How Does Mitochondrial Protein-Coding Gene Expression in Fejervarya kawamurai (Anura: Dicroglossidae) Respond to Extreme Temperatures?
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Treatments
2.2. DNA Extraction, PCR, and Sequencing
2.3. Sequence Assembly and Analysis
2.4. Molecular Phylogenetic Analyses
2.5. RNA Extraction and cDNA Synthesis
2.6. RT-qPCR Primer Design
2.7. Relative mRNA Quantification
2.8. Data Analysis
3. Results
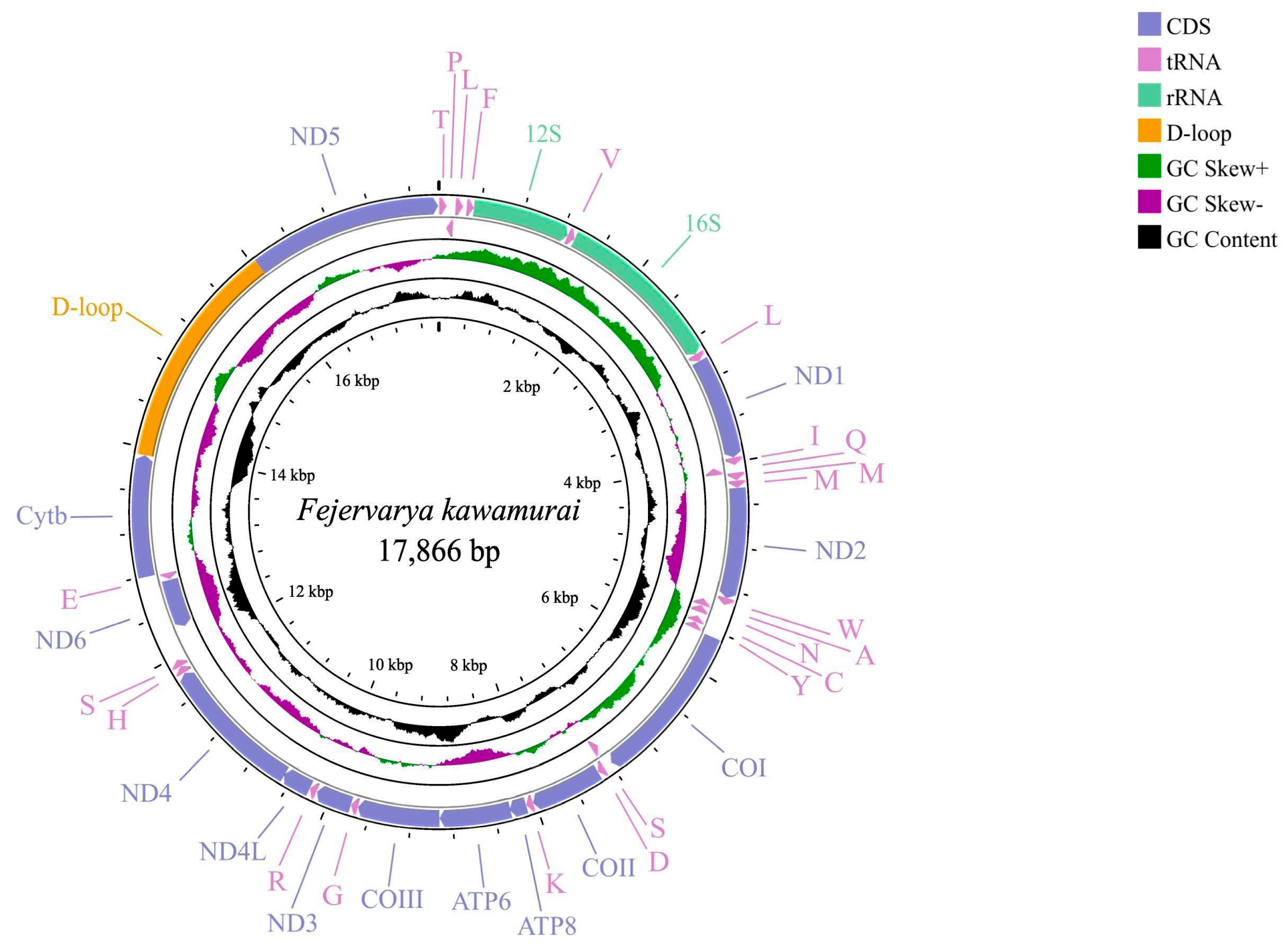
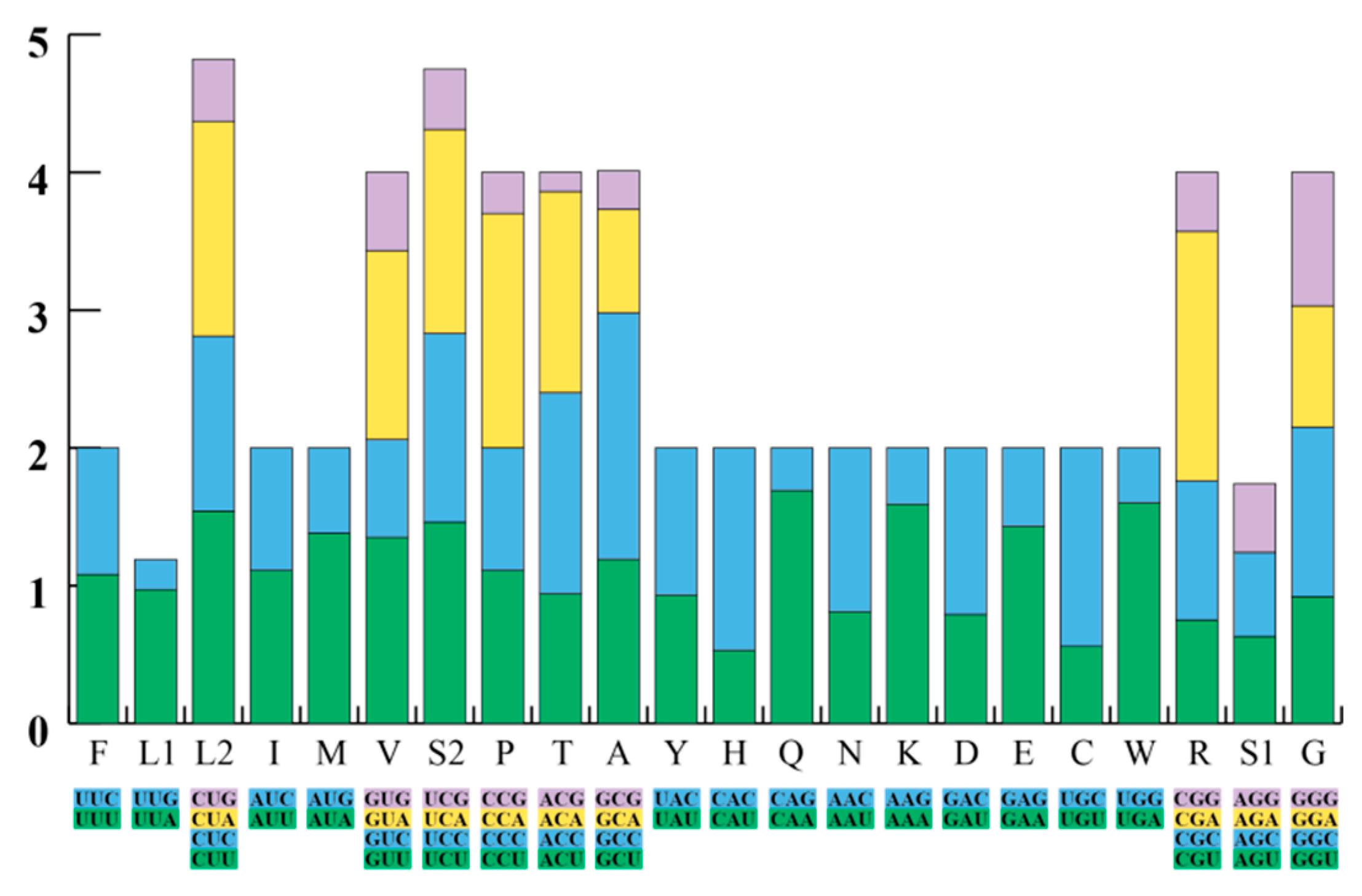
3.1. General Features of the F. kawamurai Mitogenome
3.2. Phylogenetic Relationships of F. kawamurai
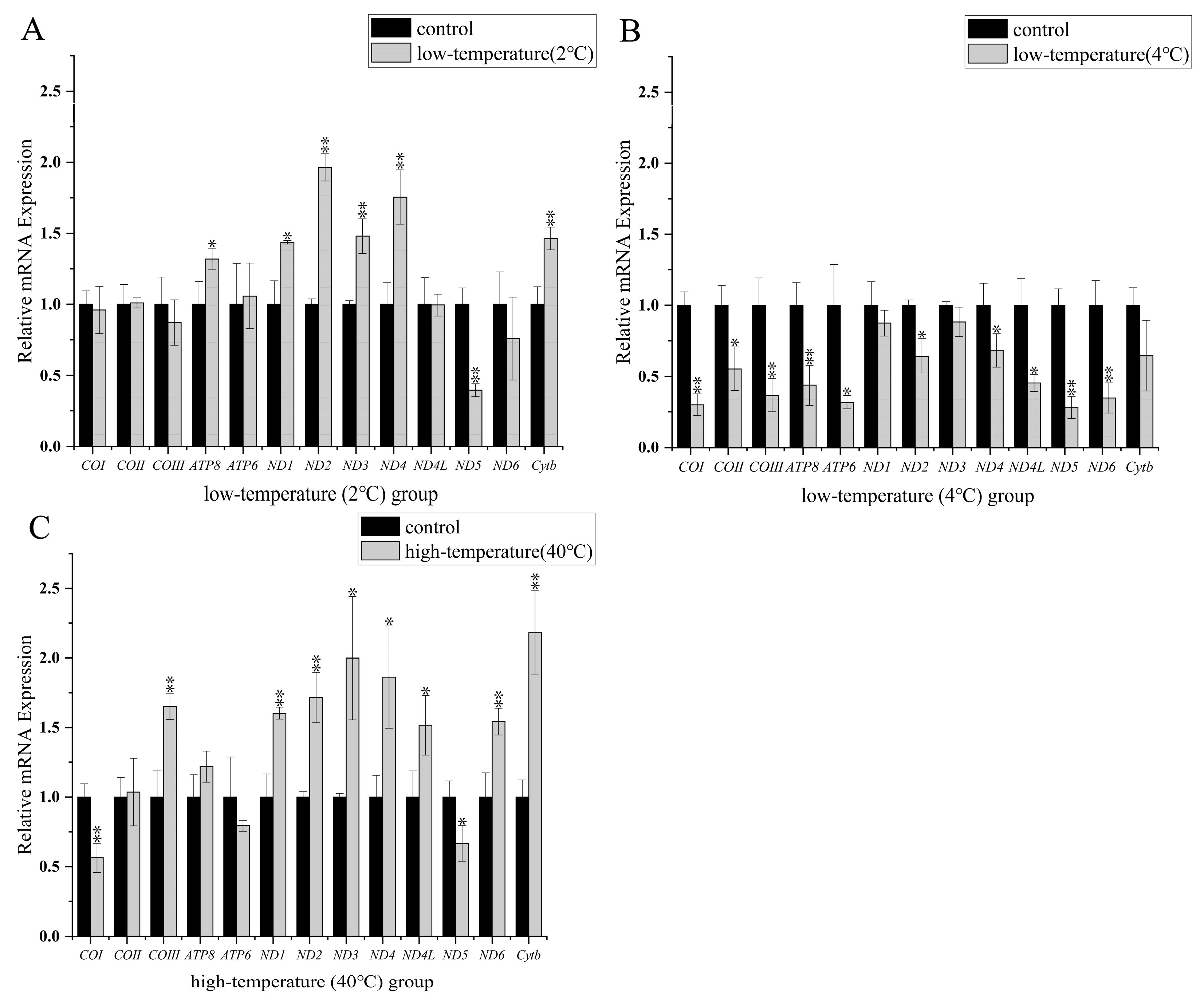
3.3. Transcript Levels of Protein-Coding Mitochondrial Genes
4. Discussion
4.1. Phylogenetic Relationships
4.2. Mitochondrial Transcript Level Analyses at Low Temperature
4.3. Mitochondrial Transcript Level Analyses at High Temperature
4.4. Characteristics of ND5 Gene Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Mitogenome | mitochondrial genome |
| PCR | polymerase chain reaction |
| NCBI | National Center for Biotechnology Information; |
| bp | base pair |
| PCGs | protein-coding genes |
| CRs | control regions |
| BI | Bayesian inference |
| ML | maximum likelihood. |
References
- Rome, L.C.; Choi, I.H.; Lutz, G.; Sosnicki, A. The influence of temperature on muscle function in the fast swimming scup: I. Shortening velocity and muscle recruitment during swimming. J. Exp. Biol. 1992, 163, 259–279. [Google Scholar] [CrossRef]
- Telemeco, R.S.; Gangloff, E.J. Introduction to the special issue-Beyond CTMAX and CTMIN: Advances in studying the thermal limits of reptiles and amphibians. J. Exp. Zool. Ecol. A Integr. Physiol. 2021, 335, 5–12. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Metabolic rate depression and biochemical adaptation in anaerobiosis, hibernation and estivation. Q. Rev. Biol. 1990, 65, 145–174. [Google Scholar] [CrossRef]
- Jackson, D.C. Hibernating without oxygen: Physiological adaptations of the painted turtle. J. Physiol. 2002, 543, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Krivoruchko, A.; Storey, K.B. Turtle anoxia tolerance: Biochemistry and gene regulation. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 1188–1196. [Google Scholar] [CrossRef] [PubMed]
- Ruf, T.; Geiser, F. Daily torpor and hibernation in birds and mammals. Biol. Rev. 2015, 90, 891–926. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Metabolic rate depression in animals: Transcriptional and translational controls. Biol. Rev. 2004, 79, 207–233. [Google Scholar] [CrossRef]
- Storey, K.B. Anoxia tolerance in turtles: Metabolic regulation and gene expression. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2007, 147, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Hochachka, P.W.; Somero, G.N. Biochemical Adaptation: Mechanism and Process in Physiological Evolution; Oxford University Press: Oxford, UK, 2002. [Google Scholar]
- Cheng, C.H.; Yang, F.F.; Liao, S.A.; Miao, Y.T.; Ye, C.X.; Wang, A.L.; Tan, J.W.; Chen, X.Y. High temperature induces apoptosis and oxidative stress in pufferfish (Takifugu obscurus) blood cells. J. Therm. Biol. 2015, 53, 172–179. [Google Scholar] [CrossRef]
- Cheng, C.H.; Ye, C.X.; Guo, Z.X.; Wang, A.L. Immune and physiological responses of pufferfish (Takifugu obscurus) under cold stress. Fish Shellfish Immunol. 2017, 64, 137–145. [Google Scholar] [CrossRef]
- Paital, B.; Panda, S.K.; Hati, A.K.; Mohanty, B.; Mohapatra, M.K.; Kanungo, S.; Chainy, G.B.N. Longevity of animals under reactive oxygen species stress and disease susceptibility due to global warming. World J. Biol. Chem. 2016, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Visser, M.E.; Caro, S.P.; Van Oers, K.; Schaper, S.V.; Helm, B. Phenology, seasonal timing and circannual rhythms: Towards a unified framework. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 3113–3127. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Jin, C.N.; Llusia, D.; Li, Y.M. Temperature-induced shifts in hibernation behavior in experimental amphibian populations. Sci. Rep. 2015, 5, 11580. [Google Scholar] [CrossRef]
- Wells, K.D. Complex life cycles and the ecology of amphibian metamorphosis. In The Ecology and Behavior of Amphibians; Wells, K.D., Ed.; University of Chicago Press: Chicago, IL, USA, 2007; pp. 599–644. [Google Scholar]
