSC17) for four MUFAs (Figure S2). H3GA0046208 explained the largest portion of the phenotypic variance (45.24%) for C18:1n-9(t) (Table S2).
3.3.3. PUFA
The Manhattan plots showed that 16 genome-wide significant loci on seven chromosomes (SSC1, SSC2, SSC5, SSC8, SSC9, SSC13, and SSC16) were identified for five PUFAs (Figure S3).
3.4. Identification of Candidate Genes
Four hundred and fifty-three genes were identified within a 500 kb region upstream and downstream of the significant SNPs (Table S2). For SFAs, 354 genes were found in a 1 Mb genomic region; 40 genes were close to 37 loci, of which 11 SNPs (WU_10.2_3_116903421, ASGA0074106, WU_10.2_4_119395133, M1GA0024654, WU_10.2_3_142168876, ALGA0010606, WU_10.2_11_3591593, ALGA0080940, H3GA0041501, MARC0054269, and WU_10.2_16_59778879) were located within 11 genes (ALK, MFAP3, CEPT1, NPEPPS, ENSSSCG00000008655, SWI5, CDK8, AVPI1, ALOX5, ITGA1, and SLIT3). For MUFAs, six SNPs (ALGA0015731, ASGA0064960, H3GA0025990, H3GA0046208, WU_10.2_5_13180559, and ASGA0031521) were intragenic variants. For PUFAs, 16 SNP loci were identified in 16 genes.
3.5. Functional Enrichment of Candidate Genes
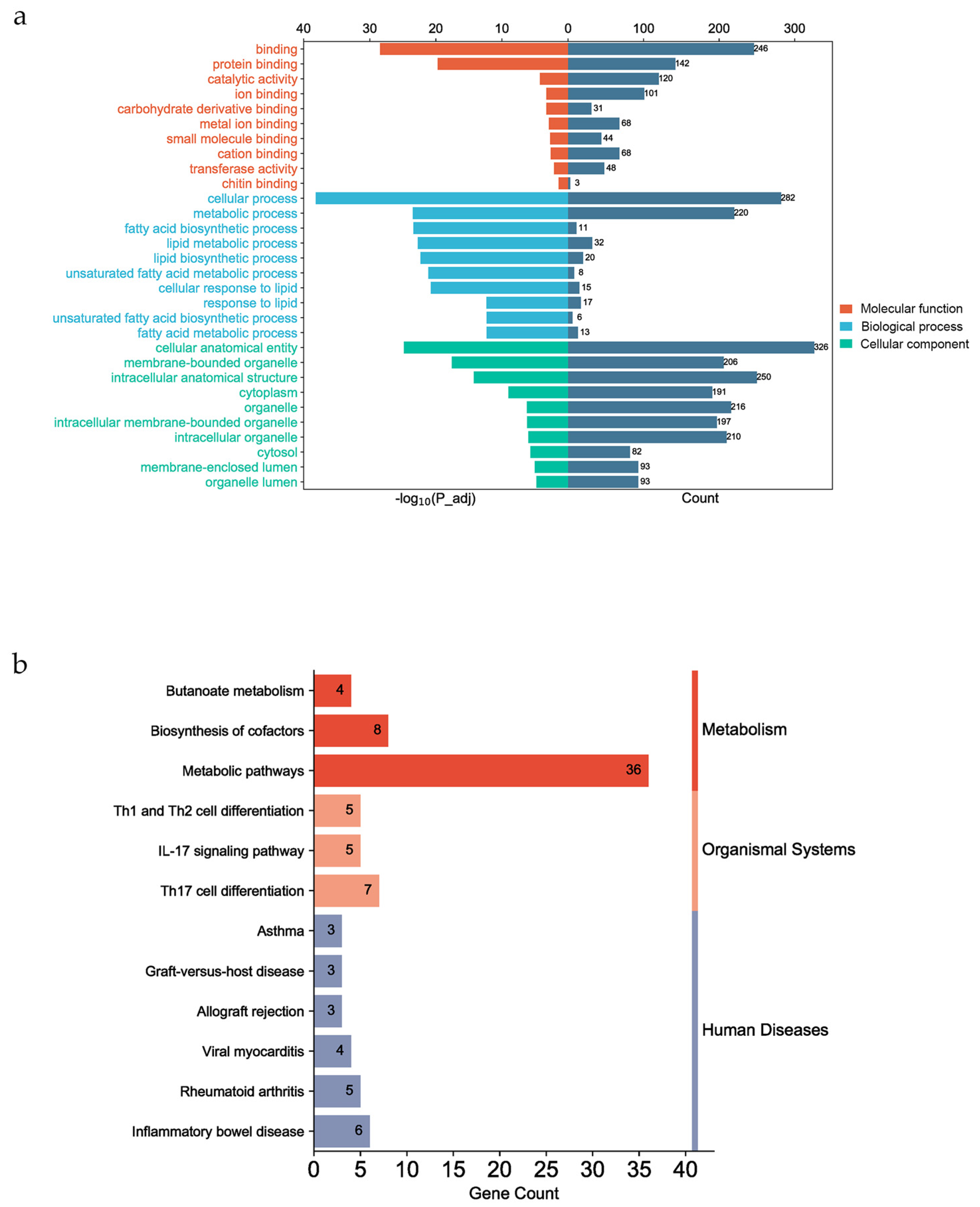
The GO and KEGG enrichment analyses were performed using the g:Profiler website for all fatty acid traits. The functional genes were significantly enriched in 262 GO terms (p_adj < 0.05) (see File S1). The top 10 molecular functions were involved in binding. The top 10 cellular components were cellular anatomical entities (GO:0110165) and membrane-bound organelles (GO:0043227). There were some GO terms associated with lipid and fatty acid metabolism in the biological process category (Figure 4a), such as fatty acid biosynthetic process (GO:0006633), unsaturated fatty acid metabolic process (GO:0033559), lipid biosynthetic process (GO:0008610), and lipid metabolic process (GO:0006629). In this study, annotated genes were significantly enriched in 12 KEGG pathways, such as metabolic pathways (KEGG:01100), inflammatory bowel disease (KEGG:05321), and Th17 cell differentiation (KEGG:04659) (Figure 4b). These KEGG pathways can be categorized into three groups on the KEGG website: Metabolism, Organismal Systems, and Human Diseases (https://www.kegg.jp/kegg/pathway.html (accessed on 1 October 2023)).
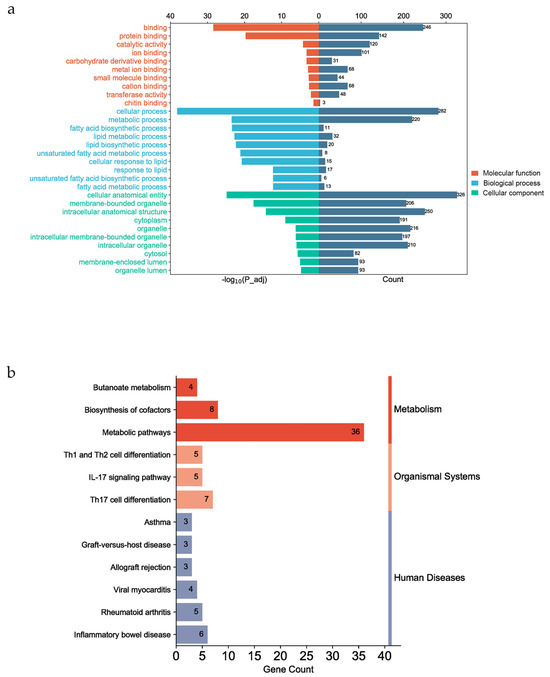
Figure 4.
Gene functional enrichment analysis. (a) The top 10 enriched molecular functions and cellular components and 10 biological processes associated with fatty acid and lipid metabolism. (b) The 12 enriched KEGG pathways, which can be divided into three categories on the KEGG website (Metabolism, Organismal Systems, and Human Diseases).
4. Discussion
4.1. Phenotypic and Genetic Correlations
In this study, a total of 25 fatty acid species were detected in the longissimus dorsi of Ningxiang pigs, with 14 species commonly found in the population. SFA was the most abundant among them, while the MUFA relative content was the highest. According to research, the distribution pattern of fatty acids in different varieties of pork is similar [25]. However, there are significant differences in the composition of fatty acids in different tissues [26], which is also influenced by sex [27]. Interestingly, docosahexaenoic acid (DHA) has only been found in the tissues of Ningxiang pigs [26,28], but not in any other pig breeds or tissues [29,30]. DHA belongs to the omega-3 family alongside alpha-linolenic acid (ALA) and eicosapentaenoic acid (EPA), which are essential nutrients integral to human life. Both linoleic acid (h2 = 0.88) and eicosa-11,14-dienoic acid (h2 = 0.63) from the omega-3 family exhibit high heritability in the longissimus dorsi of Ningxiang pigs, which is an interesting phenomenon [31]. To ensure the safety and health of pork products, pig industry breeders and researchers have long sought to enhance the PUFA content and distribution in pork [32,33].