- Sinclair, B.J.; Stinziano, J.R.; Williams, C.M.; MacMillan, H.A.; Marshall, K.E.; Storey, K.B. Real-time measurement of metabolic rate during freezing and thawing of the wood frog, Rana sylvatica: Implications for overwinter energy use. J. Exp. Biol. 2013, 216, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Storey, K.B.; Storey, J.M. Freeze tolerance in animals. Physiol. Rev. 1988, 68, 27–84. [Google Scholar] [CrossRef] [PubMed]
- Storey, K.B.; Storey, J.M. Molecular physiology of freeze tolerance in vertebrates. Physiol. Rev. 2017, 97, 623–665. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, J.P.; Lee, R.E., Jr. Cryoprotection by urea in a terrestrially hibernating frog. J. Exp. Biol. 2005, 208, 4079–4089. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, J.P.; do Amaral, M.C.F.; Rosendale, A.J.; Lee, R.E., Jr. Hibernation physiology, freezing adaptation and extreme freeze tolerance in a northern population of the wood frog. J. Exp. Biol. 2013, 216, 3461–3473. [Google Scholar] [CrossRef] [PubMed]
- Layne, J.R.; Stapleton, M.G. Annual variation in glycerol mobilization and effect of freeze rigor on post-thaw locomotion in the freeze-tolerant frog Hyla versicolor. J. Comp. Physiol. B 2009, 179, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.L.; Frisbie, J.; Goldstein, D.L.; West, J.; Rivera, K.; Krane, C.M. Excretion and conservation of glycerol, and expression of aquaporins and glyceroporins, during cold acclimation in Cope’s gray tree frog Hyla chrysoscelis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 292, R544–R555. [Google Scholar] [CrossRef]
- Weinbach, A.; Cayuela, H.; Grolet, O.; Besnard, A.; Joly, P. Resilience to climate variation in a spatially structured amphibian population. Sci. Rep. 2018, 8, 14607. [Google Scholar] [CrossRef]
- Storey, J.M.; Wu, S.; Storey, K.B. Mitochondria and the frozen frog. Antioxidants 2021, 10, 543. [Google Scholar] [CrossRef] [PubMed]
- Hittel, D.S.; Storey, K.B. Differential expression of mitochondria-encoded genes in a hibernating mammal. J. Exp. Biol. 2002, 205, 1625–1631. [Google Scholar] [CrossRef]
- Chong, R.A.; Mueller, R.L. Low metabolic rates in salamanders are correlated with weak selective constraints on mitochondrial genes. Evolution 2013, 67, 894–899. [Google Scholar] [CrossRef]
- Tzameli, I. The evolving role of mitochondria in metabolism. Trends Endocrinol. Metab. 2012, 23, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.J.; Sparagna, G.C.; Chicco, A.J.; Schulte, P.M. Patterns of mitochondrial membrane remodeling parallel functional adaptations to thermal stress. J. Exp. Biol. 2018, 221, jeb174458. [Google Scholar] [CrossRef] [PubMed]
- Luu, B.E.; Wijenayake, S.; Zhang, J.; Tessier, S.N.; Quintero-Galvis, J.F.; Gaitán-Espitia, J.D.; Nespolo, R.F.; Storey, K.B. Strategies of biochemical adaptation for hibernation in a South American marsupial, Dromiciops gliroides: 2. Control of the Akt pathway and protein translation machinery. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2018, 224, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Villarin, J.J.; Schaeffer, P.J.; Markle, R.A.; Lindstedt, S.L. Chronic cold exposure increases liver oxidative capacity in the marsupial Monodelphis domestica. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2003, 136, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Ste-Marie, E.; Watanabe, Y.Y.; Semmens, J.M.; Marcoux, M.; Hussey, N.E. A first look at the metabolic rate of Greenland sharks (Somniosus microcephalus) in the Canadian Arctic. Sci. Rep. 2020, 10, 19297. [Google Scholar] [CrossRef]
- Speers-Roesch, B.; Norin, T.; Driedzic, W.R. The benefit of being still: Energy savings during winter dormancy in fish come from inactivity and the cold, not from metabolic rate depression. Proc. R. Soc. B Biol. Sci. 2018, 285, 20181593. [Google Scholar] [CrossRef] [PubMed]
- Guppy, M.; Withers, P. Metabolic depression in animals: Physiological perspectives and biochemical generalizations. Biol. Rev. 1999, 74, 1–40. [Google Scholar] [CrossRef]
- Storey, K.B. Turning down the fires of life: Metabolic regulation of hibernation and estivation. In Molecular Mechanisms of Metabolic Arrest; Storey, K.B., Ed.; BIOS Scientific Publishers: Oxford, UK, 2000; pp. 1–21. [Google Scholar]