As an important indicator of pork quality, IMF has consistently garnered significant attention. Numerous studies have provided evidence for the impact of IMF on meat quality [34]. In recent years, the hypothesis that IMF deposition in muscle is impacted by fatty acid structure was verified [35]. By conducting a correlation analysis between IMF and various fatty acid components in the longissimus dorsi of Ningxiang pigs (Figure 3), it was observed that IMF exhibits a notably low correlation with SFAs. Furthermore, IMF exhibited a significant positive correlation with MUFAs and a significant negative correlation with PUFAs. Realini et al. [36] investigated the relationship between IMF deposition and fatty acid composition in New Zealand sheep and found results consistent with those of our study, revealing a negative correlation between MUFAs and IMF deposition, a positive correlation between PUFAs and IMF deposition, and a correlation coefficient of −0.72 between linoleic acid and IMF deposition. This phenomenon might be attributed to the endogenous synthesis rate and desaturation sequence of SFAs.
4.2. Candidate Genes for Fatty Acid Composition
Currently, the molecular mechanisms underlying fat deposition are a topic of interest. Fat deposition not only directly impacts the growth, development, and meat production traits of animals but also holds valuable implications for addressing human diseases. Ningxiang pigs are an excellent model for obesity research, and an increasing number of genes, regulatory factors, and metabolic pathways related to fat deposition in Ningxiang pigs have been identified [37]. In this study, we performed a comprehensive GWAS focusing on 14 fatty acids in the longissimus dorsi of Ningxiang pigs, and most of them exhibited significant correlations with IMF content. Seventy-four genome-wide significant SNPs were identified in this study, most of which were intronic and intergenic variations (Table S2). Only WU_10.2_2_9630034 is a missense mutation, and there is no research on the gene (SDHAF2) related to fatty acids.
A total of 40 genes were identified from 37 significant loci associated with SFAs which included hnRNPU [38], CEPT1 [39], ATP1B1, DPT, DKK1 [40], PRKG1, EXT2 [41], MEF2C, IL17RA [42], ITGA1 [43] and ALOX5. Among these, heteronuclear heterogeneous ribonucleoprotein particles (hnRNPs) represent a group of proteins with diverse functions, playing pivotal roles in RNA biogenesis, cellular localization, and transport [44]. Specifically, hnRNP U is involved in the Blnc1/hnRNPU/EBF2 heterogeneous ribonucleoprotein particle complex, thus promoting the expression of brown adipocyte genes [38]. Additionally, Dickkopf WNT signaling pathway inhibitor 1 (DKK1) can regulate placental lipid metabolism through the WNT signaling pathway [40]. Surprisingly, muscle cell enhancer factor 2C (MEF2C) not only plays a crucial regulatory role in skeletal muscle cells but also exerts significant regulatory effects on adipose tissue deposition [45]. Elias et al. found that both ALOX5 and ALOX5AP are involved in the browning of white adipose tissue through lipotoxin A4 [46]. In our study, arachidonic acid 5-nenenebb lipoxygenase (ALOX5) and its partner, arachidonic acid 5-lipoxygenase activator protein (ALOX5AP) were genes near the C20:0 and C16:1 significant SNP loci, respectively. In Ossabaw pig epicardial adipose tissue, ALOX5 is positively correlated with n-3 PUFAs [47].
Twenty-five genes were annotated in the upstream and downstream regions of twenty-one loci related to MUFAs. Among them, genes such as ALOX5AP [46], MEDAG [48], ISL1 [49], RXRB [50], CRY1 [51], and CDKAL1 [52] were found to be related to lipid synthesis and metabolism. Interestingly, ALOX5AP and MEDAG are two genes upstream and downstream of the MARC0099145 locus. Additionally, MEDAG is involved in fat metabolism, playing a crucial role in backfat deposition in most Western pig breeds [48]. Retinol-X receptor β (RXRB) is a member of the nuclear receptor superfamily of retinoic acid X receptors (RXRs) and is expressed in almost all tissues. In the liver, the activation of RXRB leads to increased expression of stearyl CoA desaturation (SCD) and CD36 fatty acid transferase [50], RXRB is consistent with the results of this study and a study on the black Iberian pig [53]. Cryptochrome gene 1 (CRY1) is a member of the circadian clock gene family and plays a vital role in adipocyte biology. CRY1 is regulated by the classic Wnt/β-catenin signaling pathway, which influences fat differentiation [54]. Moreover, CRY1 contains two interaction regions that regulate its degradation to achieve diurnal blood glucose control [55], further affecting conditions such as obesity [51,56].