- Hand, S.C.; Hardewig, I. Downregulation of cellular metabolism during environmental stress: Mechanisms and implications. Annu. Rev. Physiol. 1996, 58, 539–563. [Google Scholar] [CrossRef] [PubMed]
- Storey, K.B.; Storey, J.M. Tribute to P.L. Lutz: Putting life on ‘pause’-molecular regulation of hypometabolism. J. Exp. Biol. 2007, 210, 1700–1714. [Google Scholar] [CrossRef] [PubMed]
- Trzcionka, M.; Withers, K.W.; Klingenspor, M.; Jastroch, M. The effects of fasting and cold exposure on metabolic rate and mitochondrial proton leak in liver and skeletal muscle of an amphibian, the cane toad Bufo marinus. J. Exp. Biol. 2008, 211, 1911–1918. [Google Scholar] [CrossRef]
- Zhang, J.F.; Nie, L.W.; Wang, Y.; Hu, L.L. The complete mitochondrial genome of the large-headed frog, Limnonectes bannaensis (Amphibia: Anura), and a novel gene organization in the vertebrate mtDNA. Gene 2009, 442, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.T.; Guan, J.Y.; Dai, X.Y.; Wu, G.J.; Zhang, L.P.; Storey, K.B.; Zhang, J.Y.; Zheng, R.Q.; Yu, D.N. Mitochondrial gene expression in different organs of Hoplobatrachus rugulosus from China and Thailand under low-temperature stress. BMC Zool. 2022, 7, 24. [Google Scholar] [CrossRef]
- Baker, E.P.; Peris, D.; Moriarty, R.V.; Li, X.C.; Fay, J.C.; Hittinger, C.T. Mitochondrial DNA and temperature tolerance in lager yeasts. Sci. Adv. 2019, 5, eaav1869. [Google Scholar] [CrossRef]
- Camus, M.F.; Wolff, J.N.; Sgrò, C.M.; Dowling, D.K. Experimental support that natural selection has shaped the latitudinal distribution of mitochondrial haplotypes in Australian Drosophila melanogaster. Mol. Biol. Evol. 2017, 34, 2600–2612. [Google Scholar] [CrossRef]
- Mishmar, D.; Ruiz-Pesini, E.; Golik, P.; Macaulay, V.; Clark, A.G.; Hosseini, S.; Brandon, M.; Easley, K.; Chen, E.; Brown, M.D. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. USA 2003, 100, 171–176. [Google Scholar] [CrossRef]
- Pichaud, N.; Ballard, J.W.O.; Tanguay, R.M.; Blier, P.U. Mitochondrial haplotype divergences affect specific temperature sensitivity of mitochondrial respiration. J. Bioenerg. Biomembr. 2013, 45, 25–35. [Google Scholar] [CrossRef]
- Willett, C.S. The nature of interactions that contribute to postzygotic reproductive isolation in hybrid copepods. Genetica 2011, 139, 575–588. [Google Scholar] [CrossRef]
- Dingley, S.D.; Polyak, E.; Ostrovsky, J.; Srinivasan, S.; Lee, I.; Rosenfeld, A.B.; Tsukikawa, M.; Xiao, R.; Selak, M.A.; Coon, J.J. Mitochondrial DNA variant in COX1 subunit significantly alters energy metabolism of geographically divergent wild isolates in Caenorhabditis elegans. J. Mol. Biol. 2014, 426, 2199–2216. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.B. Differential Gene Expression under Environmental Stress in the Freeze Tolerant Wood Frog, Rana sylvatica. Ph.D. Thesis, Carleton University, Ottawa, ON, Canada, 1999. [Google Scholar]
- Zhang, J.Y.; Luu, B.E.; Yu, D.N.; Zhang, L.P.; Al-Attar, R.; Storey, K.B. The complete mitochondrial genome of Dryophytes versicolor: Phylogenetic relationship among Hylidae and mitochondrial protein-coding gene expression in response to freezing and anoxia. Int. J. Biol. Macromol. 2019, 132, 461–469. [Google Scholar] [CrossRef]
- Paital, B.; Chainy, G. Biology and conservation of the genus Scylla in India subcontinent. J. Environ. Biol. 2012, 33, 871. [Google Scholar]
- Paital, B. Antioxidant and oxidative stress parameters in brain of Heteropneustes fossilis under air exposure condition; role of mitochondrial electron transport chain. Ecotoxicol. Environ. Saf. 2013, 95, 69–77. [Google Scholar] [CrossRef]
- Parmesan, C. Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 637–669. [Google Scholar] [CrossRef]
- Root, T.L.; Price, J.T.; Hall, K.R.; Schneider, S.H.; Rosenzweig, C.; Pounds, J.A. Fingerprints of global warming on wild animals and plants. Nature 2003, 421, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.; Morice, C.; Parker, D.; Kendon, M. Global and regional climate in 2015. Weather 2016, 71, 185–192. [Google Scholar] [CrossRef][Green Version]
- Noyes, P.D.; McElwee, M.K.; Miller, H.D.; Clark, B.W.; Van Tiem, L.A.; Walcott, K.C.; Erwin, K.N.; Levin, E.D. The toxicology of climate change: Environmental contaminants in a warming world. Environ. Int. 2009, 35, 971–986. [Google Scholar] [CrossRef]
- Rahmstorf, S.; Foster, G.; Cahill, N. Global temperature evolution: Recent trends and some pitfalls. Environ. Res. Lett. 2017, 12, 054001. [Google Scholar] [CrossRef]
- Sinervo, B.; Mendez-De-La-Cruz, F.; Miles, D.B.; Heulin, B.; Bastiaans, E.; Villagrán-Santa Cruz, M.; Lara-Resendiz, R.; Martínez-Méndez, N.; Calderón-Espinosa, M.L.; Meza-Lázaro, R.N. Erosion of lizard diversity by climate change and altered thermal niches. Science 2010, 328, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.P.; Crump, M.L.; Lovejoy III, T.E. Extinction in Our Times: Global Amphibian Decline; Oxford University Press: Oxford, UK, 2009. [Google Scholar]
- Wake, D.B.; Vredenburg, V.T. Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc. Natl. Acad. Sci. USA 2008, 105, 11466–11473. [Google Scholar] [CrossRef]
- Duarte, H.; Tejedo, M.; Katzenberger, M.; Marangoni, F.; Baldo, D.; Beltrán, J.F.; Martí, D.A.; Richter-Boix, A.; Gonzalez-Voyer, A. Can amphibians take the heat? Vulnerability to climate warming in subtropical and temperate larval amphibian communities. Glob. Chang. Biol. 2012, 18, 412–421. [Google Scholar] [CrossRef]
- Fields, P.A. Protein function at thermal extremes: Balancing stability and flexibility. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2001, 129, 417–431. [Google Scholar] [CrossRef]
- Liu, Z.P.; Gu, W.B.; Tu, D.D.; Zhu, Q.H.; Zhou, Y.L.; Wang, C.; Wang, L.Z.; Shu, M.A. Effects of both cold and heat stress on the liver of the giant spiny frog (Quasipaa spinosa): Stress response and histological changes. J. Exp. Biol. 2018, 221, jeb186379. [Google Scholar] [CrossRef]
- Liu, B.; Xu, P.; Brown, P.B.; Xie, J.; Ge, X.P.; Miao, L.H.; Zhou, Q.L.; Ren, M.C.; Pan, L.K. The effect of hyperthermia on liver histology, oxidative stress and disease resistance of the Wuchang bream, Megalobrama amblycephala. Fish Shellfish Immunol. 2016, 52, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Rupik, W.; Jasik, K.; Bembenek, J.; Widłak, W. The expression patterns of heat shock genes and proteins and their role during vertebrate’s development. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2011, 159, 349–366. [Google Scholar] [CrossRef]
- Fernando, P.; Heikkila, J.J. Functional characterization of Xenopus small heat shock protein, Hsp30C: The carboxyl end is required for stability and chaperone activity. Cell Stress Chaperones 2000, 5, 148. [Google Scholar] [CrossRef]
- Daugaard, M.; Rohde, M.; Jäättelä, M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007, 581, 3702–3710. [Google Scholar] [CrossRef]
- Heikkila, J.; Schultz, G.; Iatrou, K.; Gedamu, L. Expression of a set of fish genes following heat or metal ion exposure. J. Biol. Chem. 1982, 257, 12000–12005. [Google Scholar] [CrossRef]
- Morimoto, R.I. Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998, 12, 3788–3796. [Google Scholar] [PubMed]
- Parsell, D.A.; Lindquist, S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993, 27, 437–496. [Google Scholar] [CrossRef]
- Chai, L.; Chen, A.; Luo, P.; Zhao, H.; Wang, H. Histopathological changes and lipid metabolism in the liver of Bufo gargarizans tadpoles exposed to Triclosan. Chemosphere 2017, 182, 255–266. [Google Scholar] [PubMed]
- Wu, C.; Zhang, Y.H.; Chai, L.H.; Wang, H.Y. Histological changes, lipid metabolism and oxidative stress in the liver of Bufo gargarizans exposed to cadmium concentrations. Chemosphere 2017, 179, 337–346. [Google Scholar] [PubMed]
- Gangloff, E.J.; Holden, K.G.; Telemeco, R.S.; Baumgard, L.H.; Bronikowski, A.M. Hormonal and metabolic responses to upper temperature extremes in divergent life-history ecotypes of a garter snake. J. Exp. Biol. 2016, 219, 2944–2954. [Google Scholar]
- Fobian, D.; Overgaard, J.; Wang, T. Oxygen transport is not compromised at high temperature in pythons. J. Exp. Biol. 2014, 217, 3958–3961. [Google Scholar] [CrossRef]
- Michaelsen, J.; Fago, A.; Bundgaard, A. High temperature impairs mitochondrial function in rainbow trout cardiac mitochondria. J. Exp. Biol. 2021, 224, jeb242382. [Google Scholar]
- Frost, D.R. Amphibian Species of the World: An Online Reference Version 6.1. 2023. Available online: https://amphibiansoftheworld.amnh.org/index.php (accessed on 31 March 2023).