Nineteen genes were annotated in the upstream and downstream regions of 16 loci related to PUFAs. Genes such as NDUFA4L2, SLC16A7, OTUB1, EIF4E [57], and ROBO2 [58] were found to be associated with lipid synthesis and metabolism. Mitochondrial dysfunction can cause an increase in NDUFA4L2 expression, leading to lipid accumulation in renal cells [59]. In addition, ssc-mir-708 is associated with fatty acids, but miR-708 has only been reported in the regulation of cardiomyocyte proliferation to date [60]. Finally, eukaryotic translation initiation factor 4E (EIF4E) enhances the translation of various messenger RNAs involved in lipid metabolism processing and storage pathways, leading to weight gain after a high-fat diet [57]. These genes may influence the fatty acid composition. The majority of the fatty acids in Ningxiang pigs exhibit medium to high heritability, indicating that candidate genes are likely to impact the fatty acid composition of the longissimus dorsi. Therefore, the candidate genes related to fatty acids identified in this study can be considered for use in enhancing the fatty acid composition of imported or commercial pig meat, thereby improving the meat quality of commercial pigs.
5. Conclusions
This GWAS identified 74 genome-wide SNPs associated with 14 fatty acids in the longissimus dorsi. Some SNPs were located within or near reported genes, but some were novel for fatty acid composition. In total, twenty-two genes, such as hnRNPU, ALOX5AP, and NDUFA4L2, can be used as candidate genes for fatty acid composition in Ningxiang pigs. Our findings will be helpful for understanding the genetic basis of fatty acid composition and providing new targets for further breeding of pigs.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ani13203192/s1, Table S1: Detailed information on 25 fatty acids present in IMF of the longissimus dorsi of Ningxiang pigs; Table S2. Genome-wide significant SNPs loci for SFAs, MUFAs and PUFAs in Ningxiang pigs; File S1: GO and KEGG enrichment analysis; Figure S1. Manhattan plot for five SFAs (C14:0, C16:0, C17:0, C18:0, and C20:0). The x-axis represents the chromosomes, and y-axis represents the −log10 (p_value). The green line is genome-wide level threshold, the green dashed line is chromosome-level threshold; Figure S2. Manhattan plot for four MUFAs (C16:1, C18:1n-9(c), C18:1n-9(t), and C20:1). The x-axis represents the chromosomes, and y-axis represents the −log10 (p_value). The green line is genome-wide level threshold, the green dashed line is chromosome-level threshold; Figure S3. Manhattan plot for five PUFAs (C18:2n-6(c), C18:3n-3, C20:2, C20:3n-6, and C20:4n-6). The x-axis represents the chromosomes, and y-axis represents the −log10 (p_value). The green line is genome-wide level threshold, the green dashed line is chromosome-level threshold.
Author Contributions
Conceptualization, Q.Z., H.G. and Y.Y.; methodology, H.G. and K.X.; software, Q.Z., G.S. and X.D.; validation, H.G. and S.Y.; formal analysis, Q.Z. and F.Y.; investigation, Y.P., Y.C. and X.H.; resources, J.H., Y.Y. and K.X.; data curation, F.Y., Y.F., Q.W., S.Y. and Z.J.; writing—original draft preparation, Q.Z., H.G., S.Y. and F.Y.; writing—review and editing, J.H., Y.P., Y.Y. and K.X. visualization, H.G. and S.Y.; supervision, Y.Y. and K.X.; project administration, Y.Y. and K.X.; funding acquisition, Y.Y. and K.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Laboratory of Lingnan Modern Agriculture Project (NT2021005), the Strategic Priority Research Program of Chinese Academy of Sciences (Grant No. XDA24030204), the Special Funds for the Construction of Innovative Provinces in Hunan (Grant numbers 2021NK1009 and 2021NK1012); the Natural Science Foundation of Hunan Province Project (2023JJ20043 and 2020JJ5635).