- Djong, H.T.; Matsui, M.; Kuramoto, M.; Nishioka, M.; Sumida, M. A new species of the Fejervarya limnocharis complex from Japan (Anura, Dicroglossidae). Zool. Sci. 2011, 28, 922–929. [Google Scholar]
- Cheng, J.X.; Cai, Y.T.; Zheng, Y.J.; Zhang, J.Y.; Storey, K.B.; Bao, Y.X.; Yu, D.N. The complete mitochondrial genome of Fejervarya kawamurai (Anura: Dicroglossidae) and its phylogeny. Mitochondrial DNA Part B 2018, 3, 551–553. [Google Scholar]
- Buckley, L.B.; Hurlbert, A.H.; Jetz, W. Broad-scale ecological implications of ectothermy and endothermy in changing environments. Glob. Ecol. Biogeogr. 2012, 21, 873–885. [Google Scholar]
- Li, A.J.; Leung, P.T.; Bao, V.W.; Lui, G.C.; Leung, K.M. Temperature-dependent physiological and biochemical responses of the marine medaka Oryzias melastigma with consideration of both low and high thermal extremes. J. Therm. Biol. 2015, 54, 98–105. [Google Scholar]
- Yu, D.; Zhang, J.; Zheng, R.; Shao, C. The complete mitochondrial genome of Hoplobatrachus rugulosus (Anura: Dicroglossidae). Mitochondrial DNA 2012, 23, 336–337. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Q.; Wang, Y.Q.; Su, B. The mitochondrial genome organization of the rice frog, Fejervarya limnocharis (Amphibia: Anura): A new gene order in the vertebrate mtDNA. Gene 2005, 346, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.H.; Tu, F.Y. Mitogenome of Fejervarya multistriata: A novel gene arrangement and its evolutionary implications. Genet. Mol. Res. 2016, 15, gmr.15038302. [Google Scholar] [CrossRef]
- Zhang, P.; Papenfuss, T.J.; Wake, M.H.; Qu, L.H.; Wake, D.B. Phylogeny and biogeography of the family Salamandridae (Amphibia: Caudata) inferred from complete mitochondrial genomes. Mol. Phylogenet. Evol. 2008, 49, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liang, D.; Mao, R.L.; Hillis, D.M.; Wake, D.B.; Cannatella, D.C. Efficient sequencing of anuran mtDNAs and a mitogenomic exploration of the phylogeny and evolution of frogs. Mol. Biol. Evol. 2013, 30, 1899–1915. [Google Scholar] [CrossRef]
- Zhang, L.P.; Yu, D.N.; Storey, K.B.; Cheng, H.Y.; Zhang, J.Y. Higher tRNA gene duplication in mitogenomes of praying mantises (Dictyoptera, Mantodea) and the phylogeny within Mantodea. Int. J. Biol. Macromol. 2018, 111, 787–795. [Google Scholar]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. In Bioinformatics Methods and Protocols; Humana Press: Totowa, NJ, USA, 1999; pp. 71–91. [Google Scholar]
- Donath, A.; Jühling, F.; Al-Arab, M.; Bernhart, S.H.; Reinhardt, F.; Stadler, P.F.; Middendorf, M.; Bernt, M. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 2019, 47, 10543–10552. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Zhang, J.; Miao, G.; Hu, S.; Sun, Q.; Ding, H.; Ji, Z.; Guo, P.; Yan, S.; Wang, C.; Kan, X. Quantification and evolution of mitochondrial genome rearrangement in Amphibians. BMC Ecol. Evol. 2021, 21, 19. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.M.; Zhu, B.; Ma, E.B.; Wen, J.; Tu, T.Y.; Cao, Y.; Hasegawa, M.; Zhong, Y. Complete nucleotide sequence and gene arrangement of the mitochondrial genome of the crab-eating frog Fejervarya cancrivora and evolutionary implications. Gene 2009, 441, 148–155. [Google Scholar] [CrossRef]
- Jiang, L.C.; Lv, G.H.; Liu, L.; Wu, B.X.; Xu, Z.W.; Li, Y. Characterization of the complete mitochondrial genome of the paddy frog Fejervarya multistriata (Anura: Dicroglossidae) and its phylogeny. Mitochondrial DNA Part B 2020, 5, 1248–1250. [Google Scholar] [CrossRef]
- Yu, D.N.; Zhang, J.Y.; Li, P.; Zheng, R.Q.; Shao, C. Do cryptic species exist in Hoplobatrachus rugulosus? An examination using four nuclear genes, the Cyt b gene and the complete MT genome. PLoS ONE 2015, 10, e0124825. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.S.; Kurabayashi, A.; Hayashi, Y.; Sano, N.; Khan, M.M.R.; Fujii, T.; Sumida, M. Complete mitochondrial genomes and novel gene rearrangements in two dicroglossid frogs, Hoplobatrachus tigerinus and Euphlyctis hexadactylus, from Bangladesh. Genes Genet. Syst. 2010, 85, 219–232. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jiang, L.; Ruan, Q.; Chen, W. The complete mitochondrial genome sequence of the Xizang Plateau frog, Nanorana parkeri (Anura: Dicroglossidae). Mitochondrial DNA Part A 2016, 27, 3184–3185. [Google Scholar] [CrossRef]
- Chen, G.Y.; Wang, B.; Liu, J.Y.; Xie, F.; Jiang, J.P. Complete mitochondrial genome of Nanorana pleskei (Amphibia: Anura: Dicroglossidae) and evolutionary characteristics. Curr. Zool. 2011, 57, 785–805. [Google Scholar] [CrossRef]
- Jiang, L.; You, Z.; Yu, P.; Ruan, Q.; Chen, W. The first complete mitochondrial genome sequence of Nanorana parkeri and Nanorana ventripunctata (Amphibia: Anura: Dicroglossidae), with related phylogenetic analyses. Ecol. Evol. 2018, 8, 6972–6987. [Google Scholar] [CrossRef]
- Chen, Z.; Zhai, X.F.; Zhang, J.; Chen, X.H. The complete mitochondrial genome of Feirana taihangnica (Anura: Dicroglossidae). Mitochondrial DNA 2015, 26, 485–486. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Zhang, L.P.; Yu, D.N.; Storey, K.B.; Zheng, R.Q. Complete mitochondrial genomes of Nanorana taihangnica and N. yunnanensis (Anura: Dicroglossidae) with novel gene arrangements and phylogenetic relationship of Dicroglossidae. BMC Evol. Biol. 2018, 18, 26. [Google Scholar] [CrossRef]