Institutional Review Board Statement
The experimental protocol used in this study was reviewed and approved by the Animal Experimental Ethical Inspection of Laboratory Animal Centre, Hunan Agricultural University. All sample collection was conducted under a permit (No. 2020047) approved by the Attitude of the Animal Management and Ethics Committee.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chen, J.; Chen, F.; Lin, X.; Wang, Y.; He, J.; Zhao, Y. Effect of Excessive or Restrictive Energy on Growth Performance, Meat Quality, and Intramuscular Fat Deposition in Finishing Ningxiang Pigs. Animals 2020, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Uemoto, Y.; Abe, T.; Tameoka, N.; Hasebe, H.; Inoue, K.; Nakajima, H.; Shoji, N.; Kobayashi, M.; Kobayashi, E. Whole-genome association study for fatty acid composition of oleic acid in Japanese Black cattle. Anim. Genet. 2011, 42, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Fortin, A.; Robertson, W.M.; Tong, A.K. The eating quality of Canadian pork and its relationship with intramuscular fat. Meat Sci. 2005, 69, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Michas, G.; Micha, R.; Zampelas, A. Dietary fats and cardiovascular disease: Putting together the pieces of a complicated puzzle. Atherosclerosis 2014, 234, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.; Jo, C.; Tariq, M.R. Meat flavor precursors and factors influencing flavor precursors—A systematic review. Meat Sci. 2015, 110, 278–284. [Google Scholar] [CrossRef]
- Grassi, S.; Benedetti, S.; Opizzio, M.; Nardo, E.D.; Buratti, S. Meat and Fish Freshness Assessment by a Portable and Simplified Electronic Nose System (Mastersense). Sensors 2019, 19, 3225. [Google Scholar] [CrossRef]
- Melton, S.L.; Amiri, M.; Davis, G.W.; Backus, W.R. Flavor and chemical characteristics of ground beef from grass-, forage-grain- and grain-finished steers. J. Anim. Sci. 1982, 55, 77–87. [Google Scholar] [CrossRef]
- Westerling, D.B.; Hedrick, H.B. Fatty acid composition of bovine lipids as influenced by diet, sex and anatomical location and relationship to sensory characteristics. J. Anim. Sci. 1979, 48, 1343–1348. [Google Scholar] [CrossRef]
- de Oliveira Otto, M.C.; Wu, J.H.; Baylin, A.; Vaidya, D.; Rich, S.S.; Tsai, M.Y.; Jacobs, D.R., Jr.; Mozaffarian, D. Circulating and dietary omega-3 and omega-6 polyunsaturated fatty acids and incidence of CVD in the Multi-Ethnic Study of Atherosclerosis. J. Am. Heart Assoc. 2013, 2, e000506. [Google Scholar] [CrossRef]
- Merino, J.; Guasch-Ferré, M.; Ellervik, C.; Dashti, H.; Sharp, S.; Wu, P.; Overvad, K.; Sarnowski, C.; Kuokkanen, M.; Lemaitre, R.; et al. Quality of dietary fat and genetic risk of type 2 diabetes: Individual participant data meta-analysis. BMJ (Clin. Res. Ed.) 2019, 366, l4292. [Google Scholar] [CrossRef]
- Lands, B. Historical perspectives on the impact of n-3 and n-6 nutrients on health. Prog. Lipid Res. 2014, 55, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Lands, B.; Bibus, D.; Stark, K.D. Dynamic interactions of n-3 and n-6 fatty acid nutrients. Prostaglandins Leukot. Essent. Fat. Acids 2018, 136, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Bibus, D.; Lands, B. Balancing proportions of competing omega-3 and omega-6 highly unsaturated fatty acids (HUFA) in tissue lipids. Prostaglandins Leukot. Essent. Fat. Acids 2015, 99, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Davoli, R.; Luise, D.; Mingazzini, V.; Zambonelli, P.; Braglia, S.; Serra, A.; Russo, V. Genome-wide study on intramuscular fat in Italian Large White pig breed using the PorcineSNP60 BeadChip. J. Anim. Breed. Genet. 2016, 133, 277–282. [Google Scholar] [CrossRef]
- van Son, M.; Enger, E.G.; Grove, H.; Ros-Freixedes, R.; Kent, M.P.; Lien, S.; Grindflek, E. Genome-wide association study confirm major QTL for backfat fatty acid composition on SSC14 in Duroc pigs. BMC Genom. 2017, 18, 369. [Google Scholar] [CrossRef]
- Viterbo, V.S.; Lopez, B.I.M.; Kang, H.; Kim, H.; Song, C.W.; Seo, K.S. Genome wide association study of fatty acid composition in Duroc swine. Asian-Australas J. Anim. Sci. 2018, 31, 1127–1133. [Google Scholar] [CrossRef]
- GB/T 9695.1-2008; Meat and Meat Products—Determination of Free Fat Content. AQSIQ, SAC: Beijing, China, 2008.
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Pook, T.; Mayer, M.; Geibel, J.; Weigend, S.; Cavero, D.; Schoen, C.C.; Simianer, H. Improving Imputation Quality in BEAGLE for Crop and Livestock Data. G3 (Bethesda) 2020, 10, 177–188. [Google Scholar] [CrossRef]
- Teslovich, T.; Musunuru, K.; Smith, A.; Edmondson, A.; Stylianou, I.; Koseki, M.; Pirruccello, J.; Ripatti, S.; Chasman, D.; Willer, C.; et al. Biological, Clinical, and Population Relevance of 95 Loci for Blood Lipids. Nature 2010, 466, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Zhang, H.; Tang, Z.; Yin, D.; Fu, Y.; Yuan, X.; Li, X.; Liu, X.-L.; Zhao, S. HIBLUP: An integration of statistical models on the BLUP framework for efficient genetic evaluation using big genomic data. Nucleic Acids Res. 2023, 51, 3501–3512. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Piazuelo, D.; Criado-Mesas, L.; Revilla, M.; Castello, A.; Noguera, J.L.; Fernandez, A.I.; Ballester, M.; Folch, J.M. Identification of strong candidate genes for backfat and intramuscular fatty acid composition in three crosses based on the Iberian pig. Sci. Rep. 2020, 10, 13962. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Li, C.; Yu, Y.; Xing, Y.; Xiao, D.; Zhang, B. Comparison of fatty acid profile of three adipose tissues in Ningxiang pigs. Anim. Nutr. 2018, 4, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Zhang, J.; Ma, J.; Guo, Y.; Li, L.; Xiao, S.; Ren, J.; Yang, B.; Huang, L. Sexually dimorphic genetic architecture of complex traits in a large-scale F2 cross in pigs. Genet. Sel. Evol. 2014, 46, 76. [Google Scholar] [CrossRef]
- Lee, J.B.; Kang, Y.J.; Kim, S.G.; Woo, J.H.; Shin, M.C.; Park, N.G.; Yang, B.C.; Han, S.H.; Han, K.M.; Lim, H.T.; et al. GWAS and Post-GWAS High-Resolution Mapping Analyses Identify Strong Novel Candidate Genes Influencing the Fatty Acid Composition of the Longissimus dorsi Muscle in Pigs. Genes 2021, 12, 1323. [Google Scholar] [CrossRef]
- Popova, T.; Givko, N. Fatty acid profile of the backfat layers in four pig breeds. Food Sci. Appl. Biotechnol. 2019, 2, 24. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, J.; Cui, L.; Ma, J.; Chen, C.; Ai, H.; Xie, X.; Li, L.; Xiao, S.; Huang, L.; et al. Genetic architecture of fatty acid composition in the longissimus dorsi muscle revealed by genome-wide association studies on diverse pig populations. Genet. Sel. Evol. 2016, 48, 5. [Google Scholar] [CrossRef]
- Demets, R.; Gheysen, L.; Van Loey, A.; Foubert, I. Antioxidative capacity of microalgal carotenoids for stabilizing n-3LC-PUFA rich oil: Initial quantity is key. Food Chem. 2023, 406, 135044. [Google Scholar] [CrossRef]
- Mayer, C.; Côme, M.; Ulmann, L.; Martin, I.; Zittelli, G.C.; Faraloni, C.; Ouguerram, K.; Chénais, B.; Mimouni, V. The Potential of the Marine Microalga Diacronema lutheri in the Prevention of Obesity and Metabolic Syndrome in High-Fat-Fed Wistar Rats. Molecules 2022, 27, 4246. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Very long-chain n-3 fatty acids and human health: Fact, fiction and the future. Proc. Nutr. Soc. 2018, 77, 52–72. [Google Scholar] [CrossRef] [PubMed]
- Waszkiewicz-Robak, B.; Szterk, A.; Rogalski, M.; Rambuszek, M.; Kruk, M.; Rokowska, E. Nutritional value of raw pork depending on the fat type contents in pigs feed. Acta Sci. Pol. Technol. Aliment. 2015, 14, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Z.; Oyelami, F.O.; Sun, H.; Xu, Z.; Ma, P.; Wang, Q.; Pan, Y. Identification of genes related to intramuscular fat independent of backfat thickness in Duroc pigs using single-step genome-wide association. Anim. Genet. 2021, 52, 108–113. [Google Scholar] [CrossRef]