- Shan, X.; Xia, Y.; Zheng, Y.C.; Zou, F.D.; Zeng, X.M. The complete mitochondrial genome of Quasipaa boulengeri (Anura: Dicroglossidae). Mitochondrial DNA 2014, 25, 83–84. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Q.; Chen, Q.E.; Wu, J. The complete mitochondrial genome of Quasipaa exilispinosa (Anura: Dicroglossidae). Mitochondrial DNA Part B 2020, 5, 2705–2706. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, J.Y.; Zheng, R.Q.; Yu, B.G.; Yang, G. Complete nucleotide sequence and gene organization of the mitochondrial genome of Paa spinosa (Anura: Ranoidae). Gene 2009, 447, 86–96. [Google Scholar] [CrossRef]
- Chen, Z.; Zhai, X.F.; Zhu, Y.J.; Chen, X.H. Complete mitochondrial genome of the Ye’s spiny-vented frog Yerana yei (Anura: Dicroglossidae). Mitochondrial DNA 2015, 26, 489–490. [Google Scholar] [CrossRef]
- Li, E.; Li, X.Q.; Wu, X.B.; Feng, G.; Zhang, M.; Shi, H.T.; Wang, L.J.; Jiang, J.P. Complete nucleotide sequence and gene rearrangement of the mitochondrial genome of Occidozyga martensii. J. Genet. 2014, 93, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Xie, Z. DAMBE: Software package for data analysis in molecular biology and evolution. J. Hered. 2001, 92, 371–373. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Calcott, B.; Ho, S.Y.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, O.A.; Hadj-Moussa, H.; Storey, K.B. Freeze-responsive regulation of MEF2 proteins and downstream gene networks in muscles of the wood frog, Rana sylvatica. J. Therm. Biol. 2017, 67, 1–8. [Google Scholar] [CrossRef]
- Zhang, C.; Qin, L.; Dou, D.C.; Li, X.N.; Ge, J.; Li, J.L. Atrazine induced oxidative stress and mitochondrial dysfunction in quail (Coturnix C. coturnix) kidney via modulating Nrf2 signaling pathway. Chemosphere 2018, 212, 974–982. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Ji, Y.C.; Lu, Y.Y.; Fu, R.N.; Xu, M.; Liu, X.L.; Guan, M.X. Leber’s hereditary optic neuropathy (LHON)-associated ND5 12338T> C mutation altered the assembly and function of complex I, apoptosis and mitophagy. Hum. Mol. Genet. 2018, 27, 1999–2011. [Google Scholar] [CrossRef]
- Du, C.C.; Li, X.Y.; Wang, H.X.; Liang, K.; Wang, H.Y.; Zhang, Y.H. Identification of thyroid hormone receptors α and β genes and their expression profiles during metamorphosis in Rana chensinensis. Turk. J. Zool. 2017, 41, 454–463. [Google Scholar] [CrossRef]
- Curole, J.P.; Kocher, T.D. Mitogenomics: Digging deeper with complete mitochondrial genomes. Trends Ecol. Evol. 1999, 14, 394–398. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Chen, Z.; Li, H.Y.; Zhu, Y.J.; Feng, Q.Q.; He, Y.X.; Chen, X.H. Molecular phylogeny of the family Dicroglossidae (Amphibia: Anura) inferred from complete mitochondrial genomes. Biochem. Syst. Ecol. 2017, 71, 1–9. [Google Scholar] [CrossRef]
- Matsui, M.; Toda, M.; Ota, H. A new species of frog allied to Fejervarya limnocharis from the southern Ryukyus, Japan (Amphibia: Ranidae). Curr. Herpetol. 2007, 26, 65–79. [Google Scholar]
- Yang, K.; Wo, Y.; Shao, G.; Liao, P.; Tong, H.; Brown, R.P.; Jin, Y. Phylogenetic Relationships among Chinese Rice Frogs within the Fejervarya limnocharis Species Complex (Amphibia: Dicroglossidae). Asian Herpetol. Res. 2022, 13, 232–241. [Google Scholar]
- Somero, G.N. Linking biogeography to physiology: Evolutionary and acclimatory adjustments of thermal limits. Front. Zool. 2005, 2, 1. [Google Scholar] [CrossRef]
- Kerscher, S.; Dröse, S.; Zickermann, V.; Brandt, U. The three families of respiratory NADH dehydrogenases. Results Probl. Cell Differ. 2008, 45, 185–222. [Google Scholar] [PubMed]
- Xia, D.; Yu, C.A.; Kim, H.; Xia, J.Z.; Kachurin, A.M.; Zhang, L.; Yu, L.; Deisenhofer, J. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science 1997, 277, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Guppy, M.; Fuery, C.; Flanigan, J. Biochemical principles of metabolic depression. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1994, 109, 175–189. [Google Scholar] [CrossRef]
- Tattersall, G.J.; Ultsch, G.R. Physiological ecology of aquatic overwintering in ranid frogs. Biol. Rev. 2008, 83, 119–140. [Google Scholar] [CrossRef] [PubMed]
- Paladino, F.V. Temperature effects on locomotion and activity bioenergetics of amphibians, reptiles, and birds. Am. Zool. 1985, 25, 965–972. [Google Scholar] [CrossRef][Green Version]
- Lesser, M.P. Oxidative stress in marine environments: Biochemistry and physiological ecology. Annu. Rev. Physiol. 2006, 68, 253–278. [Google Scholar] [CrossRef]
- Halliwell, B. Antioxidant defence mechanisms: From the beginning to the end (of the beginning). Free Radic. Res. 1999, 31, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Stadtman, E.R.; Levine, R.L. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids 2003, 25, 207–218. [Google Scholar] [CrossRef]
- An, M.I.; Choi, C.Y. Activity of antioxidant enzymes and physiological responses in ark shell, Scapharca broughtonii, exposed to thermal and osmotic stress: Effects on hemolymph and biochemical parameters. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2010, 155, 34–42. [Google Scholar] [CrossRef]