- Realini, C.E.; Pavan, E.; Purchas, R.W.; Agnew, M.; Johnson, P.L.; Bermingham, E.N.; Moon, C.D. Relationships between intramuscular fat percentage and fatty acid composition in M. longissimus lumborum of pasture-finished lambs in New Zealand. Meat Sci. 2021, 181, 108618. [Google Scholar] [CrossRef]
- Criado-Mesas, L.; Ballester, M.; Crespo-Piazuelo, D.; Castello, A.; Benitez, R.; Fernandez, A.I.; Folch, J.M. Analysis of porcine IGF2 gene expression in adipose tissue and its effect on fatty acid composition. PLoS ONE 2019, 14, e0220708. [Google Scholar] [CrossRef]
- Mi, L.; Zhao, X.Y.; Li, S.; Yang, G.; Lin, J.D. Conserved function of the long noncoding RNA Blnc1 in brown adipocyte differentiation. Mol. Metab. 2017, 6, 101–110. [Google Scholar] [CrossRef]
- Zayed, M.A.; Jin, X.; Yang, C.; Belaygorod, L.; Engel, C.; Desai, K.; Harroun, N.; Saffaf, O.; Patterson, B.W.; Hsu, F.F.; et al. CEPT1-Mediated Phospholipogenesis Regulates Endothelial Cell Function and Ischemia-Induced Angiogenesis Through PPARα. Diabetes 2021, 70, 549–561. [Google Scholar] [CrossRef]
- Strakovsky, R.S.; Pan, Y.X. A decrease in DKK1, a WNT inhibitor, contributes to placental lipid accumulation in an obesity-prone rat model. Biol. Reprod. 2012, 86, 81. [Google Scholar] [CrossRef]
- Pedrosa, V.B.; Schenkel, F.S.; Chen, S.Y.; Oliveira, H.R.; Casey, T.M.; Melka, M.G.; Brito, L.F. Genomewide Association Analyses of Lactation Persistency and Milk Production Traits in Holstein Cattle Based on Imputed Whole-Genome Sequence Data. Genes 2021, 12, 1830. [Google Scholar] [CrossRef]
- Shinjo, T.; Iwashita, M.; Yamashita, A.; Sano, T.; Tsuruta, M.; Matsunaga, H.; Sanui, T.; Asano, T.; Nishimura, F. IL-17A synergistically enhances TNFα-induced IL-6 and CCL20 production in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 2016, 477, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.S.; Kang, L.; Zheng, J.; Grueter, C.; Bracy, D.P.; James, F.D.; Pozzi, A.; Wasserman, D.H. Integrin α1-null mice exhibit improved fatty liver when fed a high fat diet despite severe hepatic insulin resistance. J. Biol. Chem. 2015, 290, 6546–6557. [Google Scholar] [CrossRef] [PubMed]
- Hitachi, K.; Kiyofuji, Y.; Nakatani, M.; Tsuchida, K. Myoparr-Associated and -Independent Multiple Roles of Heterogeneous Nuclear Ribonucleoprotein K during Skeletal Muscle Cell Differentiation. Int. J. Mol. Sci. 2021, 23, 108. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wu, J.; Zhou, J.; Zhang, Y.; Qiao, M.; Sun, H.; Li, Z.; Li, L.; Chen, N.; Oyelami, F.O.; et al. Integration of ATAC-seq and RNA-seq analysis identifies key genes affecting intramuscular fat content in pigs. Front. Nutr. 2022, 9, 1016956. [Google Scholar] [CrossRef] [PubMed]
- Elias, I.; Ferré, T.; Vilà, L.; Muñoz, S.; Casellas, A.; Garcia, M.; Molas, M.; Agudo, J.; Roca, C.; Ruberte, J.; et al. ALOX5AP Overexpression in Adipose Tissue Leads to LXA4 Production and Protection Against Diet-Induced Obesity and Insulin Resistance. Diabetes 2016, 65, 2139–2150. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.E.; Matthan, N.R.; Goldbaum, A.; Meng, H.; Lamon-Fava, S.; Lakshman, S.; Jang, S.; Molokin, A.; Solano-Aguilar, G.; Urban, J.F., Jr.; et al. Dietary patterns influence epicardial adipose tissue fatty acid composition and inflammatory gene expression in the Ossabaw pig. J. Nutr. Biochem. 2019, 70, 138–146. [Google Scholar] [CrossRef]
- Gozalo-Marcilla, M.; Buntjer, J.; Johnsson, M.; Batista, L.; Diez, F.; Werner, C.R.; Chen, C.Y.; Gorjanc, G.; Mellanby, R.J.; Hickey, J.M.; et al. Genetic architecture and major genes for backfat thickness in pig lines of diverse genetic backgrounds. Genet. Sel. Evol. 2021, 53, 76. [Google Scholar] [CrossRef]
- Li, H.; Heilbronn, L.K.; Hu, D.; Poynten, A.M.; Blackburn, M.A.; Shirkhedkar, D.P.; Kaplan, W.H.; Kriketos, A.D.; Ye, J.; Chisholm, D.J. Islet-1: A potentially important role for an islet cell gene in visceral fat. Obesity 2008, 16, 356–362. [Google Scholar] [CrossRef]
- Singh Ahuja, H.; Liu, S.; Crombie, D.L.; Boehm, M.; Leibowitz, M.D.; Heyman, R.A.; Depre, C.; Nagy, L.; Tontonoz, P.; Davies, P.J. Differential effects of rexinoids and thiazolidinediones on metabolic gene expression in diabetic rodents. Mol. Pharmacol. 2001, 59, 765–773. [Google Scholar] [CrossRef]
- Griebel, G.; Ravinet-Trillou, C.; Beeské, S.; Avenet, P.; Pichat, P. Mice deficient in cryptochrome 1 (cry1(−/−)) exhibit resistance to obesity induced by a high-fat diet. Front. Endocrinol. 2014, 5, 49. [Google Scholar] [CrossRef]
- Choi, W.J.; Jin, H.S.; Kim, S.S.; Shin, D. Dietary Protein and Fat Intake Affects Diabetes Risk with CDKAL1 Genetic Variants in Korean Adults. Int. J. Mol. Sci. 2020, 21, 5607. [Google Scholar] [CrossRef] [PubMed]
- Pena, R.N.; Noguera, J.L.; García-Santana, M.J.; González, E.; Tejeda, J.F.; Ros-Freixedes, R.; Ibáñez-Escriche, N. Five genomic regions have a major impact on fat composition in Iberian pigs. Sci. Rep. 2019, 9, 2031. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Zhou, L.; Yu, Y.; Zhang, T.; Wang, M. Knocking down clock control gene CRY1 decreases adipogenesis via canonical Wnt/β-catenin signaling pathway. Biochem. Biophys. Res. Commun. 2018, 506, 746–753. [Google Scholar] [CrossRef] [PubMed]
- Toledo, M.; Batista-Gonzalez, A.; Merheb, E.; Aoun, M.L.; Tarabra, E.; Feng, D.; Sarparanta, J.; Merlo, P.; Botrè, F.; Schwartz, G.J.; et al. Autophagy Regulates the Liver Clock and Glucose Metabolism by Degrading CRY1. Cell Metab. 2018, 28, 268–281.e264. [Google Scholar] [CrossRef] [PubMed]
- Sardon Puig, L.; Pillon, N.J.; Näslund, E.; Krook, A.; Zierath, J.R. Influence of obesity, weight loss, and free fatty acids on skeletal muscle clock gene expression. Am. J. Physiol. Endocrinol. Metab. 2020, 318, E1–E10. [Google Scholar] [CrossRef]
- Conn, C.S.; Yang, H.; Tom, H.J.; Ikeda, K.; Oses-Prieto, J.A.; Vu, H.; Oguri, Y.; Nair, S.; Gill, R.M.; Kajimura, S.; et al. The major cap-binding protein eIF4E regulates lipid homeostasis and diet-induced obesity. Nat. Metab. 2021, 3, 244–257. [Google Scholar] [CrossRef]
- Mohammadi, H.; Farahani, A.H.K.; Moradi, M.H.; Mastrangelo, S.; Di Gerlando, R.; Sardina, M.T.; Scatassa, M.L.; Portolano, B.; Tolone, M. Weighted Single-Step Genome-Wide Association Study Uncovers Known and Novel Candidate Genomic Regions for Milk Production Traits and Somatic Cell Score in Valle del Belice Dairy Sheep. Animals 2022, 12, 1155. [Google Scholar] [CrossRef]
- Laursen, K.B.; Chen, Q.; Khani, F.; Attarwala, N.; Gross, S.S.; Dow, L.; Nanus, D.M.; Gudas, L.J. Mitochondrial Ndufa4l2 Enhances Deposition of Lipids and Expression of Ca9 in the TRACK Model of Early Clear Cell Renal Cell Carcinoma. Front. Oncol. 2021, 11, 783856. [Google Scholar] [CrossRef]
- Deng, S.; Zhao, Q.; Zhen, L.; Zhang, C.; Liu, C.; Wang, G.; Zhang, L.; Bao, L.; Lu, Y.; Meng, L.; et al. Neonatal Heart-Enriched miR-708 Promotes Proliferation and Stress Resistance of Cardiomyocytes in Rodents. Theranostics 2017, 7, 1953–1965. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).