- England, K.; O’Driscoll, C.; Cotter, T. Carbonylation of glycolytic proteins is a key response to drug-induced oxidative stress and apoptosis. Cell Death Differ. 2004, 11, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.L.; Fujiwara, Y.; Kondo, T. Mechanism of cell death induction by nitroxide and hyperthermia. Free Radic. Biol. Med. 2006, 40, 1131–1143. [Google Scholar] [CrossRef]
- Brand, M.D.; Chien, L.F.; Ainscow, E.K.; Rolfe, D.F.; Porter, R.K. The causes and functions of mitochondrial proton leak. Biochim. Biophys. Acta 1994, 1187, 132–139. [Google Scholar] [CrossRef]
- Downs, C.A.; Heckathorn, S.A. The mitochondrial small heat-shock protein protects NADH: Ubiquinone oxidoreductase of the electron transport chain during heat stress in plants. FEBS Lett. 1998, 430, 246–250. [Google Scholar] [CrossRef]
- Pörtner, H.O. Climate variations and the physiological basis of temperature dependent biogeography: Systemic to molecular hierarchy of thermal tolerance in animals. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2002, 132, 739–761. [Google Scholar] [CrossRef] [PubMed]
- Pörtner, H.O.; Knust, R. Climate change affects marine fishes through the oxygen limitation of thermal tolerance. Science 2007, 315, 95–97. [Google Scholar] [CrossRef] [PubMed]
- Pörtner, H.O.; Bock, C.; Mark, F.C. Oxygen-and capacity-limited thermal tolerance: Bridging ecology and physiology. J. Exp. Biol. 2017, 220, 2685–2696. [Google Scholar] [CrossRef]
- Frederich, M.; Pörtner, H.O. Oxygen limitation of thermal tolerance defined by cardiac and ventilatory performance in spider crab, Maja squinado. Am. J. Physiol. 2000, 279, R1531–R1538. [Google Scholar] [CrossRef]
- Dawson, N.J.; Storey, K.B. A hydrogen peroxide safety valve: The reversible phosphorylation of catalase from the freeze-tolerant North American wood frog, Rana sylvatica. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 476–485. [Google Scholar] [CrossRef]
- Li, B.; Ma, Y.; Zhang, Y.H. Oxidative stress and hepatotoxicity in the frog, Rana chensinensis, when exposed to low doses of trichlorfon. J. Environ. Sci. Health B 2017, 52, 476–482. [Google Scholar] [CrossRef]
- Herrmann, J.M.; Funes, S. Biogenesis of cytochrome oxidase-sophisticated assembly lines in the mitochondrial inner membrane. Gene 2005, 354, 43–52. [Google Scholar] [CrossRef]
- Sousa, J.S.; D’Imprima, E.; Vonck, J. Mitochondrial respiratory chain complexes. Membr. Protein Complexes Struct. Funct. 2018, 87, 167–227. [Google Scholar]
- Wirth, C.; Brandt, U.; Hunte, C.; Zickermann, V. Structure and function of mitochondrial complex I. Biochim. Biophys. Acta Bioenerg. 2016, 1857, 902–914. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.P.; Vinothkumar, K.R.; Hirst, J. Structure of mammalian respiratory complex I. Nature 2016, 536, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Nakhle, J.; Rodriguez, A.M.; Vignais, M.L. Multifaceted roles of mitochondrial components and metabolites in metabolic diseases and cancer. Int. J. Mol. Sci. 2020, 21, 4405. [Google Scholar] [CrossRef] [PubMed]
- Pörtner, H.O.; Hardewig, I.; Sartoris, F.J.; Van Dijk, P. Energetic aspects of cold adaptation: Critical temperatures in metabolic, ionic and acid-base regulation? In Cold Ocean Physiology; Portner, H.O., Playle, R.C., Eds.; Cambridge University Press: Cambridge, UK, 1998; pp. 88–120. [Google Scholar]
- Brand, M.D. Uncoupling to survive? The role of mitochondrial inefficiency in ageing. Exp. Gerontol. 2000, 35, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Stier, A.; Massemin, S.; Criscuolo, F. Chronic mitochondrial uncoupling treatment prevents acute cold-induced oxidative stress in birds. J. Comp.Physiol. B 2014, 184, 1021–1029. [Google Scholar] [CrossRef] [PubMed]
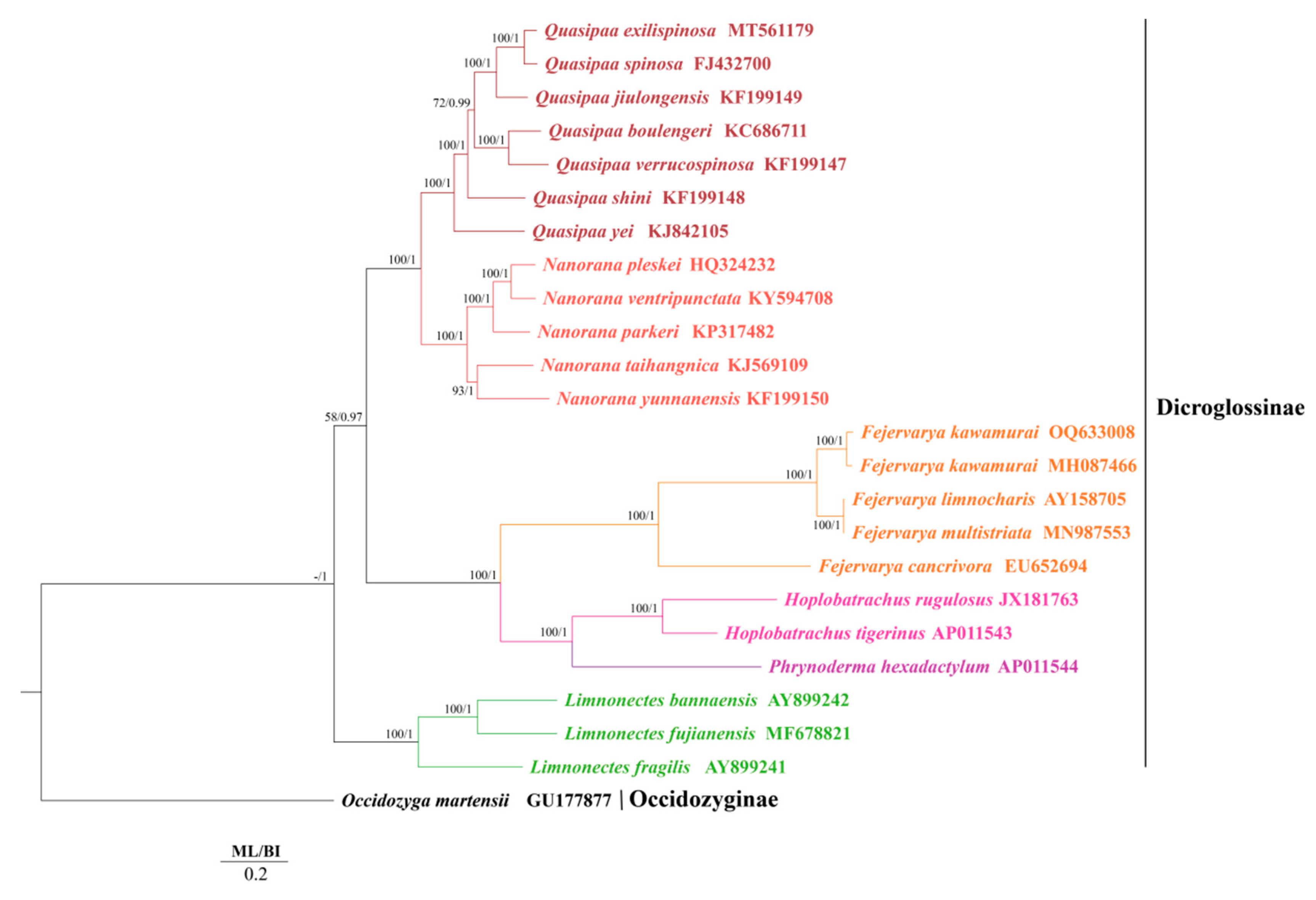
| Family | Subfamily | Genus | Species | Genome Length | GenBank No. | References |
|---|---|---|---|---|---|---|
| Dicroglossidae | Dicroglossinae | Limnonectes | Limnonectes bannaensis | 16,867 bp | AY899242 | [38] |
| Limnonectes fujianensis | 18,154 bp | MF678821 | [90] | |||
| Limnonectes fragilis | 16,640 bp | AY899241 | Unpublished | |||
| Fejervarya | Fejervarya cancrivora | 17,843 bp | EU652694 | [91] | ||
| Fejervarya kawamurai_GDGZ | 17,866 bp | OQ633008 | This study | |||
| Fejervarya kawamurai | 17,650 bp | MH087466 | [75] | |||
| Fejervarya limnocharis | 17,717 bp | AY158705 | [79] | |||
| Fejervarya multistriata | 17,759 bp | MN987553 | [92] | |||
| Hoplobatrachus | Hoplobatrachus rugulosus | 20,926 bp | JX181763 | [93] | ||
| Hoplobatrachus tigerinus | 20,462 bp | AP011543 | [94] | |||
| Phrynoderma | Phrynoderma hexadactylum | 20,280 bp | AP011544 | [94] | ||
| Nanorana | Nanorana parkeri | 17,837 bp | KP317482 | [95] | ||
| Nanorana pleskei | 17,660 bp | HQ324232 | [96] | |||
| Nanorana ventripunctata | 18,373 bp | KY594708 | [97] | |||
| Nanorana taihangnica | 17,412 bp | KJ569109 | [98] | |||
| Nanorana yunnanensis | 23,685 bp | KF199150 | [99] | |||
| Quasipaa | Quasipaa boulengeri | 17,741 bp | KC686711 | [100] | ||
| Quasipaa verrucospinosa | 15,063 bp | KF199147 | [99] | |||
| Quasipaa exilispinosa | 17,046 bp | MT561179 | [101] | |||
| Quasipaa spinosa | 18,012 bp | FJ432700 | [102] | |||
| Quasipaa jiulongensis | 15,072 bp | KF199149 | [99] | |||
| Quasipaa shini | 14,943 bp | KF199148 | [99] | |||
| Quasipaa yei | 17,072 bp | KJ842105 | [103] | |||
| Occidozyginae | Occidozyga | Occidozyga martensii | 18,321 bp | GU177877 | [104] |
| Gene Name | Forward Primers (5′-3′) | Reverse Primers (5′-3′) |
|---|---|---|
| COI | GDCC-COI-J TTGTTCACTGATTCCCACTTT | GDCC-COI-N GAGGTATCCCCGCTAAACCA |
| COII | GDCC-COII-J ATGGACGAGTTAGGTGCC | GDCC-COII-N AAGGTCATTTGTGGGGAT |
| COIII | GDCC-COIII-J GGCATCTACGGAACCACA | GDCC-COIII-N AAGCCGAAGTGGTGTTGA |
| ATP8 | GDCC-ATP8-J ATGCCTCAATTACTACCT | GDCC-ATP8-N GCTTCAGGTTACAGAGTT |
| ATP6 | GDCC-ATP6-J AATAAGTATTAACCTTCTCGG | GDCC-ATP6-N TACGGAGGCCGATAAGGACTG |
| ND1 | GDCC-ND1-J CTTGCGGTAGCATTCCTCA | GDCC-ND1-N AGGATTTGCGAGGAGGTTG |
| ND2 | GDCC-ND2-J TCAGGAGAATGGTCCATCG | GDCC-ND2-N ATGTTGAGAGGATTAGTCCA |
| ND3 | GDCC-ND3-J CTCATTGCCTCTGCCCTA | GDCC-ND3-N GGAAGAAGCGTATGGAAT |
| ND4 | GDCC-ND4-J GGCACTATTTTCCAACCC | GDCC-ND4-N AAGCAAGTAAAGAGGGAGTT |
| ND4L | GDCC-ND4L-J GGCCTATCTTTCCACCGTAT | GDCC-ND4L-N AAGGGGGATAGGACAAAAGA |
| ND5 | GDCC-ND5-J TGCTGTGAAACACAACGACA | GDCC-ND5-N TGATTATTCCCGAGATTATGA |
| ND6 | GDCC-ND6-J TTCTAATCCGTCACCATACT | GDCC-ND6-N TCCCACCTAAATACACTAGC |
| Cytb | GDCC-CYTB-J TCATCTAATCCAACAGGGCT | GDCC-CYTB-N GTGAAGTTATCTGGGTCTCC |
| β-actin | GDCC-Actin-J GTGCGTGACATCAAGGAG | GDCC-Actin-N GGCTTCTGGACATCTGAAC |
| Feature | Start Position | Stop Position | Intergenic Nucleotide | Length (bp) | Start Codon | Stop Codon | Anticodon | Strand |
|---|---|---|---|---|---|---|---|---|
| tRNAThr | 1 | 72 | −1 | 72 | TGT | H | ||
| tRNAPro | 72 | 140 | 17 | 69 | TAG | H | ||
| tRNALeu(CUN) | 158 | 229 | 33 | 72 | TGG | L | ||
| tRNAPhe | 263 | 330 | 68 | GAA | H | |||
| 12S rRNA | 331 | 1264 | −1 | 934 | H | |||
| tRNAVal | 1264 | 1335 | 72 | TAC | H | |||
| 16S rRNA | 1336 | 2927 | −1 | 1592 | H | |||
| tRNALeu(UUR) | 2927 | 2999 | 73 | TAA | H | |||
| ND1 | 3000 | 3957 | 958 | ATG | T | H | ||
| tRNAIle | 3958 | 4028 | 71 | GAT | H | |||
| tRNAGln | 4029 | 4099 | −1 | 71 | TTG | L | ||
| tRNAMet | 4099 | 4169 | 3 | 71 | CAT | H | ||
| tRNAMet | 4173 | 4241 | 69 | CAT | H | |||
| ND2 | 4242 | 5276 | −2 | 1035 | ATT | TAG | H | |
| tRNATrp | 5275 | 5343 | 69 | TCA | H | |||
| tRNAAla | 5344 | 5412 | 2 | 69 | TGC | L | ||
| tRNAAsn | 5415 | 5487 | 2 | 73 | GTT | L | ||
| tRNACys | 5525 | 5590 | 66 | GCA | L | |||
| tRNATyr | 5591 | 5657 | 4 | 67 | GTA | L | ||
| COI | 5662 | 7192 | 11 | 1531 | ATA | T | H | |
| tRNASer(UCN) | 7204 | 7274 | 71 | TGA | L | |||
| tRNAAsp | 7275 | 7342 | 2 | 68 | GTC | H | ||
| COII | 7345 | 8026 | 682 | ATG | T | H | ||
| tRNALys | 8027 | 8094 | 1 | 70 | TTT | H | ||
| ATPase8 | 8096 | 8257 | −7 | 162 | ATG | TAA | H | |
| ATPase6 | 8251 | 8932 | 682 | ATG | T | H | ||
| COIII | 8933 | 9716 | 784 | ATG | T | H | ||
| tRNAGly | 9717 | 9785 | 69 | TCC | H | |||
| ND3 | 9786 | 10,130 | 4 | 345 | GTG | TAA | H | |
| tRNAArg | 10,135 | 10,203 | 1 | 69 | TCG | H | ||
| ND4L | 10,205 | 10,483 | −7 | 279 | ATG | TAA | H | |
| ND4 | 10,477 | 11,829 | 3 | 1353 | ATG | TAA | H | |
| tRNAHis | 11,833 | 11,901 | 69 | GTG | H | |||
| tRNASer(AGY) | 11,902 | 11,969 | 259 | 68 | GCT | H | ||
| ND6 | 12,229 | 12,717 | 5 | 489 | ATG | AGG | L | |
| tRNAGlu | 12,723 | 12,790 | 5 | 68 | TTC | L | ||
| Cytb | 12,796 | 13,932 | 1137 | ATG | TAA | H | ||
| D-loop | 13,933 | 16,039 | 2107 | H | ||||
| ND5 | 16,040 | 17,857 | 1818 | GTA | TAA | H |
| Region | A (%) | T (%) | C (%) | G (%) | A + T (%) | C + G (%) | AT Skew | GC Skew | |
|---|---|---|---|---|---|---|---|---|---|
| Mito (H strand) | 27.8 | 29.4 | 27.5 | 15.4 | 57.2 | 42.9 | −0.028 | −0.282 | |
| PCGs | J | 25.3 | 31.2 | 28.7 | 14.7 | 56.5 | 43.4 | −0.104 | −0.322 |
| N | 17.2 | 34.8 | 12.1 | 36.0 | 52.0 | 48.1 | −0.339 | 0.498 | |
| tRNAs (H strand) | 29.3 | 27.1 | 23.4 | 20.2 | 56.4 | 43.6 | 0.039 | −0.072 | |
| rRNAs (H strand) | 33.3 | 24.4 | 23.7 | 18.6 | 57.7 | 42.3 | 0.154 | −0.119 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.-Y.; Zhang, L.-H.; Hong, Y.-H.; Cai, L.-N.; Storey, K.B.; Zhang, J.-Y.; Zhang, S.-S.; Yu, D.-N. How Does Mitochondrial Protein-Coding Gene Expression in Fejervarya kawamurai (Anura: Dicroglossidae) Respond to Extreme Temperatures? Animals 2023, 13, 3015. https://doi.org/10.3390/ani13193015
Wang J-Y, Zhang L-H, Hong Y-H, Cai L-N, Storey KB, Zhang J-Y, Zhang S-S, Yu D-N. How Does Mitochondrial Protein-Coding Gene Expression in Fejervarya kawamurai (Anura: Dicroglossidae) Respond to Extreme Temperatures? Animals. 2023; 13(19):3015. https://doi.org/10.3390/ani13193015
Chicago/Turabian StyleWang, Jing-Yan, Li-Hua Zhang, Yue-Huan Hong, Ling-Na Cai, Kenneth B. Storey, Jia-Yong Zhang, Shu-Sheng Zhang, and Dan-Na Yu. 2023. "How Does Mitochondrial Protein-Coding Gene Expression in Fejervarya kawamurai (Anura: Dicroglossidae) Respond to Extreme Temperatures?" Animals 13, no. 19: 3015. https://doi.org/10.3390/ani13193015
APA StyleWang, J.-Y., Zhang, L.-H., Hong, Y.-H., Cai, L.-N., Storey, K. B., Zhang, J.-Y., Zhang, S.-S., & Yu, D.-N. (2023). How Does Mitochondrial Protein-Coding Gene Expression in Fejervarya kawamurai (Anura: Dicroglossidae) Respond to Extreme Temperatures? Animals, 13(19), 3015. https://doi.org/10.3390/ani13193015






